Abstract
The Mdm2 oncoprotein promotes cell survival and cell cycle progression by inhibiting the p53 tumor suppressor protein. To regulate p53, Mdm2 must gain nuclear entry, and the mechanism that induces this is now identified. Mitogen-induced activation of phosphatidylinositol 3-kinase (PI3-kinase) and its downstream target, the Akt/PKB serine-threonine kinase, results in phosphorylation of Mdm2 on serine 166 and serine 186. Phosphorylation on these sites is necessary for translocation of Mdm2 from the cytoplasm into the nucleus. Pharmacological blockade of PI3-kinase/Akt signaling or expression of dominant-negative PI3-kinase or Akt inhibits nuclear entry of Mdm2, increases cellular levels of p53, and augments p53 transcriptional activity. Expression of constitutively active Akt promotes nuclear entry of Mdm2, diminishes cellular levels of p53, and decreases p53 transcriptional activity. Mutation of the Akt phosphorylation sites in Mdm2 produces a mutant protein that is unable to enter the nucleus and increases p53 activity. The demonstration that PI3-kinase/Akt signaling affects Mdm2 localization provides insight into how this pathway, which is inappropriately activated in many malignancies, affects the function of p53.
The p53 tumor suppressor protein regulates the response of mammalian cells to stresses and damage through transcriptional activation of genes involved in cell cycle control, DNA repair, senescence, angiogenesis, and apoptosis (1–3). Disruption of any of these processes can allow cells to escape from growth constraints, thereby permitting passage of mutations from one generation to the next. Evidence for the tumor suppressor function of p53 comes from studies showing that mice in which the p53 gene has been “knocked out” develop tumors with high frequency (4, 5) and from data showing that deletions or point mutations in the p53 gene are prevalent in a majority of human cancers (6, 7).
The p53 protein is activated in response to environmental stresses, which can be transient, and need not irrevocably direct cells to a pathological course. Thus, the level and activity of p53 are regulated. The Mdm2 protein accomplishes both functions (8). The Mdm2 gene is induced by p53 and the protein binds the transcriptional activation domain of p53, thereby blocking recruitment of factors necessary for induction of gene expression (9–11). Once formed, the Mdm2/p53 complex shuttles from the nucleus to the cytoplasm (12–14), where Mdm2 targets p53 for degradation by acting as an E3 ubiquitin ligase (15–17). Thus, p53 and Mdm2 form an autoregulatory feedback loop in which p53 positively regulates Mdm2 expression and Mdm2 negatively regulates p53 (18).
Interactions between p53, Mdm2, and a product of the INK4a-ARF tumor suppressor locus, p19ARF (19, 20), stabilize p53 by blocking the nuclear to cytoplasmic movement of Mdm2 (21). However, although much is known of how p53/Mdm2 complexes may be retained in the nucleus, the mechanism that induces Mdm2 to enter the nucleus has remained unidentified.
Among the proteins that transmit signals from receptors that promote cell survival is phosphatidylinositol 3-kinase (PI3-kinase) (22), which catalyzes phosphorylation of membrane-bound inositol at the D-3 position, thus producing phospholipid second messengers required for activation of downstream targets. One of these, the Akt/PKB serine-threonine kinase (23–25), transmits survival signals into cells (26–28) and protects cells from anoikis, a form of apoptosis initiated by detachment of cells from the extracellular matrix (29), growth factor deprivation (30), and UV radiation (31). The ability of Akt to suppress apoptosis initiated by so many stresses led us to consider that it might affect the ability of p53 to activate the cell-death machinery. This led us to test whether PI3-kinase/Akt signaling might act on Mdm2 and thereby negatively regulate p53. The present study shows that PI3-kinase/Akt signaling initiated by survival factors promotes nuclear entry of Mdm2.
Materials and Methods
Reagents.
We thank Richard Roth for Akt constructs, Masato Kasuga for dominant-negative and constitutively active p85 PI3-kinase, Gerry Zambetti and Steven Berberich for p53 and Mdm2 constructs, and Tori Ouchi for the mdm2luc reporter. The IF-2 mouse monoclonal antibody to Mdm2 was from Oncogene Science. Mouse monoclonal antibody to the insulin receptor was from Santa Cruz Biotechnology. Rabbit Akt antibodies were from Cell Signaling (Beverly, MA). Activated, recombinant Akt was from Upstate Biotechnology (Lake Placid, NY). Unless otherwise noted, wortmannin was used at 100 nM and LY294002 at 20 μM.
Cell Culture and Transfections.
MCF-7, Saos-2, and human embryonic kidney (HEK) 293 cells were grown in DMEM, 10% FBS, and 10 μg/ml gentamicin. Human keratinocytes were isolated and propagated in keratinocyte media in a humidified incubator under 5% CO2 at 37°C. HEK 293 cells were transfected by using the calcium phosphate procedure and keratinocytes by using the Geneporter system. Lipofectamine was used to transfect Saos-2 cells.
Site-Directed Mutagenesis and Cloning.
The Quik change site directed mutagenesis kit (Stratagene) was used to mutate Akt phosphorylation sites in pRSETAMdm2 to alanines. Oligonucleotides used for mutagenesis were GGAGAGCAATTGCTGAGACAGAAG and CTTCTGTCTCAGCAATTGCTCTCC (serine 166) and CGCCACAAAGCTGATAGTATTTCCC and GGGAAATACTATCAGCTTTGTGGCG (serine 186). Dideoxynucleotide sequencing ensured successful mutagenesis for 2xSA. pRSETA wild-type mdm2 was digested with BamHI and PvuII followed by ligation into FLAG vector digested with BamHI and SmaI.
Mass Spectrometry.
Mdm2 was immunoprecipitated from serum-starved MCF-7 cells stimulated with vehicle or insulin-like growth factor (IGF-1) (50 ng/ml, 30 min, 37°C). Alternatively, recombinant Mdm2 was incubated with or without recombinant, active Akt in vitro. After SDS/PAGE and Coomassie staining, the portion of the gel containing Mdm2 was excised and digested with trypsin. The peptides produced were fractionated by liquid chromatography. Ionizing spray–mass spectrometry was conducted by using a Thermo Finnigan (San Jose, CA) LCQ mass spectrometer programmed to trap masses of predicted target ions containing single or multiple phosphorylations, trypsin miscleavages, and multiple charge states, as well as unphosphorylated Mdm2. The trapped ion targets recovered from experiments conducted in vivo were fragmented by collision-induced dissociation energy of 40. Data were analyzed by using Thermo Finnigan BIOWORKS software.
Confocal Microscopy.
Cells were transfected with 1 μg of FLAG-Mdm2. Cotransfections used 1 μg of mdm2 and 3 μg of constitutively active Akt (CA-Akt), Δp85, or kinase dead Akt (KD-Akt) expression plasmids. A 1:3 ratio of Mdm2 to other plasmids ensured coexpression of both plasmids. Twenty-four hours after transfections cells were fixed, permeated with 0.1% Triton X-100, and blocked with 2% BSA. A Flag primary antibody was added, followed by a Texas red-conjugated sheep anti-mouse secondary antibody. The nucleus was detected by staining with a nuclear dye, Syto 16. Excitation of the stains was performed on a Bio-Rad MRC 1024 Krypton/Argon laser confocal imaging system under 60× magnification and analyzed by using the METAMORPH imaging program (Universal Imaging Corp., Downingtown, PA). Double-blind analysis of cells in numerous fields demonstrated consistent cytoplasmic or nuclear localization of Mdm2 in the presence of constructs or treatment conditions.
Gene Reporter Assays.
Cells were cotransfected with Rous sarcoma virus (RSV) β-galactosidase (β-gal) and a p53 luciferase reporter construct containing 13 copies of the p53 responsive element or the P2 promoter of the mdm2 gene together with Mdm2, p53, CA-Akt, KD-Akt, or combinations of these constructs. Gene reporter activity was then determined as described in the figure legends. Control experiments showed that CA-Akt, KD-Akt, and Mdm2 had no effect on RSV β-gal or the p53 reporter.
Results and Discussion
Mdm2 Is a Substrate for Akt.
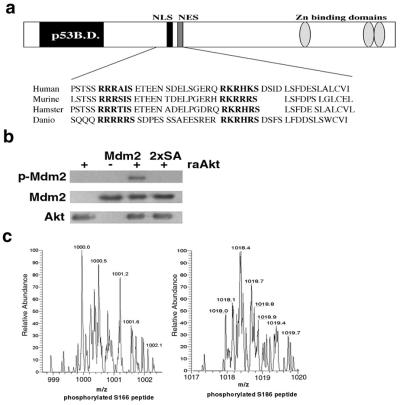
We identified two putative Akt phosphorylation sites (serines 166 and 186) in RxRxxS/T motifs (32) in human Mdm2. The motifs contain an acidic amino acid (+1) following the phosphorylation sites and threonine and serine at the +2 in the position of the respective sites. The motifs are proximal to the nuclear localization signal (NLS; amino acids 181–185) and nuclear export signal (NES; amino acids 190–200) sequences of Mdm2 and are conserved among human, mouse, hamster, and zebra fish Mdm2, indicating functional importance (Fig. 1a). The Akt phosphorylation motif, RxRxxS/T, has been refined to include amino acids that contribute to its three-dimensional structure. Downstream of the phospho-acceptor serine or threonine is a structural amino acid, serine, threonine, glycine, or proline that induces a tight turn in the peptide. This provides the optimal structure for Akt recognition and distinguishes the motif from that recognized by PKC, which selects for basic or aromatic amino acids at the + 2 position (33). Peptide library analysis demonstrates selection for phenylalanine immediately following the phosphorylated serine in the Akt motif. However, substitution of glutamic acid for phenylalanine only modestly decreases peptide phosphorylation by Akt. Thus, for optimal recognition and phosphorylation of an Akt substrate, acidic or aliphatic amino acids are permissible at the +1 position, and an amino acid that induces a turn in the structure is required at the +2 position. The sites in Mdm2 fit the criteria that define an Akt phosphorylation site.
Figure 1.
Akt phosphorylation sites in Mdm2. (a) Schematic of the Akt phosphorylation sites (bold) containing serine 166 and 186 in human Mdm2 and their relationship to the domains in other species. (b) In vitro phosphorylation of human Mdm2 and 2xSA by recombinant activated Akt (raAkt). One microgram of each recombinant protein was incubated with 150 ng of raAkt and 12 μCi (1 Ci = 37 GBq) γ 32P ATP for 30 min at 30°C and a Western blot was prepared. Autoradiography detected phosphorylation of Mdm2 (Top). The blot was probed with anti-Mdm2 (IF-2; Middle) and then anti-Akt (Bottom) to show the presence of equal amounts of each protein. (c) Calculated target ions for serine 166 (mass 1,001) and serine 186 (mass 1,018) from His-Mdm2 incubated in vitro with activated Akt were isolated by mass spectrometry using a quadrapole ion trap with a width of 3 m/z. The high-resolution scan is shown.
The presence of putative Akt phosphorylation sites in Mdm2 led us to determine whether Mdm2 is a substrate for Akt. Human Mdm2, or an Mdm2 mutant in which serines 166 and 186 were mutated to nonphosphorylatable alanine residues (2xSA), was used as a substrate for Akt. Recombinant (34) His-Mdm2 or 2xSA was mixed with recombinant, activated Akt (raAkt) in the presence of 32P gamma ATP, and assayed for phosphorylation. Akt phosphorylated Mdm2, but not 2xSA (Fig. 1b).
Mass spectrometry established that serines 166 and 186 are Akt phosphorylation sites. His-Mdm2 was incubated in vitro in the absence or presence of raAkt. Phosphopeptides from Mdm2 were subjected to electrospray ionization mass spectrometry. For the sample incubated with raAkt, target ions were calculated as charge to mass ratios of 1,001 and 1,018 with a charge state of m+2 for phosphorylated serine 166 and 186, respectively. Consistent with the predicted values, phosphorylated serine 166 and serine 186 were trapped (Fig. 1c). Collision-induced fragmentation of these peptides yielded predicted products (data not shown). These phosphopeptides were not recovered from Mdm2 not incubated with raAkt (data not shown). The results from mass spectrometry are consistent with Mdm2 phosphorylation observed after incubation with raAkt, and with the absence of such phosphorylation in control samples of Mdm2 (Fig. 1b).
PI3-Kinase/Akt Signaling Mediates Mdm2 Phosphorylation.
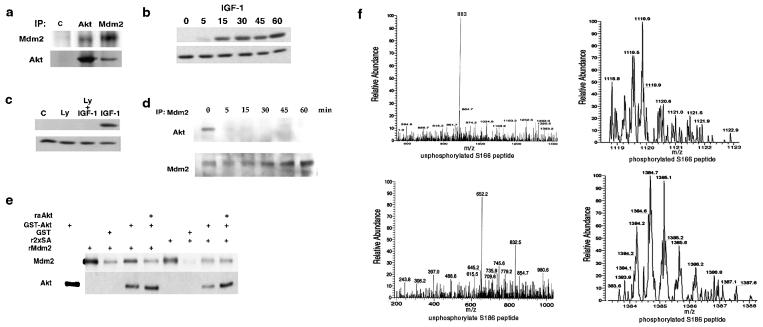
Phosphorylation of Mdm2 by Akt in vitro prompted a test for interaction between the proteins. Western blot analysis showed that under serum-free conditions Akt and Mdm2 associate in MCF-7 cells (Fig. 2a). Interaction between Akt and Mdm2 led us to characterize the pathway that leads to Akt activation. Western blot analysis using an antibody to phosphorylated (activated) Akt established that IGF-1 (Fig. 2b) and insulin (data not shown) activate Akt. To show that Akt activation occurs in a PI3-kinase-dependent manner, serum-deprived cells were treated with LY294002, a PI3-kinase inhibitor, and then with IGF-1. Ablation of PI3-kinase activity abrogated IGF-1-induced Akt phosphorylation (Fig. 2c).
Figure 2.
Phosphorylation of Mdm2. (a) Western blot analysis of insulin receptor (control, 2 μg), Akt, and Mdm2 (2 μg) immunoprecipitated from serum-starved MCF-7 cells probed with anti-Mdm2 (IF-2) or anti-Akt. (b) Serum-starved MCF-7 cells were stimulated with 50 ng/ml IGF-1 and a Western blot was probed with anti-phospho (activated)-Akt (Upper). The blot was stripped and reprobed with anti-Akt (Lower). (c) Serum-starved MCF-7 cells were treated with medium or LY294002 for 30 min and then IGF-1 for 30 min. Western blots were probed with anti-phospho Akt (Upper) or anti-Akt (Lower). (d) MCF-7 cells were stimulated with insulin before immunoprecipitation of Mdm2. A Western blot was probed with anti-Akt and anti-Mdm2. (e) Recombinant Mdm2 or 2xSA was incubated with GST or GST-Akt-agarose for 30 min in vitro, centrifuged, and washed three times. Vehicle or raAkt was incubated with the agarose complexes for 30 min at 37°C and these were centrifuged and washed three times. A Western blot probed with anti-Mdm2 or anti-Akt quantitated the amount of Mdm2 or 2xSA that had associated with GST-Akt agarose. (f) Mdm2 immunoprecipitated from MCF-7 cells incubated with vehicle or IGF-1 was analyzed by mass spectrometry. Predicted parent ions for unphosphorylated serine 166 and serine 186 were recovered from the control incubates (Left), whereas parent ions predicted for phosphorylated serine 166 and serine 186 were recovered from cells treated with IGF-1 (Right).
Because Akt and Mdm2 associate under serum-free conditions, we determined whether stimulation of the Akt pathway would affect the Akt/Mdm2 interaction. Mdm2 was immunoprecipitated from serum-starved cells incubated with medium or insulin. The association of Mdm2 and Akt was lost within 5 min of insulin treatment (Fig. 2d Upper). Fig. 2d Lower shows that the failure to detect Akt association with Mdm2 after insulin treatment did not result from the absence of Mdm2 in immunoprecipitates. To demonstrate by an independent procedure that Akt activation and Mdm2 phosphorylation result in dissociation Akt/Mdm2 complexes, recombinant His-Mdm2 and glutathione S-transferase (GST)-Akt (inactive) immobilized on glutathione beads were mixed and allowed to complex. Addition of raAkt dramatically decreased the amount of Mdm2 associated with GST-Akt, whereas a GST-Akt/2xSA complex was unaffected by raAkt (Fig. 2e).
The demonstration of Akt activation by IGF-1 led us to test whether Mdm2 phosphorylation was promoted by IGF-1. Serum-starved MCF-7 cells were treated with vehicle or IGF-1 and then mass spectrometry analyzed endogenous Mdm2 (Fig. 2f). From cells incubated with vehicle, unphosphorylated parent peptides with a mass to charge ratio of 883 and 652 and a charge state of m+3 were predicted to contain serine 166 and serine 186, respectively, and these were trapped. From cells incubated with IGF-1, phosphorylated parent peptides with a mass to charge ratio of 1,119 and 1,385 and a charge state of m+2 were predicted to contain serine 166 and serine 186, and these were trapped. Unphosphorylated serine 166 and serine 186 were not detected in IGF-1-stimulated cells, nor were phosphorylated parent peaks trapped from unstimulated cells, showing that activation of PI3-kinase/Akt signaling increases Mdm2 phosphorylation.
Confirmation that phosphorylation of Mdm2 occurs on the serine 166 and serine 186 was derived from collision-induced fragmentation of parent ions recovered from stimulated MCF-7 cells (see Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org). All of the recovered peptide fragments (only some of which are listed) were predicted to be present in digests of phosphorylated Mdm2. Our results show that Akt and Mdm2 associate in vivo and in vitro. Akt activation leads to Mdm2 phosphorylation on serines 166 and 186 and dissociation of the Mdm2/Akt complex.
PI3-Kinase/Akt Signaling Induces Localization of Mdm2 to the Nucleus.
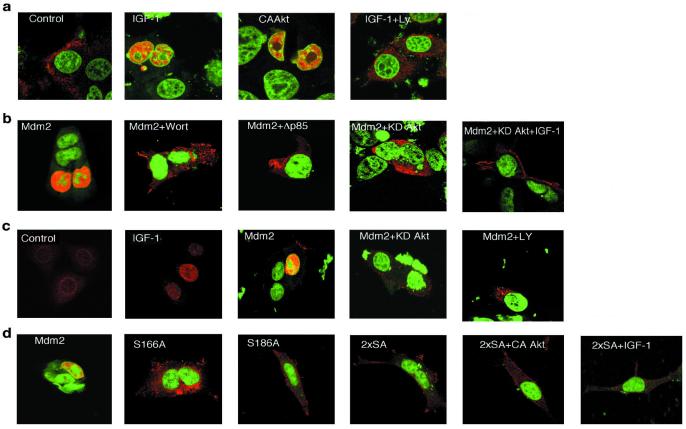
Because the Akt phosphorylation sites in Mdm2 are near the nuclear localization sequence, we tested whether a growth factor or CA-Akt would affect the localization of endogenous Mdm2. Confocal microscopy was performed on serum starved MCF-7 cells transfected with CA-Akt or incubated in the absence or presence of LY294002 and then treated with IGF-1. The cell nucleus was detected with syto-16, a green fluorescent dye, and endogenous Mdm2 with a Texas red-conjugated secondary antibody that recognized an Mdm2 monoclonal antibody. Mdm2 registered as a red signal in the cytoplasm of serum-starved MCF-7 cells (Fig. 3a). CA-Akt or IGF-1 induced translocation of Mdm2 (an orange signal due to superimposition of green and red fluorescence) into the nucleus, and this was blocked by LY294002.
Figure 3.
Localization of Mdm2 characterized by confocal microscopy. (a) Endogenous Mdm2 in serum-starved MCF-7 cells transfected with constitutively active Akt (CAAkt) or incubated in the absence or presence of LY294002 and then treated with IGF-1 (5 min). (b) HEK 293 cells transiently transfected with human Flag-Mdm2 and Flag-Mdm2, then treated with wortmannin (1 h), Flag-Mdm2 and KD-Akt, Flag-Mdm2 and Δp85 PI3-kinase, and Flag-Mdm2 and KD-Akt, and then treated with IGF-1. (c) Primary, serum-starved human keratinocytes were treated with growth factor-free medium (control) or medium containing IGF-1. Keratinocytes were also transfected with Flag-Mdm2, Flag-Mdm2, and KD-Akt, or treated with LY294002 (1 h). (d) HEK 293 cells transfected with Flag-Mdm2, Flag-S166A, Flag-186A, and Flag-2xSA. Additionally, cells were cotransfected with Flag-2xSA and CA-Akt or were transfected with Flag-2xSA and then treated with IGF-1.
The role of PI3-kinase/Akt signaling in the redistribution of Mdm2 was also studied in HEK 293 cells transfected with dominant negative PI3-kinase or Akt. Transiently expressed Flag-tagged Mdm2 was in the nucleus of HEK 293 cells cultured in the presence of growth factors (serum) that activate Akt (Fig. 3b), an observation consistent with the ability of IGF-1 to induce translocation of Mdm2 from the cytoplasm into the nucleus (Fig. 3a). Wortmannin, a PI3-kinase inhibitor, dominant-negative PI3-kinase (Δp85), or KD-Akt led to Flag-Mdm2 localization in the cytoplasm (red fluorescence). KD-Akt also blocked IGF-1-induced nuclear localization of Mdm2, showing that the PI3-kinase/Akt pathway induces nuclear localization of Mdm2 and inhibition of this pathway prevents such localization.
Our observations were extended to primary cells (Fig. 3c). Endogenous Mdm2 was largely cytoplasmic in serum-starved human keratinocytes. Treatment of these cells with IGF-1 led to relocalization of Mdm2 to the nucleus. Quantitation of localization in the primary, serum-starved keratinocytes revealed that among 46 cells no Mdm2 could be detected in the nucleus (100% cytoplasmic localization). Among cells treated with IGF-1 for 15 min, Mdm2 was cytoplasmic in 11 cells and nuclear in 42 cells (80% nuclear localization). In growing keratinocytes transiently expressing Flag-Mdm2, Mdm2 was predominantly in the nucleus, but was restricted to the cytoplasm by treatment with LY294002 or expression of KD-Akt (Fig. 3c).
To demonstrate that the Akt phosphorylation sites in Mdm2 are obligate for entry into the nucleus, Mdm2, S166A, S186A, and 2xSA were transfected into HEK 293 cells. In contrast with the nuclear localization of Mdm2 in proliferating cells, the mutants remained in the cytoplasm (Fig. 3d). Furthermore, CA-Akt, or IGF-1, was not able to alter the cytoplasmic localization of 2xSA, showing that PI3-kinase/Akt could not affect Mdm2 lacking the Akt phosphorylation sites. The fact that mutation of serine 166 or serine 186 to alanine, or blockade of the Akt pathway, prevented nuclear entry shows that each Akt phosphorylation site is important for defining Mdm2 localization. Both serines and the nuclear localization and nuclear export sequences are in a region of Mdm2 that contains predicted helical and coiled structures. Thus, phosphorylation of both sites may be necessary for conformational changes that unmask amino acids in the nuclear localization sequence that permits nuclear entry of Mdm2.
Pharmacological inhibitors determined whether signaling molecules other than Akt might promote nuclear entry of Mdm2. Inhibitors of PKC, p90 RSK, or p38 MAPK did not block mitogen-induced translocation (data not shown). S6 kinase was activated by IGF-1 and this was blocked by inhibition of PI 3 kinase. However, in contrast with the time course of mitogen-induced Akt activation, S6 kinase was not activated rapidly enough to account for the nuclear translocation of Mdm2 (data not shown). We cannot rule out the possibility that an unidentified kinase activated by Akt promotes nuclear entry of Mdm2 in vivo. However, our observations show that a PI3-kinase/Akt pathway regulates the localization of Mdm2 in primary, immortalized, and transformed cells.
Effect of PI3-Kinase/Akt Signaling on p53 Protein Levels.
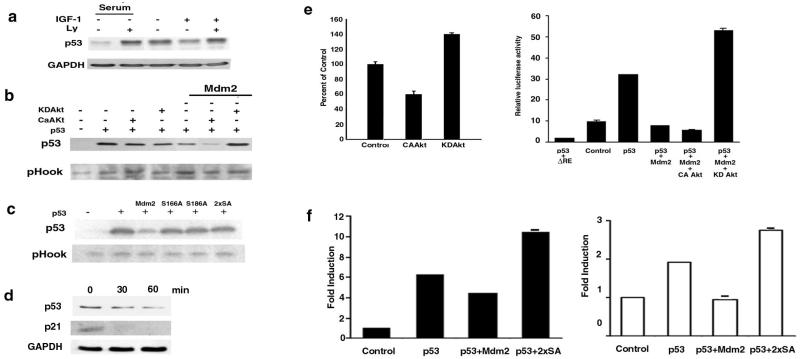
MCF-7 cells were serum starved or incubated with LY294002. These conditions increased endogenous levels of p53, whereas treatment of cells with IGF-1, which promotes nuclear entry of Mdm2, decreased p53 and this was blocked by LY294002 (Fig. 4a). Similar observations were made in experiments with primary human keratinocytes, primary human foreskin fibroblasts, and NIH 3T3 cells (data not shown).
Figure 4.
Regulation of p53 by Akt. (a) MCF-7 cells were cultured in the absence or presence of serum for 24 h and then incubated in the absence or presence of LY294002 for 1 h before treatment with medium or IGF-1 (30 min). A Western blot was then probed for p53 expression. (b) Saos-2 cells were transiently transfected with pHook, p53 in the absence or presence of Mdm2, or Mdm2 with CA-Akt. After 24 h, p53 expression relative to pHook expression was determined by Western blotting. (c) Saos-2 cells were transiently transfected with pHook and p53 alone, or in combination with Mdm2 or mutants of Mdm2 (S166A, S186A, 2xSA). p53 was detected by probing a Western blot with anti-p53. (d) Serum-starved keratinocytes were incubated with IGF-1 for various times. Western blot analysis assayed the effect of such treatment on p53 and p21 protein levels. (e) MCF-7 cells (Left) were transiently transfected with 1 μg β-gal, 2 μg of the p53 luc reporter plasmid, and 7 μg of CA-Akt or KD-Akt. HEK 293 cells were transfected with 0.3 μg β-gal, 1 μg of the mutant or functional mdm2luc reporter plasmid, 1 μg p53, and 1 μg p53 and 5 μg Mdm2 in the absence or presence of 2.4 μg CA-Akt or KD-Akt. After 24 h, β-gal and luciferase activities were measured. Transfections were conducted in triplicate and two aliquots from each cell population were used for β-gal and luciferase assays. Relative luciferase activity was calculated from three independent experiments, normalized to β-gal expression, and standard deviation is derived from the mean. (f) MCF-7 (Left) or HEK 293 (Right) cells were transiently transfected with the mdm2luc reporter, the β-gal expression plasmid, and empty vector (control) or 1 μg of p53 alone, with 5 μg of Mdm2, or with 2xSA Mdm2. Relative luciferase activity was calculated and fold induction is reported relative to control.
p53 null Saos-2 osteosarcoma cells were transiently transfected with KD-Akt or CA-Akt and p53 in the absence or presence of Mdm2 (Fig. 4b). Without ectopic expression of Mdm2, neither KD-Akt nor CA-Akt had a significant effect on p53 protein expression, when normalized for transfection efficiency. However, when expressed with Mdm2, CA-Akt diminished p53, an effect not observed with KD-Akt.
Mdm2 mutants lacking the Akt phosphorylation sites were tested for their ability to stabilize p53. Mdm2 overexpression, but not expression of the S166A, S186A, or 2xSA, diminished p53 protein levels (Fig. 4c). These observations show that the ability of Mdm2 to enter the nucleus plays a fundamental role in regulating the cellular level of p53. Consequently, blockade of PI3-kinase/Akt signaling, or inhibition of Mdm2 phosphorylation by mutation, abrogates mitogen-induced down-regulation of p53.
The observations detailed above were recapitulated in serum-starved keratinocytes that were treated with IGF-1. p53 expression was diminished in a time-dependent manner by IGF-1 (Fig. 4d), a condition that activates PI3-kinase/Akt signaling and Mdm2 redistribution to the nucleus in these primary cells. Furthermore, loss of p53 was accompanied by a decrease in the cellular level of p21, which is transcriptionally regulated by p53.
Effect of PI3-Kinase/Akt Signaling on p53 Function.
Because Mdm2 suppresses the expression of p53 responsive target genes, such as p21 in keratinocytes (Fig. 4d), we used the p53 responsive promoter placed upstream of luciferase as a reporter, and assessed the effect of CA-Akt and KD-Akt on endogenous p53 transactivation. CA-Akt diminished and KD-Akt increased transactivation of endogenous p53 in MCF-7 breast cancer cells (Fig. 4e Left). Additionally, HEK 293 cells (Fig. 4e Right) were transfected with p53 and Mdm2 in the absence or presence of CA-Akt or KD-Akt. Mdm2 suppressed p53 transactivation and this effect was augmented by CA-Akt. In contrast with this latter observation, KD-Akt reversed the ability of Mdm2 to decrease p53 transactivation. These data, and the results from confocal microscopy, show that, in various cell types, CA-Akt induces translocation of Mdm2 into the nucleus and suppresses p53 transactivation function. KD-Akt prevents nuclear localization of Mdm2 and increases p53 activity.
We analyzed the effect of the 2xSA Mdm2 mutant on p53 transcriptional activity (35). In MCF-7 (Fig. 4f Left) and HEK 293 (Fig. 4f Right) cells, Mdm2 diminished p53 activity, whereas 2xSA had the opposite effect. As MCF-7 and HEK 293 cells contain endogenous Mdm2, the 2xSA Mdm2 mutant, by potentiating p53 activity, acted in a dominant-negative manner, which may result from the capacity of Mdm2 to self- associate (36). Binding of Mdm2 to mutant Mdm2 may block nuclear localization of endogenous Mdm2 and its ability to inhibit p53.
Our observations indicate that PI3-kinase/Akt signaling is necessary for nuclear entry of Mdm2 and plays a role in p53 degradation and suppression of p53 function. However, analysis of parent peptides from MCF-7 cells treated with IGF-1 that were trapped by mass spectrometry indicated that threonine 168 was phosphorylated, as were serines 166 and 186. This result may indicate either that Akt could phosphorylate this additional site or that other kinases downstream of PI3-kinase or Akt can modify Mdm2. Additionally, export of the p53/Mdm2 complex from the nucleus, a process regulated by nuclear proteins (19–21) is necessary for p53 degradation (14, 17, 18). Thus, PI3-kinase/Akt signaling appears to be required, but may not be sufficient, to entirely account for the role of Mdm2 in regulating p53 protein levels.
Activation of PI3-kinase and Akt by cytokines or growth factors promotes cell survival through multiple mechanisms (26–31). Phosphorylation of proapoptotic Bad by Akt prevents Bad from interacting with and inhibiting Bcl-x, an antiapoptotic protein (37). Akt phosphorylates and inhibits caspase 9, which initiates an enzyme cascade that promotes apoptosis (38), and FKHRL1, a member of the forkhead family of transcription factors, suppressing its ability to induce expression of genes that promote apoptosis (39). Akt also promotes activation of NF-κB (40–42), which up-regulates antiapoptotic genes (43). The present study shows that PI3-kinase/Akt signaling promotes nuclear localization of Mdm2. This is a crucial step in the mechanism through which Mdm2 inhibits the transcriptional activity of p53, targets p53 for degradation, and provides a mechanism whereby Akt can delay p53-dependent apoptosis in cells (44).
Experimental overexpression of Mdm2 immortalizes and transforms cells and ectopic expression of an mdm2 transgene leads to tumor formation in mice (45–47). A significant fraction of human tumors overexpress Mdm2 and this is predictive of high-grade, aggressive, metastatic malignancies that tend to be refractory to chemotherapy, leading to poor prognosis. Mdm2 overexpression is most often detected in soft tissue sarcomas, osteosarcomas, and leukemias. A smaller subset of esophageal carcinomas, gliomas, anaplastic astrocytomas, and neuroblastomas overexpress Mdm2 (8). The best-defined mechanism through which Mdm2 overexpression leads to uncontrolled cell growth is through negative regulation of p53.
Definition of a PI3-kinase/Akt/Mdm2 pathway that promotes interaction of p53 with Mdm2 shows how mitogens can modulate p53 function and thereby the growth and viability of normal cells, as exemplified by our studies with keratinocytes. Characterization of the role of PI3-kinase/Akt signaling in regulating the Mdm2/p53 interaction, and thereby p53 function, also explains how tumor-derived mitogens can down-regulate p53, which is deplorable in pathologies associated with unregulated cell growth. Targeting components of this pathway may increase the efficacy of cancer therapies that rely on p53.
Supplementary Material
Acknowledgments
We thank Ruben Sandoval for help with confocal microscopy, John Hawes for assistance with mass spectrometry, Ezra Toussaint and Ann Roman for primary keratinocytes, and Mark Kaplan and Jason Gustin for review of the manuscript. L.D.M. is supported by a Hematology Oncology Training Grant from the National Institutes of Health. This work was supported by National Institutes of Health Grants CA67891 and CA73023 (to D.B.D.).
Abbreviations
- PI3-kinase
phosphatidylinositol 3-kinase
- CA-Akt
constitutively active Akt
- KD-Akt
kinase dead Akt
- IGF-1
insulin-like growth factor
- GST
glutathione S-transferase
- β-gal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 10983.
References
- 1.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. J Biol Chem. 1997;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K H. Cell Mol Life Sci. 1999;55:28–37. doi: 10.1007/s000180050267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 5.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 6.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 7.Greenblatt M S, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 8.Freedman D A, Wu L, Levine A J. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 11.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 12.Kubbutat M H G, Ludwig R L, Levine A J, Vousden K H. Cell Growth Differ. 1999;10:87–92. [PubMed] [Google Scholar]
- 13.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao W, Levine A J. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 16.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 17.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Bayle H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 19.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 21.Tao W, Levine A J. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toker A, Cantley L C. Nature (London) 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- 23.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 24.Coffer P J, Woodgett J R. Eur J Biochem. 1991;201:475–481. doi: 10.1111/j.1432-1033.1991.tb16305.x. [DOI] [PubMed] [Google Scholar]
- 25.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsischlis P N, Hay N. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 28.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Science. 1997;275:628–630. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 31.Kulik G, Klippel A, Weber M J. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alessi D R, Caudwell F B, Andjelkovic M, Hemmings B A, Cohen P. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 33.Obata T, Yaffe M B, Leparc G G, Piro E T, Maegawa H, Hashiwagi A, Kikkawa R, Cantley L C. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 34.Mayo L D, Turchi J J, Berberich S J. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 35.Ouchi T, Monteiro A N A, August A, Aaronson S A, Hanafusa H. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 37.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 38.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 39.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 40.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. Nature (London) 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 41.Romashkova J A, Makarov S S. Nature (London) 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 42.Kane L P, Shapiro V S, Stokoe D, Weiss A. Curr Biol. 1999;9:610–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang C-Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 44.Sabbatini P, McCormick F. J Biol Chem. 1999;274:24263–24269. doi: 10.1074/jbc.274.34.24263. [DOI] [PubMed] [Google Scholar]
- 45.Fakharzadeh S S, Trusko S P, George D L. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finlay C A. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones S N, Sands A T, Hancock A R, Vogel H, Donehower L A, Linke S P, Wahl G M, Bradley A. Proc Natl Acad Sci USA. 1996;93:14106–14111. doi: 10.1073/pnas.93.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.