Abstract
Identification of previously unrecognized viral agents in serum or plasma samples is of great medical interest but remains a major challenge, primarily because of abundant host DNA. The current methods, library screening or representational difference analysis (RDA), are very laborious and require selected sample sets. We have developed a simple and reproducible method for discovering viruses in single serum samples that is based on DNase treatment of the serum followed by restriction enzyme digestion and sequence-independent single primer amplification (SISPA) of the fragments, and have evaluated its performance on known viruses. Both DNA viruses and RNA viruses at a concentration of ≈106 genome equivalents per ml were reproducibly identified in 50 μl of serum. While evaluating the method, two previously unknown parvoviruses were discovered in the bovine sera used as diluent. The near complete genome sequence of each virus was determined; their classification as two species (provisionally named bovine parvoviruses 2 and 3) was confirmed by phylogenetic analysis. Both viruses were found to be frequent contaminants of commercial bovine serum. DNase treatment of serum samples may prove to be a very useful tool for virus discovery. The DNase-SISPA method is suitable for screening of a large number of samples and also enables rapid sequence determination of high-titer viruses.
Many human diseases of unknown etiology may be caused by unrecognized viruses. Frozen serum and plasma samples are widely available, which makes such samples a prime target for virus discovery efforts. The successful cloning of hepatitis C virus in 1989 triggered the use of molecular methods to search for unknown viral agents (in particular, additional hepatitis agents) in serum samples (1).
The main obstacle to the identification of viruses in serum and plasma is the extremely small quantity of viral nucleic acids present, in combination with a relatively high level of host genomic DNA. Therefore, molecular virus discovery methods generally need to have two components: amplification and selection (2). Nucleic acids of unknown sequence can be amplified with sequence-independent PCR methods, in which a primer-binding sequence is ligated to both ends of a cDNA fragment that can then be PCR amplified with a single primer. Several variants of this procedure have been described under different names (2), but it is probably most well known as sequence-independent single primer amplification (SISPA; ref. 3). A slightly different approach is random PCR (4), in which the primer-binding sequence is introduced during cDNA synthesis by using a primer-binding extension sequence on random primers. Both methods are efficient but nonselectively amplify essentially all DNA present in the sample. For selection of viral sequences, library construction followed by colony screening [immunoscreening (1) or differential hybridization (5)] has been the most commonly used approach. SISPA followed by immunoscreening enabled cloning of the Norwalk agent genome from stool (6) and “hepatitis G virus” (identical to GB virus C) from plasma (7). It is notable that hepatitis C virus and hepatitis E virus, two medically important viruses, were identified by screening of libraries constructed without any of the amplification techniques (1, 5). Instead, experimentally infected animals provided optimized cloning sources. Regardless of amplification method, the low relative abundance of viral nucleic acids generally requires screening of a very large number of clones.
Recently, representational difference analysis (RDA; ref. 8) has been widely used in the search for new viruses. RDA combines sequence-independent PCR amplification with subtractive hybridization, and thereby addresses the need for both amplification and selection, without library screening. RDA has been applied to identify the tamarin GB viruses and the human TT virus in plasma and serum, respectively (9, 10). However, RDA was originally designed for comparison of two cellular genomes, and its suitability for identifying viruses in serum samples is questionable. First, the original RDA as described by Lisitsyn et al. (8) uses a set of restriction enzymes that cut relatively infrequently, and small virus genomes are likely to have too few restriction sites for these enzymes. This problem has been overcome by using an enzyme that recognizes a four-base pair sequence (9). However, this solution leads to an increased DNA complexity and the risk of inefficient subtractive hybridization (8). Second, serum and plasma samples contain variable amounts of host DNA. Serum in particular may contain high and variable levels of DNA released during clotting (11). If the serum DNA is to be removed by subtractive hybridization, this requires that the DNA composition be similar in the tester and driver samples, which is unlikely. These limitations may explain why RDA has yielded limited results for virus discovery despite its impressive performance in model systems (8). Thus, available molecular methods for virus discovery are labor-intensive, applicable only to selected samples, and not very reproducible.
The host DNA in serum or plasma is present in the form of DNA–histone complexes (oligonucleosomes), which are likely also associated with serum proteins (12, 13). Pharmacodynamic studies suggest that most of the serum DNA can, despite its protein association, be degraded by DNaseI under the proper conditions (14). Viral nucleic acids will generally be protected from DNase degradation by the more stable protein capsids and sometimes also by a lipid envelope.
We developed a method for virus discovery based on DNase treatment of serum, followed by nucleic acid extraction, restriction enzyme digestion, and SISPA of the restriction fragments. The amplification method is derived from the RDA protocol and takes advantage of the fact that virus genomes are numerous, small, and have a low complexity compared with the host genome, and are thus more likely to produce multiple copies of the same fragments, leading to visible bands on gels. Here we describe the performance of this method, called DNase-SISPA, and the characterization of two unknown parvoviruses discovered while evaluating the method.
Methods
Pretreatment of Serum and Extraction of Nucleic Acids.
For each sample analyzed, two 50 μl of serum aliquots (for DNA and RNA virus detection, respectively) were each diluted with 150 μl of H2O and filtered through a 0.22 μm filter (Ultrafree MC, Millipore) at 2,000 × g in a microcentrifuge. DNaseI (100 units; Stratagene) was added to each aliquot, and samples were incubated for 2 h at 37°C. DNA was extracted from one aliquot with a QIAamp blood mini kit (Qiagen, Valencia, CA). RNA was extracted from the other aliquot with Trizol reagent (Life Technologies, Rockville, MD).
Restriction Enzyme-Defined SISPA.
Extracted DNA and RNA were processed separately. For DNA virus detection, a second strand of DNA was synthesized from all extracted DNA with 10 pmol of random hexamers and 5 units of 3′-5′ exo− Klenow fragment of DNA polymerase (New England Biolabs) for 1h at 37°C in 60 μl of a buffer containing 80 nM each dNTP, 10 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, and 7.5 mM DTT. For RNA detection, all extracted RNA was converted to double-stranded cDNA (cDNA synthesis module, Amersham Pharmacia Biotech). First-strand synthesis was carried out in a 20 μl reaction containing 50 mM Tris⋅HCl, pH 8.3, 50 mM KCl, 10 mM MgCl2, 4 mM sodium pyrophosphate, 1 mM each dNTP, 10 pmol of random hexamers, 20 units of RNase inhibitor, and 6 units of avian myeloblastosis virus (AMV) reverse transcriptase at 42°C for 1 h. The second-strand cDNA synthesis reaction mix (40 mM Tris⋅HCl (pH 7.5)/100 mM KCl/5 mM MgCl2/0.06 mg/ml BSA/0.16 units RNaseH/4 units Escherichia coli DNA polymerase I) of 80 μl was added to the first-strand reaction mix and incubated for 1 h at 12°C followed by 1 h at 22°C.
After the second-strand DNA synthesis (DNA protocol) or the complete cDNA synthesis (RNA protocol), the reaction mixtures were heat-inactivated at 72°C for 10 min, and 10 units of Csp6.I (Fermentas, Hanover, MD) was added and incubated for 1 h at 37°C. Then the DNA fragments were extracted, precipitated, dissolved in H2O, and ligated to the adaptor NCsp [hybridized oligonucleotides NBam24 (8), AGG CAA CTG TGC TAT CCG AGG GAG; and NCsp11, TAC TCC CTC GG) in a 10 μl reaction containing 20 pmol of adaptor and 6 units of T4 DNA ligase (New England Biolabs) in a standard ligation buffer at 4°C for 1 h followed by 16°C for 4 h. Two microliters of the ligation reaction was used as a template for PCR, using the oligonucleotide NBam24 as a primer (8). The 50 μl PCR reaction contained 10 mM Tris⋅HCl, pH 8.3, 5 mM KCl, 2.5 mM MgCl2, 0.2 mM each dNTP, and 50 pmol of NBam24. The reaction mixture was heated to 72°C for 3 min before 2.5 units of Taq DNA polymerase (Applied Biosystems) was added. After an additional 5 min at 72°C to extend the DNA and generate the primer-binding sites, 40 cycles of amplification (94°C 1 min, 72°C 3 min) were performed.
Analysis of SISPA Products.
PCR products were analyzed on a 1.5% agarose gel. Intense bands were cut out directly, extracted from the gel, and cloned. To analyze faint bands, a piece of the gel was punched out with a pipette tip, transferred to a PCR reaction tube, and the fragment was reamplified in a second PCR, using the same primer NBam24. This procedure facilitated cloning of barely visible bands. Because each fragment had the same primer sequence at both ends, it could not be used for direct sequencing, and the fragments had to be cloned into a vector. However, to avoid DNA structure-related sequencing problems, the primers were removed by digesting the PCR product with RsaI (in the PCR reaction mix) before gel purification and cloning. The RsaI-digested fragments were cloned in the vector pCR-Blunt (Invitrogen) and introduced into chemically competent E. coli TOP-10 (Invitrogen). Single colonies were cultured and sequenced by using standard techniques. Usually two clones were sequenced for each DNA band.
Sequence Analysis.
The sequences obtained were subjected to homology search with blast (http://www.ncbi.nlm.nih.gov/blast/) (15). Each sequence was analyzed for nucleotide sequence homology (blastn) by searching the standard nonredundant databases as well as the high throughput genomic sequences database. Deduced amino acid sequence homology was analyzed with the translated blast search (blastx).
Phylogenetic Analysis.
The published sequences included in the phylogenetic analysis of parvoviruses have been described previously in detail (16), and aligned sequences were kindly provided by V. Lukashov (University of Amsterdam, The Netherlands; see table 1 in ref. 16). Multiple sequence alignments were obtained by clustal x (version 1.81) or pileup (version 10.2). For ORF2 amino acid alignments, heterogeneity and uncertainty about splicing and location of the N terminus of the actual proteins prompted us to exclude the N-terminal part of the multiple alignment from the phylogenetic analysis [amino acids 1–435 in bovine parvovirus 2 (BPV-2) ORF2 and amino acids 1–364 in BPV-3 ORF2, corresponding to amino acids 1–229 in B19 ORF2]. Trees were constructed with the aid of paup (version 4.0.0d55, created by D. L. Swafford, Laboratory of Molecular Systematics, Smithsonian Institution) by means of the Sequence Analysis Suite (version 10.2; GCG). Bootstrap 70% majority rule consensus trees (midpoint rooting) were obtained by performing heuristic searches (optimality criterion, maximum parsimony; all characters equal weight; 100 replicates). Trees were displayed with the aid of treeview (version 1.6.5; ref. 17).
Results
The Effect of DNase Treatment.
A serum sample derived from an infant with acute hepatitis B virus (HBV) infection [strain HT, complete virus sequence determined (18)] was used for the initial experiments. A 10−2 dilution in bovine serum, previously stored for chimpanzee challenge experiments, was used. The HBV genome titer in this sample was, in the present study, repeatedly found to be 108 genome equivalents (GE) per ml by nested PCR, which was 5-fold higher than reported when tested by another method (18).
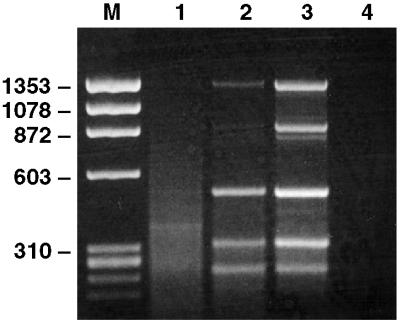
Because the HBV genome is a partially double-stranded DNA molecule, second-strand DNA synthesis was not performed. First, 50 μl of the sample containing 108 GE/ml was subjected to DNA extraction, without DNase treatment, followed by digestion with Csp6.I, adaptor ligation, and PCR, using a single adaptor-specific primer. This protocol repeatedly resulted in a smear of heterogeneous PCR products, with two faint bands visible (Fig. 1). The more intense band was found to contain 326 bp of bovine satellite DNA, whereas the fainter band contained a 462-bp fragment of HBV strain HT, a fragment expected from the Csp6.I digest. When the serum was treated with DNase before extraction, results improved dramatically, because the PCR product was dominated by a number of distinct bands (Fig. 1). The bands appeared to be more intense when the serum was filtered through a 0.22 μm filter before DNase treatment (Fig. 1), and filtering was routinely included in all of the following experiments. All bands were confirmed by sequencing to be derived from HBV strain HT. In fact, the entire HBV HT genome (3,212 bp), except for a 23-bp fragment, was represented. All predicted Csp6.I restriction products were identified, and there was also one additional band representing a minor sequence variant with a mutated restriction site. It is notable that virtually the complete genome was represented, because HBV DNA in virus particles is partially single-stranded and the second-strand synthesis was not performed. It is likely that a fraction of the HBV genomes had two complete strands. Accordingly, the most efficiently amplified fragment (462 bp) was located in the part of the genome that is always double-stranded.
Figure 1.
The effect of DNase treatment and filtering of serum on detection of HBV by SISPA. An HBV-containing serum sample diluted to 108 GE/ml was pretreated in different ways before DNA extraction and SISPA. Products were separated on an agarose gel. The six bands in lane 3 were verified to have HBV sequence. Lane 1, no treatment; Lane 2, DNase treatment only; Lane 3, filtering (0.22 μm) and DNase treatment; Lane 4, negative PCR control (no template); M, molecular weight marker PhiX 174 (HaeIII fragments).
Detection Levels of Different Virus Genomes.
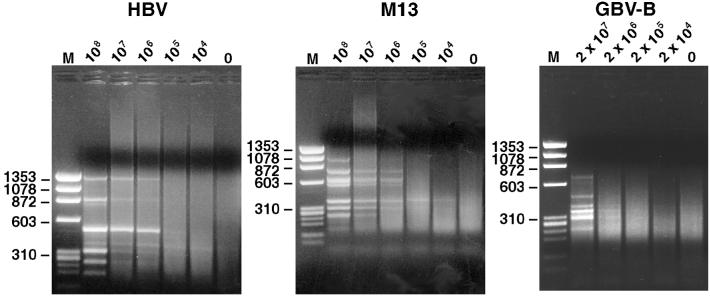
The sensitivity of virus detection was determined by diluting further the HBV HT sample in nonirradiated commercial donor calf serum. Fifty microliters of each dilution was subjected to filtering, DNase treatment, and SISPA as above. Most HBV-derived bands were visible down to the 106 GE/ml dilution, and a single HBV-specific band (462 bp) was visible also at the 105 GE/ml dilution (Fig. 2). A faint bovine satellite DNA band of 326 bp was visible at most dilutions. The experiment was repeated with normal human serum as diluent instead of bovine serum. Results were similar, and the 462-bp HBV fragment was again visible at the the105 GE/ml dilution. Host DNA bands were not visible.
Figure 2.
Detection levels of different virus genomes by DNase-SISPA. HBV, bacteriophage M13, and GBV-B were diluted in 10-fold increments in bovine serum and subjected to the DNase-SISPA procedure. Products were separated on an agarose gel. The virus titer (GE/ml) is shown above each lane. M, molecular weight marker PhiX 174 (HaeIII fragments).
The ability to detect single-stranded DNA viruses was investigated by subjecting bacteriophage M13 (VCSM13, Stratagene) to the DNase-SISPA procedure. Phage were purified by using standard precipitation methods, and the genome titer of the phage preparation was determined by measuring the optical density of extracted DNA. Phage were diluted in bovine serum and subjected to filtration, DNase treatment, and DNA extraction. Double-stranded DNA was produced by using 3′-5′ exo− Klenow fragment of DNA polymerase and random hexamers, before applying the Csp6.I digest and SISPA protocol, as for HBV. A control without the polymerase was tested to exclude the possibility that results were obtained from contaminating double-stranded phage DNA. At least six phage-derived bands were visible in the sample containing as few as 106 genomes per ml (Fig. 2). The diluent-derived bovine satellite band was visible in some of the lanes in which M13 bands were absent.
The ability to detect RNA viruses was evaluated with an acute phase serum from a tamarin experimentally infected with GB virus B (GBV-B). The titer of GBV-B was determined to be 108 GE/ml by reverse transcription (RT)-nested PCR (19). Because of limited availability of the serum, a 1:5 dilution followed by serial 10-fold dilutions in the same bovine serum as above was tested. The samples were filtered, treated with DNase, and RNA was extracted. First- and second-strand cDNA was synthesized and subjected to Csp6.I digest and SISPA. At the lowest dilution tested (1:5 or ≈2 × 107 GE/ml), 7 GBV-B bands (predicted restriction digest fragments and specificity confirmed by sequencing) were found, representing 3,162 bp (34%) of the GBV-B genome. Three of these GBV-B bands (confirmed by sequencing) were also visible at a dilution of 2 × 106 GE/ml (Fig. 2).
Cloning of BPV-2.
The seven bands seen in the lowest dilution in the GBV-B titration experiment were cloned and sequenced. Of the 12 clones, 10 had GBV-B sequence. The two other clones had approximately the correct cloned insert size of a GBV-B band but did not have GBV-B sequence. A blast search of the deduced amino acid sequence indicated that one clone could be derived from a parvovirus. Both fragments were amplified from the bovine serum used as diluent but not from the tamarin serum when specific primers were used for PCR. The two fragments could be bridged by PCR when combining primers specific for each fragment. The resulting product of 874 bp apparently overlapped the upstream terminus of a parvovirus genome. The genome titer of the virus in the source serum was determined to be 105 GE/ml by nested PCR of serially diluted DNA with two specific primer sets (Table 1). A combination of methods was applied to obtain additional fragments covering the virus genome: PCR walking according to Sørensen et al. (20), subsequent identification of one additional clone by RDA (see Discussion), and bridging of the fragments by PCR, as well as deoxycytidine tailing followed by PCR [5′ rapid amplification of cDNA ends kit Life Technologies]. The near complete consensus sequence (5,610 bp) was determined, including the two ORFs and a putative downstream terminus (Fig. 3A). We failed to extend the upstream 305-bp noncoding sequence further than the Csp6.I restriction fragment derived from SISPA. The virus was classified as a parvovirus, because the sequence recovered was of DNA origin and had the size of a parvovirus genome, and it had two large ORFs, of which the first encoded a conserved parvovirus NS-1 domain and the second encoded a conserved parvovirus capsid domain. Thus, the deduced amino acid sequence of both ORF1 and ORF2 had significant homology with a wide range of parvoviruses. However, at the nucleotide level, a blast search found no significant sequence homology with other viruses, including parvoviruses. The virus was provisionally named bovine parvovirus 2, because one bovine parvovirus has been already identified (21).
Table 1.
Primers used for detection of BPV-2 and BPV-3
| Target | 5′ Position | Sequence |
|---|---|---|
| BPV-2 NS | 528F | TAC TTT ATA CAG CTA GAG CAA G |
| BPV-2 NS | 872R | GTC TAT GTC CGG TCT AAC TG |
| BPV-2 NS | 559F | ACA AGA GGC GGC ACA TAC |
| BPV-2 NS | 828R | TTA AAC AAG CGT CTC CTA TCT T |
| BPV-2 UTR | 19F | GTA GTT CCG GGT CAA GTG |
| BPV-2 UTR | 299R | CTC ACC AAG AGA CGT CCT C |
| BPV-2 UTR | 38F | CGT AGT TCC GGT GAC GTG TC |
| BPV-2 UTR | 279R | CTC TCT GAC AGC TGG GAG AAG |
| BPV-3 capsid | 4411F | CGG AAC CGC TAA GGC ATC AG |
| BPV-3 capsid | 4829R | ACT GGT GCA GGT AAT GCT CTG A |
| BPV-3 capsid | 4692F | AGC GAC CAG CAT TGG GTG GAT |
| BPV-3 capsid | 4826R | GGT GCA GGT AAT GCT CTG AG |
NS, nonstructural gene; UTR, untranslated region.
Figure 3.
Overview of the BPV-2 (A) and BPV-3 (B) genomes as deposited in GenBank. The actual sizes and sequences of the transcripts and proteins of the indicated ORFs have not been investigated. NS, nonstructural gene.
Cloning of BPV-3.
In a search for higher sensitivity of virus detection, we applied DNase treatment followed by random PCR (4) to the serially diluted HBV sample. In brief, this was done by first-round PCR with the primer N4G4 (AGG CAA CTG TGC TAT CCG AGG GAG CTC NNN NGG GG) and 45°C annealing temperature, followed by second round PCR with the primer NBam24 as described for the SISPA, and subsequent random cloning and sequencing of the products. In this experiment, the detection limit for HBV was greater than 106 GE/ml and thus the procedure was not as sensitive as SISPA. However, at the lower dilutions we recovered not only HBV sequence, but also a number of clones having a deduced amino acid sequence homologous to parvoviruses. Interestingly, they did not share the sequence of the recently discovered BPV-2. This virus was not found by PCR with specific primers (Table 1) in the bovine serum used as diluent in the titration, but it was detected in the bovine serum used many years earlier to produce the HBV challenge pool (18). It had not been detected in the SISPA experiments with HBV, because the second-strand DNA synthesis step was not included in those experiments. This fact confirmed the single-stranded nature of the virus genome. By nested PCR, the virus was detected in 5 of 8 commercial bovine sera tested (Table 2). The titer seemed to be high in most of the positive sera and was determined to be 108 GE/ml by 10-fold dilutions and nested PCR in one serum (calf serum-1). This lot of bovine serum was used for determining the sequence of the virus. Because of the high titer, DNase-SISPA facilitated sequencing of the entire genome. Two 50 μl aliquots were filtered, DNase-treated, and subjected to DNA extraction. The SISPA procedure for single-stranded DNA was applied as described above; however, Csp6.I was used for digesting the DNA of one aliquot, and TaqI and a corresponding adaptor (hybridized oligonucleotides NTaq24, AGG CAA CTG TGC TAT CCG AGG GAT; and NTaq11,: CGA TCC CTC GG) were used for the other aliquot (Fig. 4). The purpose was to obtain overlapping clones for assembly of the whole sequence. However, the Csp6.I fragments were never analyzed; from the sequence of the 8 bands of the TaqI reaction (Fig. 4), the genome was easily assembled by homology search. PCR experiments were carried out to confirm the sequence and identified only 8 additional nucleotides at the junction of two TaqI fragments. The resulting consensus sequence of 5,276 bp included two ORFs plus over 200 bp of flanking noncoding sequence at both ends (Fig. 3B). The virus was classified as a parvovirus based on the same criteria as described above for BPV-2 and was provisionally named bovine parvovirus 3.
Table 2.
Detection of BPV-2 and BPV-3 in eight lots of commercial bovine serum
| FCS1 | FCS2 | FCS3 | FCS4 | NBCS | CS1 | CS2 | CS3 | |
|---|---|---|---|---|---|---|---|---|
| BPV-2 | − | − | − | − | + | + | + | − |
| BPV-3 | − | − | + | + | + | + | + | − |
NBCS, newborn calf serum; CS, calf serum.
Figure 4.

The use of DNase-SISPA for obtaining the sequence of unknown viruses, illustrated by SISPA products of a serum containing 108 GE/ml of BPV-3. (Left) Marker. (Center) SISPA based on digestion with Csp6.I. (Right) SISPA based on digestion with TaqI.
Phylogenetic Analysis of BPV-2 and BPV-3.
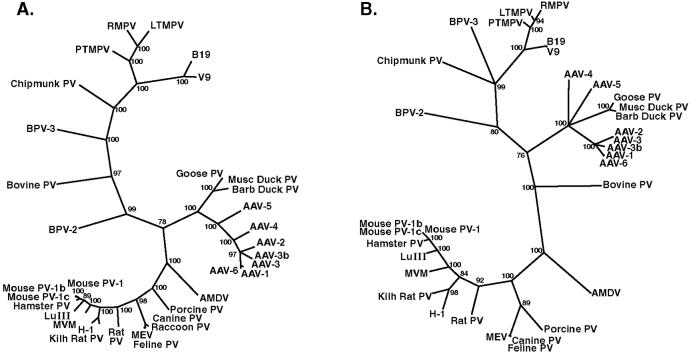
Lukashov and Goudsmit (16) have recently published a comprehensive phylogenetic analysis of full-length sequences of the known parvoviruses. We aligned BPV-2 and BPV-3 to these sequences and constructed phylogenetic trees based on multiple sequence alignments of the full-length nucleotide sequence (Fig. 5A) as well as the amino acid sequences of ORF1 (not shown) and most of ORF2 (Fig. 5B). All trees were consistent with the analysis of Lukashov and Goudsmit (16) and correctly identified the three major phylogenetic groups: (i) adeno-associated viruses and avian parvoviruses, (ii) primate and chipmunk parvoviruses, and (iii) parvoviruses from rodents, carnivores, and pigs. Lukashov and Goudsmit (16) included the bovine parvovirus as an outlier with the primate and chipmunk parvoviruses. With BPV-2 and BPV-3 added to the analysis, this grouping is questionable. BPV-3 was the bovine virus most closely related to the chipmunk and primate parvoviruses. However, bovine parvovirus as well as BPV-2 branched off variably in the different analyses, near the center of the tree. Because of considerable sequence heterogeneity, the variable results for these viruses may have resulted from alignment difficulties. Therefore, the most conserved fragment of about 500 nucleotides located in ORF1 (corresponding to nucleotides 1,147–1,670 in BPV-2 and nucleotides 1,096–1,616 in BPV-3) was selected to make an additional tree that was based on a precise alignment. In the resulting tree, the branching points for bovine parvovirus, BPV-2, and BPV-3 became insignificant (bootstrap value <70%), and they all branched off separately from a common node in the center of the tree (data not shown). In conclusion, BPV-2 and BPV-3 clearly constitute two parvovirus species that are not closely related to each other or to any other known parvovirus. Our data suggest that BPV-3 may group with the primate and chipmunk parvoviruses, but neither bovine parvovirus nor BPV-2 can be readily included in any of the three major phylogenetic groups previously recognized.
Figure 5.
Phylogenetic trees of full-length nucleotide sequences (A) and truncated ORF2 amino acid sequences (B) of parvoviruses (Parvovirinae subfamily), including BPV-2 and BPV-3. Bootstrap values >70% are indicated.
Presence of BPV-2 and BPV-3 in Commercial Bovine Serum.
Eight lots of commercial bovine serum from three suppliers were investigated for the presence of BPV-2 and BPV-3 DNA by nested PCR (Table 1). Four lots of FCS, one lot of newborn calf serum (animals 0–10 days old) and three lots of calf serum (animals 10 days to 6 months old) were tested (Table 2). BPV-2 was absent in FCS but present in 3 of 4 nonfetal sera. BPV-3 was present in 5 of 8 sera, including two fetal sera. The identity of the PCR products was confirmed by sequencing, and a high degree of conservation was seen among the various isolates of BPV-2 and BPV-3, respectively. This limited investigation suggests that both viruses are frequently present in commercial bovine sera. BPV-3 generally gave an intense band in the first PCR, suggesting a high titer, consistent with the titer of 108 GE/ml found in calf serum-1. Because BPV-3 was found in an HBV challenge pool, which had been used to inoculate chimpanzees (18), we tested four sera (weeks 1–4 after inoculation) from one of the inoculated chimpanzees by PCR. BPV-3 DNA was not detected.
Discussion
Great efforts have been made to identify new etiologic agents in serum samples. New powerful molecular techniques such as screening of expression libraries and subtractive hybridization methods (RDA) have proven very useful in this search (2). However, these methods are labor-intensive and require weeks to analyze each sample. Access to a preinfection as well as an acute phase sample may also be required. These are important limitations that inhibit analysis of multiple serum samples and thereby reduce the chance of identifying viral agents.
The DNase-SISPA method offers reproducible detection of viruses having a titer of ≈106 GE/ml or higher. The detection level is an obvious limitation to this method. However, it is not unusual that the known human pathogens found in blood (e.g., HIV and hepatitis C virus) reach titers higher than 106 GE/ml, particularly during acute infection. Moreover, RDA may not have a greater sensitivity than this for use with serum. Of the viruses discovered in serum using RDA, the GB viruses regularly reach titers of ≈108 GE/ml (19), and TTV often reaches 106–107 GE/ml (22). Because the DNase method allows for simultaneous screening of a large number of samples, the limited sensitivity can, at least in part, be overcome by analysis of consecutive samples from a potentially infected individual or samples from many individuals with the same disease. A virus with a lower titer may also occasionally be detected, as illustrated by the discovery of BPV-2 at a titer of 105 GE/ml.
Identification of new viral sequences requires both amplification and selection. In the DNase-SISPA method, the selection is based on the physical properties of virus particles: they consist of small filterable particles, in which the nucleic acid is protected from DNase, and they are present in multiple copies, resulting in visible bands after amplification of restriction enzyme fragments. The final discovery step is the database search. The current explosion in the accumulation of sequence data has made this an effective strategy. The majority of nonviral sequences could be identified as either mammalian or bacterial. In fact, the majority of such sequences were not host-derived but bacterial, and we found that a majority of the nonviral DNA was derived from the reagents. Enzymes and reagents for molecular biology do contain DNA from the bacteria in which they are produced and possibly from other sources (23). Indeed, not all cDNA synthesis reagents we tested were suitable for the DNase-SISPA procedure because they had a high content of bacterial or plasmid DNA that gave “false positive” bands.
The important selective step of the method is the DNase treatment. After this step, several approaches can be taken to amplify the virus-enriched nucleic acids. For example, we tested DNase treatment followed by random PCR (4) in an attempt to minimize the use of enzymes, and this procedure identified the first fragments of BPV-3. In another attempt to improve assay sensitivity, DNase-treated serial dilutions of GBV-B were subjected to RDA. This approach resulted in recovery of a GBV-B sequence from a sample diluted to 2 × 105 GE/ml. A fragment of the BPV-2, having a titer of 105 GE/ml in the diluent serum, was also recovered by this procedure, although only in 1 of 4 samples. Thus, DNase treatment followed by RDA may provide a more sensitive approach than DNase-SISPA. However, a large number of false positive bands with bacterial sequence were also generated, and therefore the increased sensitivity is gained at the cost of more cloning and sequencing. The virus-enriched DNase-SISPA products may of course also be subjected to cloning and library screening.
In addition to its use for virus discovery, DNase treatment of serum could prove to be a generally useful tool for virological studies of serum samples whenever backgrounds derived from host DNA is a problem, for example PCR with degenerate or random primers or determination of terminal sequences by anchored PCR. In the present study, DNase treatment of the serum facilitated the various procedures involved in determining the complete BPV-2 sequence. The DNase-SISPA method is not only suitable for virus discovery, but also for generating fragments suitable for sequencing the near complete genome of an unknown high-titer virus, as illustrated by the sequencing of BPV-3 in this report.
The viruses identified in the present report belong to the Parvoviridae family (commonly referred to as parvoviruses). The family has two subfamilies, Densovirinae, which infect arthropods, and Parvovirinae, which infect vertebrates. The Parvovirinae subfamily has three genera, Dependovirus (adeno-associated viruses), Erythrovirus (B19 and other primate parvoviruses), and Parvovirus (animal parvoviruses). The genus classification is based on questionable phenotypic properties, among them helper virus dependence in vitro. It is not necessarily consistent with the phylogenetic relationships, and has therefore been questioned (16). Because very little is known about the phenotypes of BPV-2 and BPV-3, they cannot be currently classified under this system. Until now, mainly one parvovirus per host species has been described with the exception of humans, and phylogenetic analysis suggests coevolution of virus and host in many cases (16). It is therefore interesting that three phylogenetically very different parvoviruses have been found in cattle. This finding suggests that the Parvovirinae subfamily may be substantially larger than known today, with a number of virus species per host animal.
The finding of two parvoviruses in commercial bovine serum is yet another reminder that viruses can be present in any biological material. Indeed, the serum in which BPV-2 was detected was a cell culture reagent that was carefully selected to be free of bovine viral diarrhea virus and other known viruses. Because bovine sera are used for production of pharmaceuticals, any virus in these reagents is a concern. However, we could not detect BPV-3 in the serum of a chimpanzee inoculated with this virus. It is worth noting that in particular, BPV-3 is frequently found in common laboratory reagents at a high titer, but despite this it has not been detected previously. Whether either of the two viruses causes disease in cattle remains to be determined. Our examination of commercial sera suggests that infection by both viruses is frequent. BPV-2 occurs in calves soon after birth, whereas BPV-3 may be transmitted in utero. It remains to be investigated whether BPV-2 and BPV-3 viremia is transient or persistent.
In conclusion, we have developed a relatively simple and reproducible DNase-based method for detecting unknown viruses in a small volume of serum. When evaluating the sensitivity of this method with known viruses diluted in bovine serum, two bovine parvovirus species commonly present in commercial bovine sera were identified. DNase treatment may prove to be a very useful tool for the discovery of viruses in serum samples.
Acknowledgments
We thank Dr. V. Lukashov for providing sequence alignments and Dr. R. T. Simpson (Pennsylvania State University) for valuable advice. This study was supported in part by National Institutes of Health Contract CO-56000. T.A. is the recipient of a grant from the Swedish Foundation for International Cooperation in Research and Higher Education (STINT).
Abbreviations
- SISPA
sequence-independent single primer amplification
- RDA
representational difference analysis
- HBV
hepatitis B virus
- GE
genome equivalents
- GBV-B
GB virus B
- BPV-2 or -3
bovine parvovirus 2 or 3
Footnotes
References
- 1.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Muerhoff A S, Leary T P, Desai S M, Mushahwar I K. J Med Virol. 1997;53:96–103. [PubMed] [Google Scholar]
- 3.Reyes G R, Kim J P. Mol Cell Probes. 1991;5:473–481. doi: 10.1016/s0890-8508(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 4.Froussard P. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes G R, Purdy M A, Kim J P, Luk K C, Young L M, Fry K E, Bradley D W. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 6.Matsui S M, Kim J P, Greenberg H B, Su W, Sun Q, Johnson P C, DuPont H L, Oshiro L S, Reyes G R. J Clin Invest. 1991;87:1456–1461. doi: 10.1172/JCI115152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linnen J, Wages J, Jr, Zhang-Keck Z Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, et al. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 8.Lisitsyn N, Lisitsyn N, Wigler M. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 9.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, et al. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 11.Steinman C R. J Clin Invest. 1975;56:512–515. doi: 10.1172/JCI108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumore P M, Steinman C R. J Clin Invest. 1990;86:69–74. doi: 10.1172/JCI114716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bickerstaff M C, Botto M, Hutchinson W L, Herbert J, Tennent G A, Bybee A, Mitchell D A, Cook H T, Butler P J, Walport M J, et al. Nat Med. 1999;5:694–697. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]
- 14.Prince W S, Baker D L, Dodge A H, Ahmed A E, Chestnut R W, Sinicropi D V. Clin Exp Immunol. 1998;113:289–296. doi: 10.1046/j.1365-2249.1998.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukashov V V, Goudsmit J. J Virol. 2001;75:2729–2740. doi: 10.1128/JVI.75.6.2729-2740.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page R D. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 18.Ogata N, Miller R H, Ishak K G, Purcell R H. Virology. 1993;194:263–276. doi: 10.1006/viro.1993.1257. [DOI] [PubMed] [Google Scholar]
- 19.Bukh J, Apgar C L, Yanagi M. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- 20.Sørensen A B, Duch M, Jørgensen P, Pedersen F S. J Virol. 1993;67:7118–7124. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K C, Shull B C, Moses E A, Lederman M, Stout E R, Bates R C. J Virol. 1986;60:1085–1097. doi: 10.1128/jvi.60.3.1085-1097.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto A, Yeo A E, Shih J W, Tanaka E, Kiyosawa K, Alter H J. Hepatology. 1999;30:283–288. doi: 10.1002/hep.510300118. [DOI] [PubMed] [Google Scholar]
- 23.Corless C E, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski E B, Fox A J. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]