Abstract
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is a widely cultivated and economically important vegetable crop with typical leaf curvature. The TCP (Teosinte branched1, Cycloidea, Proliferating cell factor) family proteins are plant-specific transcription factors (TFs) and play important roles in many plant biological processes, especially in the regulation of leaf curvature. In this study, 39 genes encoding TCP TFs are detected on the whole genome of B. rapa. Based on the phylogenetic analysis of TCPs between Arabidopsis thaliana and Brassica rapa, TCP genes of Chinese cabbage are named from BrTCP1a to BrTCP24b. Moreover, the chromosomal location; phylogenetic relationships among B. rapa, A. thaliana, and rice; gene structures and protein conserved sequence alignment; and conserved domains are analyzed. The expression profiles of BrTCPs are analyzed in different tissues. To understand the role of Chinese cabbage TCP members in regulating the curvature of leaves, the expression patterns of all BrTCP genes are detected at three development stages essential for leafy head formation. Our results provide information on the classification and details of BrTCPs and allow us to better understand the function of TCPs involved in leaf curvature of Chinese cabbage.
Keywords: Chinese cabbage, TCP TFs, head formation, leaf curvature
1. Introduction
The TCP family proteins are widespread and specific transcription factors (TFs) in plants; examples include teosinte branched1 (TB1) from Zea mays, cycloidea (CYC) from Antirrhinum majus, and proliferating cell factor (PCF) from Oryza sativa [1,2,3,4,5] Based on phylogenetic analysis, TCP proteins are divided into two classes: class I and class II. Class I (TCP-P) is related to PCF1 and PCF2 proteins in rice, whereas class II (TCP-C) to CIN (Cincinatta in Antirrhinum) and the CYC/TB1 subgroups [6]. The TCP family genes encode a conserved region containing 59 amino acid residues with a basic helix–loop–helix (bHLH) structure, which are involved in DNA binding, protein–protein interaction, and protein nuclear localization [3]. However, the DNA binding sites of the two TCP classes are partly overlapping. Class I TCPs bind the DNA sequence GGNCCCAC, whereas class II GTGGNCCC [6].
The TCP family TFs regulate plant growth and development and lateral branching [7,8], leaf morphogenesis [8,9], flower development [10,11], embryo growth [12,13], circadian rhythm [14], and hormonal pathways [15,16]. In Arabidopsis, class I and II TCP members are involved in leaf development. The class II CIN-type genes, AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, are regulated by miR319a and lead to serrated and crinkled leaves [17]. The repression of class I AtTCP11 in the C-terminal domain results in curling of rosette leaves in Arabidopsis [16]. AtTCP15 and its closest homologous gene AtTCP14 play a negative role in the elaboration of leaf shape [18]. Moreover, TCP TFs have attracted the attention of the scientific community because of their function in developmental processes and defense response against biotic and abiotic stresses. TCP13, TCP14, and TCP19 were remarkably found to participate directly in pathogenesis [19]. The tcp15 mutant improves disease resistance to HpaNoco2 in Arabidopsis. Furthermore, compared with other TCPs, TCP8, TCP14, and TCP15 interact more strongly with SRFR1, an adaptor protein in cytoplasmic microsomal and nuclear protein complexes. By contrast, class I TCP proteins with a conserved cysteine residue at position 20 (Cys-20) are sensitive to redox conditions, which affect the DNA-binding activity of TCP. In a previous study, a class I TCP protein OsPCF2 induced by salt stress was shown to participate in salt stress by binding the promoter of OsNHX1 to activate its expression [20]. NHX1 encodes a kind of K+-Na+/H+ antiporter, and its overexpression enhances the tolerance of salt and drought stress in Arabidopsis and rice [21]. Moreover, the overexpression of OsTCP19 improves the tolerance of Arabidopsis under drought stress by modulating the ABI4-mediated pathways [22]. Thus, based on the above findings, TCP proteins play a significant role in developmental processes and defense against biotic and abiotic stresses.
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is a widely cultivated and economically important vegetable crop in Asia. Chinese cabbage originates in China, and it has now become increasingly popular in other countries [23]. The vegetative periods of Chinese cabbage are divided into four stages: germination, seedling, rosette, and heading. Leafy head formed at the heading period is the storage organ and commonly the eaten part of the cabbage. The yield and quality of cabbage are generally measured by the size and solidity of the head. Head formation is affected by the direction of leaf curvature, and more and more research on the heading mechanism of cabbage has been made. Previous studies speculated that head formation is caused by the asymmetrical distribution of auxin [24]. In another study, some genes of the BrLAX, BrPIN, and BrPGP families have been found to play important roles in the asymmetrical auxin distribution by affecting the polar auxin transport during leafy head development [25]. Furthermore, in accordance with the work model of leaf shape diversity regulated by adaxial–abaxial (Ad–Ab) polarity in Arabidopsis [26], six leaf Ad–Ab patterning candidate genes were selected between heading and non-heading Chinese cabbages, namely BrARF3.1, BrARF4.1, BrKAN1, BrKAN2.1, BrRDR6, and BrHYL1.1, which are linked to head formation [27]. Some genes are known to participate in regulating the shape of leaves in Arabidopsis; BrpTCP4 was particularly reported to regulate the head shape of Chinese cabbage by miR319a [9].
As the whole genome of Chinese cabbages is sequenced [28], genome-wide analysis of TCP genes is performed for the first time in our study. In the present study, the TCP TF family in Chinese cabbage is comprehensively analyzed. We identify a total of 39 genes encoding TCP TFs on the whole genome of Brassica rapa. By associating the phylogenetic analysis of TCP domain proteins between Arabidopsis thaliana and Brassica rapa, BrTCP1a to BrTCP24b are named. Then, the chromosomal location; phylogenetic relationships among B. rapa, A. thaliana, and rice; gene structures and protein conserved sequence alignment; and conserved domains were analyzed. The expression profiles of BrTCPs are detected in different tissues. To determine the role of Chinese cabbage TCP members in regulating the curvature of leaves, the expression patterns of all BrTCP genes are detected at three essential development stages. The results of our study provide information on the classification and details of BrTCPs and lay the foundation for future studies on the leaf curvature mechanism of TCP proteins in Chinese cabbage.
2. Results
2.1. Identification of TCP TFs in Chinese Cabbage
A conserved TCP domain is usually found at the N terminal of TCP TFs. To obtain the TCP members of Chinese cabbage, the TCP domains (PF03634) of the B. rapa genome were searched on the Brassica Database (BRAD), and all TCP candidate sequences were examined using the domain analysis online tools: PfamScan (https://www.ebi.ac.uk/Tools/pfa/pfamscan/), InterPro (a protein sequence analysis and classification website, http://www.ebi.ac.uk/interpro/), and SMART (http://smart.embl-heidelberg.de/). As a result, 43 genes were found with a typical TCP domain, and 39 genes with no redundant sequences were screened. These genes were confirmed to contain the TCP domain with the ScanProsite tool. They were named BrTCP1 to BrTCP24 according to the TCP gene classifications of A. thaliana (Table S1). However, no homologous genes of AtTCP11, AtTCP16, and AtTCP23 were found in Chinese cabbage. The main TCP genetic characteristics of B. rapa are summarized in Table 1, including the gene names, gene ID, chromosome location, full length of cDNA, exon numbers, protein length, molecular weights, and isoelectric points. For all BrTCPs, the protein length ranged from 176 aa to 538 aa, the molecular weights from 18,601 Da to 59,180.4 Da, and the isoelectric points from 5.419 to 10.8713 as shown in Table 1.
Table 1.
TCP family transcription factor genes in Chinese cabbage.
| Gene Name | Gene ID | Chromosome Location | CDS Length (bp) | Exon | Protein length (aa) | MW (Da) | PI |
|---|---|---|---|---|---|---|---|
| BrTCP1a | Bra004212 | A07:20728546–20729668 | 1026 | 2 | 341 | 38,786.3 | 6.5294 |
| BrTCP1b | Bra004097 | A07:20111066–20112206 | 1044 | 2 | 347 | 39,380 | 6.1198 |
| BrTCP1c | Bra034010 | A02:10223409–10224449 | 1041 | 1 | 346 | 38,511.8 | 5.477 |
| BrTCP2a | Bra013304 | A01:4882809–4883528 | 720 | 1 | 239 | 39,219.7 | 8.3134 |
| BrTCP2b | Bra012600 | A03:23315387–23316535 | 1149 | 1 | 382 | 42,262.9 | 7.6883 |
| BrTCP3 | Bra030952 | A08:1031146–1032183 | 1038 | 1 | 345 | 37,595.3 | 7.2706 |
| BrTCP4a | Bra027284 | A05:20829037–20830257 | 1221 | 1 | 406 | 44,035.4 | 7.5321 |
| BrTCP4b | Bra001579 | A03:17191721–17192944 | 1224 | 1 | 407 | 44,472.9 | 8.0355 |
| BrTCP4c | Bra021586 | A01:23505568–23506620 | 1053 | 1 | 350 | 38,203.2 | 7.3461 |
| BrTCP5a | Bra012990 | A03:21199353–21200453 | 1101 | 1 | 366 | 40,698.2 | 6.6787 |
| BrTCP5b | Bra029344 | A02:25211660–25212733 | 906 | 2 | 301 | 33,796.16 | 6.5 |
| BrTCP6 | Bra025528 | A04:8476440–8477114 | 675 | 1 | 224 | 24,724.6 | 7.816 |
| BrTCP7a | Bra029366 | A02:25051573–25052103 | 531 | 1 | 176 | 18,601 | 8.2872 |
| BrTCP7b | Bra009667 | A06:17073181–17073933 | 753 | 1 | 250 | 26,968.1 | 10.3355 |
| BrTCP7c | Bra026506 | A09:3299278–3299928 | 651 | 1 | 216 | 23,116.8 | 9.867 |
| BrTCP8 | Bra027886 | A09:10035227–10036411 | 1185 | 1 | 394 | 41,385.3 | 6.5677 |
| BrTCP9a | Bra004929 | A05:2539838–2540812 | 975 | 1 | 324 | 34,510.1 | 10.1796 |
| BrTCP9b | Bra000395 | A03:10937732–10938667 | 936 | 1 | 311 | 33,174.9 | 10.2854 |
| BrTCP10 | Bra018280 | A05:7148195–7149244 | 1050 | 1 | 349 | 38,727.4 | 6.8541 |
| BrTCP12 | Bra038350 | A02:10071368–10072479 | 1008 | 1 | 335 | 38,247.8 | 8.7635 |
| BrTCP13a | Bra039158 | A05:25073263–25074192 | 930 | 1 | 309 | 35,797.8 | 7.4173 |
| BrTCP13b | Bra001032 | A03:14476673–14479645 | 1617 | 4 | 538 | 59,180.4 | 7.2645 |
| BrTCP14 | Bra018126 | A06:10447686..10449086 | 1400 | 1 | 466 | 51,551 | 7.2085 |
| BrTCP15a | Bra004407 | A07:21777258–21778223 | 966 | 1 | 321 | 34,066.1 | 7.4651 |
| BrTCP15b | Bra003986 | A07:19409073–19409810 | 738 | 1 | 245 | 33,798.6 | 7.7239 |
| BrTCP15c | Bra007875 | A02:10484033–10484773 | 741 | 1 | 246 | 33,870.94 | 7.12 |
| BrTCP17 | Bra005967 | A03:1433460–1434200 | 741 | 1 | 246 | 27,367.4 | 7.0159 |
| BrTCP18a | Bra001710 | A03:18008822–18010603 | 1278 | 4 | 425 | 48,457.2 | 8.3237 |
| BrTCP18b | Bra037579 | A01:21234671–21236261 | 1284 | 4 | 428 | 48,910.3 | 7.1925 |
| BrTCP19 | Bra022568 | A02:7842256–7845540 | 894 | 2 | 297 | 30,182.5 | 5.419 |
| BrTCP20a | Bra025244 | A06:21530080–21531003 | 924 | 1 | 307 | 32,446.8 | 7.954 |
| BrTCP20b | Bra039096 | A09:1525195–1526031 | 837 | 1 | 278 | 29,671.6 | 6.5716 |
| BrTCP21a | Bra005984 | A03:1498581–1499285 | 705 | 1 | 234 | 24,263.2 | 10.8713 |
| BrTCP21b | Bra028662 | A02:1265168–1265857 | 690 | 1 | 229 | 23,957.7 | 10.2007 |
| BrTCP21c | Bra009338 | A10:14235005–14235715 | 711 | 1 | 236 | 24,733.4 | 9.2658 |
| BrTCP22a | Bra008001 | A02:11473562–11474686 | 1125 | 1 | 374 | 39,098.2 | 8.9956 |
| BrTCP22b | Bra016090 | A07:22761310–22762380 | 1071 | 1 | 356 | 37,206.3 | 7.4347 |
| BrTCP24a | Bra010789 | A08:15682126–15683052 | 927 | 1 | 308 | 34,410.5 | 7.1472 |
| BrTCP24b | Bra032365 | A09:22194917–22195873 | 957 | 1 | 318 | 35,327.5 | 8.1305 |
2.2. Chromosomal Location and Distribution of TCP Genes in Chinese Cabbage
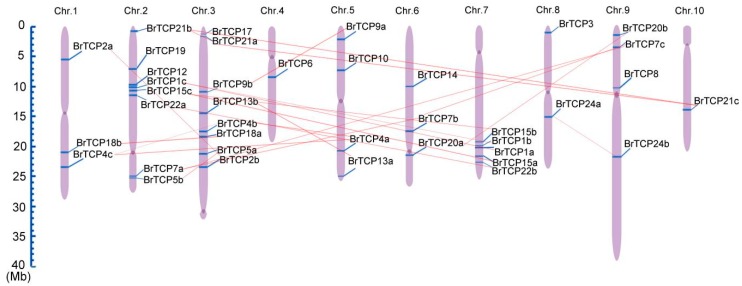
The physical chromosomal location of TCP genes was searched on BRAD (the Brassica database) by using BLASTN (http://brassicadb.org/brad/index.php), as shown in Figure 1. All 39 members of Chinese cabbage TCPs were unevenly distributed in all 10 B. rapa chromosomes, containing one to eight genes per chromosome. Four TCP genes were each found on chromosomes A5 and A9. The largest number (nine) of BrTCPs was located on chromosome A2 and A3, followed by five BrTCP genes in A7. There were two BrTCPs on A8. Only one gene was located in A4 and A10. Chromosome A1 and A6 each had seven BrTCPs. Duplication analysis showed that nearly 80% (31 out of 39) of BrTCP genes existed in two or three copies. However, we found that most paralogous genes were distributed on different chromosomes. Both BrTCP15a and BrTCP15b were located on the long arm of chromosome A7, and BrTCP1a and BrTCP1b genes were located between them, however BrTCP1a and BrTCP1b were tandem on A7, suggesting that duplicate events in Chinese cabbage were caused by both segment duplications and tandem. Eight (33%), three (13%), six (13%), one (4%), and six (13%) AtTCPs were found in Chr.1 to 5 of A. thaliana, respectively, and three (7%), eight (20%), eight (20%), one (3%), and four (10%) BrTCPs were found in Chr.1 to Chr.5, respectively. The remaining BrTCPs (38%) were found in Chr.6 to Chr.10 (Table S2 and Figure S1). This finding suggests that the duplicate events of the BrTCP family were caused by the expansion of chromosomes in Chinese cabbage.
Figure 1.
The chromosomal location of TCP genes from Chinese cabbage. The scale represents 40 Mb chromosomal distance. The chromosome numbers are labeled on the top of them. The duplicated gene are connected with red lines.
2.3. Phylogeny and Gene Structure of the BrTCP Family
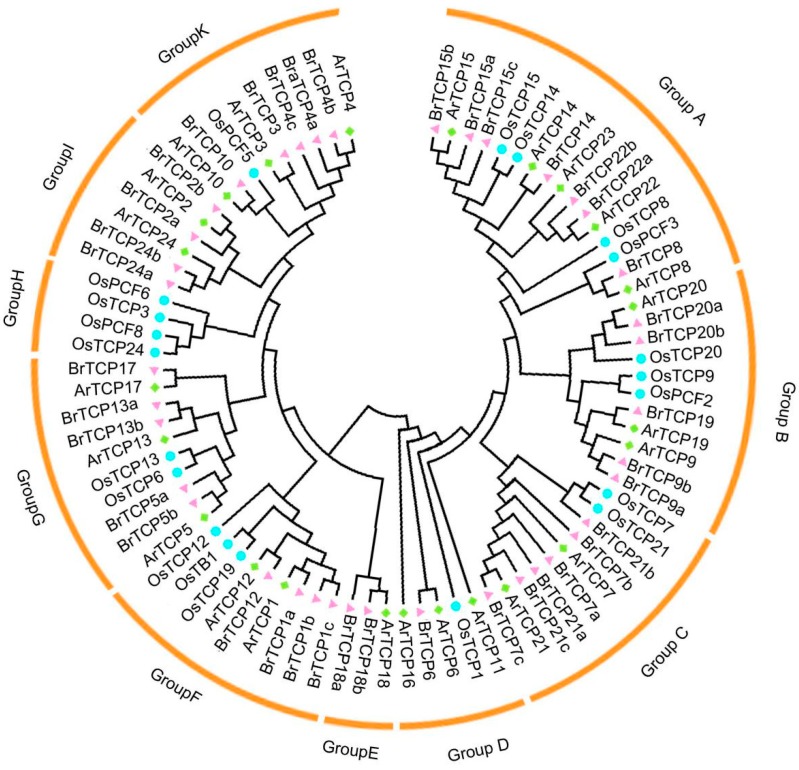
To better understand the phylogenetic relationships of TCP genes in Chinese cabbage, Arabidopsis, and rice, a neighbor-joining (NJ) phylogenetic tree was built based on multiple sequence alignment of 39 B. rapa, 24 A. thaliana, and 21 Oryza sativa genes with MEGA6 (Figure 2). All TCP genes were divided into seven subgroups based on their sequence features: group A to G. Groups A–D belonged to the class I subfamily, which is also called the PCF subfamily, and 13 Arabidopsis and 19 Brassica rapa TCP genes were found (Table 2). There were similar gene distribution in turnip, which has a close genetic relationship with Chinese cabbage. By contrast, groups E to G belonged to class II, which consists of two types: CIN and CYC/TB type. Furthermore, 29 CIN-type and 9 CYC/TB type members were found, representing 36.3% and 11.3% of all TCP members, respectively; however, 8 and 14 AtTCPs were found in CIN-type and CYC/TB type members of Arabidopsis, respectively. The largest number of TCP members belonged to the PCF type, which occupied 52.5%. Sixteen TCP proteins were found (20%) in group A, the largest subgroup, which also belonged to the PCF type. Meanwhile, the smallest subgroup was the CYC/TB type, which contained three BrTCP proteins (4%) but only one Arabidopsis TCP member. This finding suggested that the TCP family expanded before divergence of the lineages. The Chinese cabbage TCP genes were highly homologous to the TCP genes of A. thaliana. In addition, one Arabidopsis TCP gene may have at least one copy or even two or three copies in cabbage. However, BrTCP11 and BrTCP16 were not found in Chinese cabbage, which suggested that they not only underwent replication but also deletion of chromatin during evolution of Chinese cabbage.
Figure 2.
Phylogenetic relationships of TCP transcription factors among Brassica rapa, Arabidopsis thaliana and Oryza. sativa. The unroot phylogenetic tree was constructed by MEGA 6.0 (Tokyo Metropolitan University, Tokyo, Japan) using Neighbor-Joining method with 1000 bootstrap replicates. TCPs of different plants are indicated with different colors and shapes.
Table 2.
TCP family genes in Arabidopsis thaliana, Chinese cabbage and turnip.
| PCF | CIN | CYC/TB1 | Total | |
|---|---|---|---|---|
| Arabidopsis thaliana | 13 | 8 | 3 | 24 |
| Brassica rapa (Chinese cabbage) | 19 | 14 | 6 | 39 |
| Brassica rapa (turnip) | 20 | 13 | 6 | 39 |
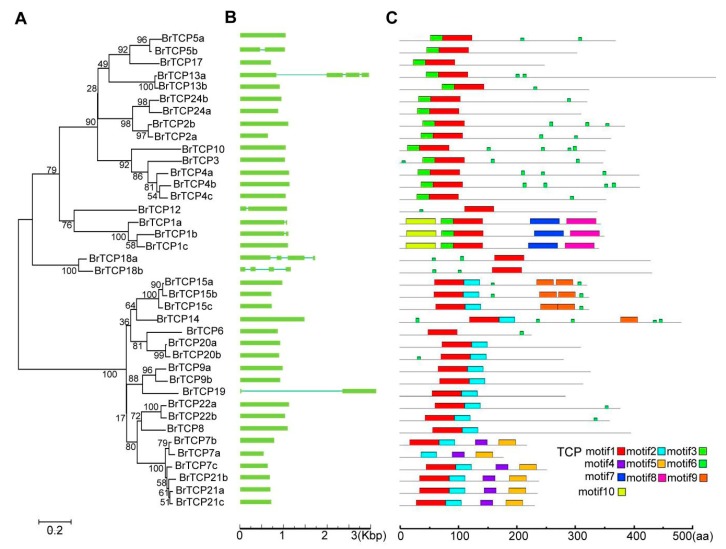
The structure of BrTCP genes was analyzed by alignment of cDNA and genomic DNA sequences, as shown in Figure 3B. Most Chinese cabbage TCP genes (31 out of 39) contained only one exon without an intron, and six BrTCP members contained two exons and one intron. In addition, BrTCP13a, BrTCP18a, and BrTCP18b contained four exons and three introns. Most BrTCP genes had similar exon/intron structures and distribution in the same phylogenetic subfamily, which were confirmed by the phylogenetic tree (Figure 3A).
Figure 3.
The Phylogenetic tree, gene structure and multiple motifs of TCP family in Chinese cabbage. (A) Phylogenetic tree was constructed based on the protein sequence of Chinese cabbage by MAGE6.0; (B) The gene structure of Chinese cabbage TCPs. Exon and intron were represented by green boxes and lines; (C) The multiple conserved motifs of Chinese cabbage TCP proteins. Motifs were identified by an online tool MEME and labeled with colored boxes. Motifs 1–3 belong to the TCP subfamily.
2.4. Conserved Domains and Motif Analysis of Chinese Cabbage
The conserved motifs of Chinese cabbage TCP proteins were confirmed by InterPro and analyzed using the MEME program (The University of Queensland, Brisbane, Australian) [29]. Ten putative motifs were identified by MEME analysis, namely, motifs 1–10. Motifs 1–3 were all identified as the TCP domain; however, motifs 4–10 were unknown domains (Figure 3A and Figure S2). Nearly all BrTCPs contained motif 1, except BrTCP7a, which should be an essential motif in BrTCPs. Motif 3 existed in almost every class I BrTCP, except BrTCP6, whereas most class II BrTCPs contained motif 2, except BrTCP12, BrTCP18a, and BrTCP18b, which belonged to the CYC/TB type. Among them, an R domain was present, which was predicted to be a coiled coil domain mediating protein–protein interactions (Figure S3). As a result of our identifications, BrTCPs of the same subfamily contained similar motif compositions, but they differed from different subfamilies, which suggested that different subfamilies have complementary functions, but the same subfamily exhibits redundancy.
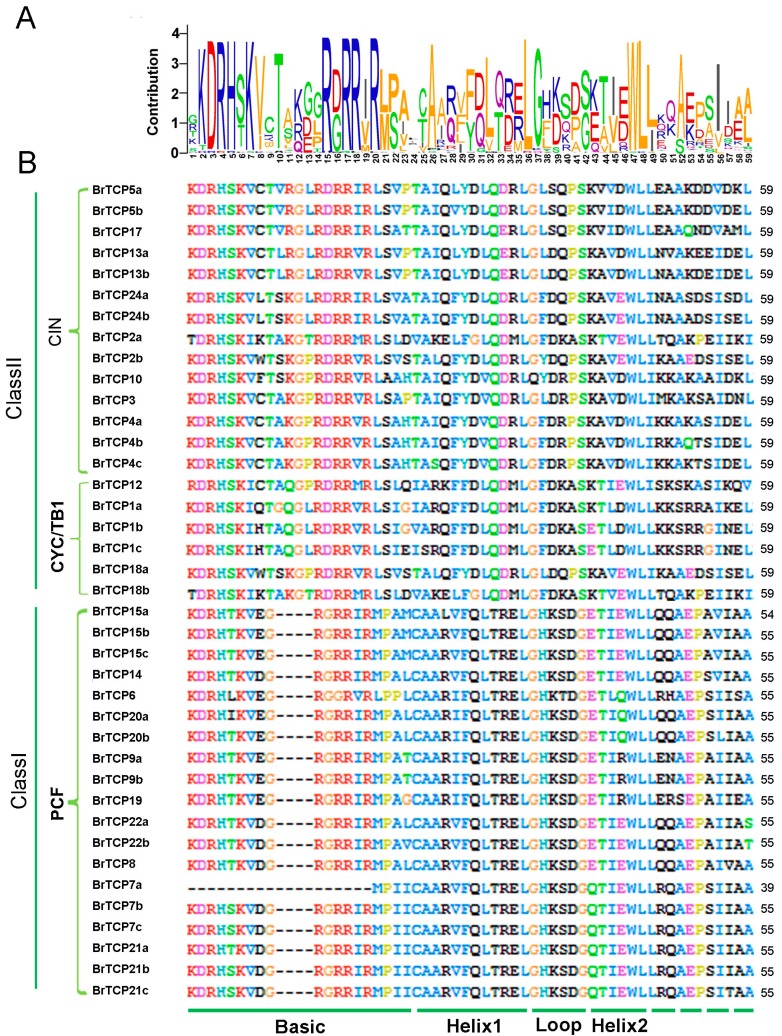
The TCP domain was reported to participate in dimerization and DNA binding by a bHLH motif with usually 59 amino acid residues [3]. TCP domains were found in all BrTCPs and consisted of 39 to 59 amino acid residues with a bHLH structure (Figure 4). In the class I BrTCP subfamily, four amino acid deletions were found relative to that in the class II subfamily at the basic region. Sequences between the class I and II subfamilies were quite different in helix I, loop, and helix II, whereas a conservative tandem of W (tryptophan) and L (leucine) was found in helix II. These results revealed that different subfamilies have complementary functions, while the same subfamilies exhibit redundancy.
Figure 4.
Multiple protein sequences alignment of TCP domain in Chinese cabbage. (A) Sequence LOGO of TCPs in Chinese cabbage. The LOGO representation created by Weblogo online software (Berkeley University, CA, USA) based on BrTCP protein sequences; (B) Multiple sequence alignment of Chinese cabbage TCP motifs by Clustal X2(University College Dublin, Dublin, Ireland).
2.5. Expression Levels of TCP Genes in Chinese Cabbage
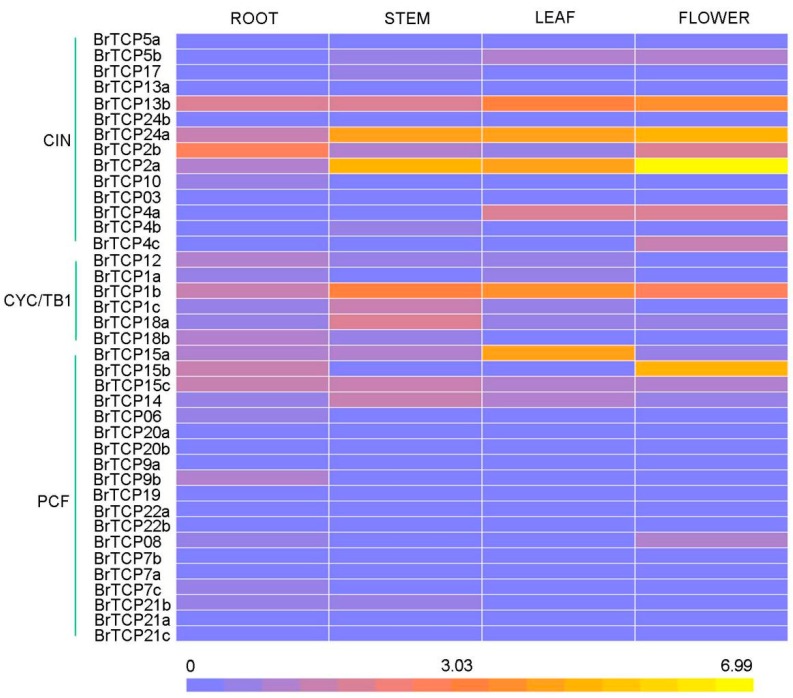
TCPs from A. thaliana were reported to be involved in the regulation of leaf morphology, including the shape and curvature of leaves [16,25]. To explore the potential function of TCP genes in Chinese cabbage, the expression levels of all 39 BrTCP genes were detected in root, stem, leaf, and flower tissues of “Dongnong A160” cabbage by real time polymerase chain reaction (RT-PCR). Total mRNAs were isolated at the reproductive stage, and all data were normalized with Actin as an internal control (Figure 5). The expression levels of different subfamily BrTCP genes varied significantly in these four tissues. BrTCP1b, BrTCP2a, BrTCP5b, BrTCP15a, BrTCP15c, and BrTCP24a were highly expressed in all four tissues. However, the expression levels of BrTCP15b were higher in flower than in other tissues, which was the same as BrTCP4c. Moreover, BrTCP4a was highly expressed in leaf and flower tissues. Most PCF subfamily genes showed low expression levels. The differential expression levels of BrTCPs in specific tissues indicated that these genes played multiple regulatory roles at different development stages.
Figure 5.
Expression patterns of Chinese cabbage TCP genes in different tissues represented by heat map. The expression level of BrTCP genes in root, stem, leaf and flower were detected by quantitative real-time PCR.
2.6. Associated Relationship between the Expression of BrTCPs and Heading of Chinese Cabbage
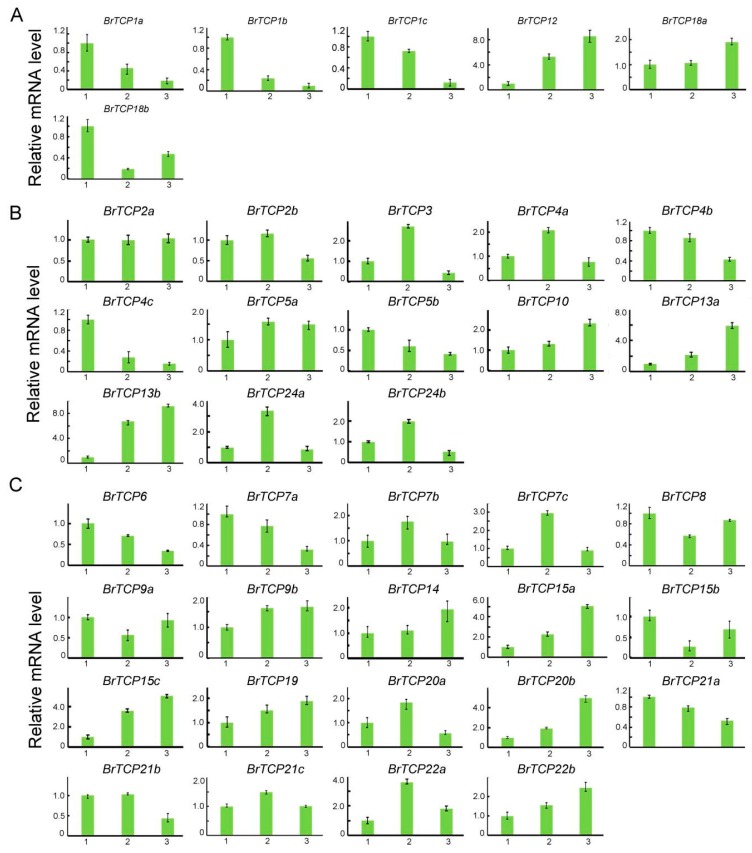
During the development of Chinese cabbage, heading is important for the formation of leafy head and cabbage yield. The TCP family is known to be involved in regulating the development of leaf curvature [16,25]. To gain insights on the relationship between heading and the TCP family in Chinese cabbage, the expression levels of BrTCPs were detected at the seeding, rosette, and heading stages of Chinese cabbage “Dongnong” A160 by RT-PCR. All BrTCPs were divided into three groups according to the subfamily, CYC/TB1, CIN, and PCF. For the CYC/TB1 type, six BrTCPs were detected, and four genes were downregulated at the rosette and heading stages compared with the seedling stage. Especially, TCP1a, -b, and -c were decreased by tenfold at the heading stage. However, BrTCP12 was upregulated by 5.3- and 8.5-fold at the rosette and heading stages, respectively (Figure 6A). In the CIN-type TCP family, BrTCP3, BrTCP4a, and BrTCP24a were significantly upregulated at the rosette leaf stage compared to the other two stages. At the rosette stage, BrTCP13a and BrTCP13b were upregulated by 2.18- and 6.7-fold, respectively, and were even higher at the heading stage (5.8- and 9.1-fold). However, the expression of BrTCP4c dramatically decreased to 6.5-fold at the heading stage (Figure 6B). For the PCF type, the expression levels of BrTCP7a decreased gradually from seeding to heading. At the same time, BrTCP7b and BrTCP7c were enhanced by up to 2.9-fold at the rosette stage. Furthermore, BrTCP15a and BrTCP15c increased by 2.3- and 3.6-fold at the rosette stage, respectively, and both increased by fivefold at the heading stage. However, BrTCP15b decreased by 3.7-fold at the rosette stage, while BrTCP22a was upregulated by 3.6-fold. In addition, BrTCP20b was increased by fivefold at the heading stage (Figure 6C).
Figure 6.
The expression level of BrTCP genes in Chinese cabbage selfing line A160 at the stage of seedings, rosette and headings: (A) the expression level of CyC/TB1 type BrTCP genes A160 at the stage of seedlings, rosette and headings were detected by quantitative real-time PCR; (B) the expression level of CIN type BrTCP genes A160 at the stage of seedlings, rosette and headings; and (C) the expression level of PCF type BrTCP genes A160 at the stage of seedlings, rosette and headings. Numbers 1,2 and 3 represent the stages of seedling, rosette and heading, respectively. The represent results performed in triplicate. Error bars represent ± SE
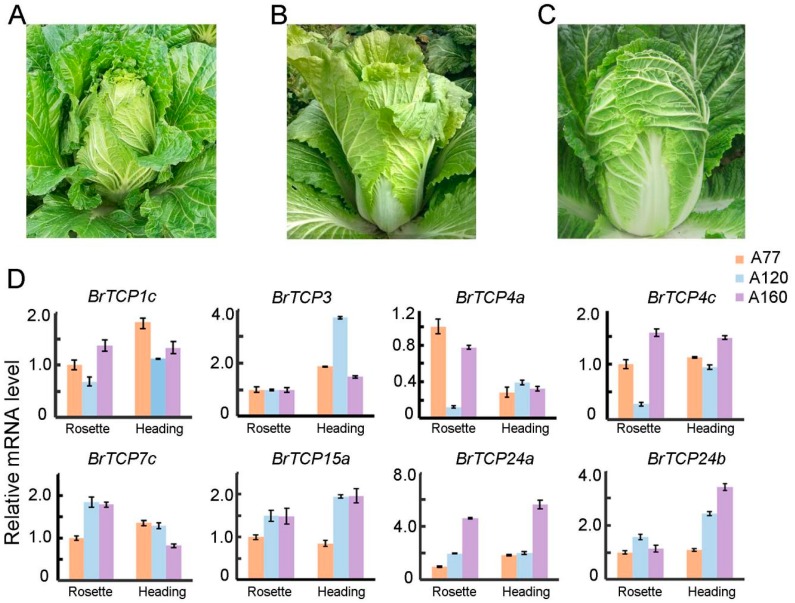
Several A. thaliana TCP members were reported to be involved in regulating the curvature of leaves, such as AtTCP2, AtTCP3, AtTCP4, AtTCP10, AtTCP24, and AtTCP15 [7,8,13,16,18,26]. In terms of phylogenetic analysis and the differential expression of BrTCPs between the rosette and heading stage, eight BrTCP genes were selected to measure the expression levels among different heading-type cabbages, namely, BrTCP1c homologous to AtTCP1, BrTCP3 homologous to AtTCP3, BrTCP4a and -4c homologous to AtTCP4, BrTCP7c homologous to AtTCP7, BrTCP15a homologous to AtTCP15, and BrTCP24a and -24b homologous to AtTCP24. Leafy heads of Chinese cabbage were generally divided into three types: the fold heading type (A77 inbred line) with the involute leaves less than the axle wire (Figure 7A), overlapping heading type (A160 inbred line) with the involute leaves exceeding the axle wire (Figure 7C), and straight heading type (A120 inbred line) with revolute leaves (Figure 7B). The expression levels of BrTCP1c, BrTCP4a, and BrTCP4c were lower in A120 than in A77 and A160, among which BrTCP4a and BrTCP4c were downregulated by 8.3- and 3.7-fold, respectively. However, BrTCP3 in A120 was upregulated by 3.7-fold at the heading stage. BrTCP24a in A160 was upregulated by 4.6- and 5.7-fold at the rosette and heading stages, respectively, and the expression of BrTCP24a in A160 was only upregulated by 3.4-fold at the heading stage (Figure 7D). No obvious changes were found in BrTCP7c and BrTCP15a. These results revealed that BrTCPs may be involved in different pathways and development stages to regulate the curvature of leaves.
Figure 7.
The expression level of BrTCP genes in leaves of different heading types of Chinese cabbages: (A) the inbred line of A120 Chinese cabbage at heading stage; (B) the inbred line of A77 Chinese cabbage at heading stage; (C) the inbred line of A160 Chinese cabbage at heading stage; and (D) the expression level of BrTCP1c, BrTCP3, BrTCP4a, BrTCP4c, BrTCP7c, BrTCP15a, BrTCP24a, and BrTCP24b in the inbred lines of A120, A77, and A160 at rosette and heading stages. The represent results performed in triplicate. Error bars represent ± SE
3. Discussion
The TCP TFs are a kind of plant-specific TF family, which participate in multiple functions in plant growth and development. A number of TCP TFs have been identified with genome-wide analysis, for example, Arabidopsis, rice, and other plants and even turnips (B. rapa ssp.), which had close relationship with Chinese cabbage [30,31,32]. However, no information on Chinese cabbage TCP TFs is available. In our study, 39 BrTCP members were identified from the Chinese cabbage genome, and the number of TCPs in Chinese cabbage was more than that of Arabidopsis, while in Arabidopsis 24 TCPs were found. Both Arabidopsis and Chinese cabbage belong to the Brassicaceae family, and Chinese cabbage is also a subspecies of B. rapa, which has undergone polyploidization, leading to additional whole genome triplication [27]. The BrTCP gene family was expanded by 1.625-fold, and two or three copies were homologous to one AtTCP protein. Eight BrTCPs contained a single duplication, BrTCPs contained a single duplication, nine sister pairs and four of three duplications. However, three BrTCPs were missing in the Chinese cabbage: BrTCP11, BrTCP16 and BrTCP23 (Table S1). All these BrTCPs were segmentally duplicated and unevenly distributed on the genome, which played a role in genomic rearrangement and diversification. In addition, turnip also had 39 TCPs, but there were some differences in duplicated copies between Chinese cabbage and turnip. TCP2, TCP5, and TCP22 were single copy in turnip, however they were two duplicated copies in cabbage. Conversely, there was only one BrTCP17 but two BrrTCP17. TCP23 was missing in cabbage, while turnip had a TCP23. There were three BrrTCP20 and two BrTCP20. These different duplicated copies indicate that TCP may play distinguishing role in biological process between Chinese cabbage and turnip even their close correlation between relatives.
TCP family TFs were divided into three subfamilies according to their conservative domains, including PCF, CIN, and CYC/TB1 [3]. The TCP family in Chinese cabbage also consisted of these three parts: PCF with 1.5-fold expansion compared to Arabidopsis and containing 19 members but 20 members in turnips, CIN with 1.8-fold expansion and 14 members but 13 members in turnips, and CYC/TB1 with twofold expansion and six members, the same as in turnips (Table 2). Almost all the BrTCPs contained the conservative motif 1, except for BrTCP7a, which was obviously shorter than other proteins. Besides motif 1, the PCF subfamily usually contains motif 2 in front of motif 1 in proteins. Motif 3 was found in both the CIN and CYC/TB1 subfamilies. Furthermore, BrTCP12, BrTCP18a, and BrTCP18b of the CYC/TB1 subfamily contained an R domain (Figure 3C and Figure 3). In addition, motifs 1–3 were specific to the TCP domain, which was formed by a bHLH motif of 54–59 amino acid residues. These indicated that BrTCPs may play different roles in growth and development, although they were in the same TCP subfamily. However, most BrTCP proteins of the same subfamily shared not only relatively similar conservative domain but also similar gene structures. Next, the expression pattern of each BrTCP subfamily was detected in root, stem, leaf, and flower tissues by qRT-PCR. Most BrTCP genes of the PCF subfamily were downregulated in these tissues, except for BrTCP15a and BrTCP15b. BrTCP1c of CYC/TB1 was apparently upregulated, and the majority of upregulated genes were found in the CIN subfamily, which indicate BrTCPs of CIN subfamily play a vital role in head formation of Chinese cabbage. The expression pattern and distribution of conserved domain were related to their molecular functions. These results indicated that single or pairs of genes of the same subfamily may play similar or complementary roles in biological processes.
TCP family TFs are involved in the regulation of leaf curvature in many plants. AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24 of the CIN subfamily were the targeted genes of miR319, and they modulate leaf development in Arabidopsis [12,13,18,29]. In terms of the close phylogenetic relationship between Arabidopsis and Chinese cabbage, BrTCP genes might play a role in leaf morphogenesis. To explore the correlation between BrTCP genes and leaf curvature, we monitored the expression levels of 39 BrTCP genes at the seedling, rosette, and heading stages in the inbred line A160, which is a typical “overlapping” cabbage. For CYC/TB1 subfamily, the expression of BrTCP1a, BrTCP1b, BrTCP1c, and BrTCP18b were remarkable downregulated at the rosette and heading than at the seedling stage, but BrTCP12 and BrTCP18a got the maximum on the heading stage. Most of the CIN subfamily members were the taget genes of miR319, such as BrTCP2, BrTCP3, BrTCP4, BrTCP10, and BrTCP24. BrTCP2, BrTCP3, BrTCP4a, BrTCP5a, BrTCP24a, and BrTCP24b upregulated to the peak on the rosette stage. However, the highest expression of BrTCP10 was observed at heading stage. BrTCPs have discrepant functions in heading formation even in the same subfamily, which indicated that these genes were involved in heading formation caused by the remarkable differential expressions BrTCP15a and BrTCP15c increased gradually during Chinese cabbage development. TCP14 and TCP15 act as repressors of cell proliferation in the developing leaf in Arabidopsis [18], and BrTCP15a and BrTCP15c increased gradually during Chinese cabbage development with no obvious expression change of BrTCP14, suggested that BrTCP15 played a dominant role in Chinese cabbage. To further analyze the relationship between the expression levels and different shape heads, eight BrTCPs were selected for further determination in the inbred lines A77, A120, and A160. It was reported that TCP1 played a positive role in BR biosynthesis pathway by regulating the expression of DWF4 [15] and involved in longitudinal elongation of leaves in Arabidopsis [33]. We showed that the expression level of BrTCP1 were higher in heading cabbage, A77 and A160, than straight cabbage in both rosette and heading stages, which indicated that BR may played a role in leaf curvature to regulate heading formation. Further studies are required. The expression of BrTCP3 was upregulated only in A120 at the heading stage. Whereas BrTCP4a in A120 was downregulated at the rosette stage, and the expression of BrTCP4c was highest in overlapping heading type (A160), secondly folding type (A77) and lowest in straight type (A120), which were agreed with the reported gene BrpTCP4 in Chinese cabbage. BrpTCP4 showed low expression level in cylindrical head shape and high in round head shape, which was reported to regulate the size and shape of leafy heads in Chinese cabbage [9]. The double mutant of tcp2 tcp4 showed enlarged flat leaves, however the tcp2 tcp3 tcp4 tcp10 mutant had strongly crinkled leaves in Arabidopsis [34]. The expression pattern of BrTCP3 and BrTCP4 seemed to be opposite among different heading types, which revealed the different BrTCPs played diverse role in leaf curvature. Moreover, the expressions of TCP24 were elevated in the p35S:mTCP24 transgenic Arabidopsis, which showed rosette leaves slightly downward [35]. BrTCP24a in overlapping heading (A160) was highest at both the rosette and heading stages; the expression level of BrTCP24b in A160 was highest compare to others on the heading stage, that suggested that the function of BrTCP24 were similar with AtTCP24 in leaf curvature. Our results revealed that BrTCP genes played either a synergistic, complementary, or opposite role in the formation of leaf curvature. Therefore, further studies are needed to determine their specific functions.
4. Materials and Methods
4.1. Identification and Analysis of TCP Family Members in Chinese Cabbage
To identify the TCP members in Chinese cabbage, we searched for a conserved TCP domain (PF03634) based on the Hidden Markov Model on BRAD (http://brassicadb.org/brad/) and verified the BrTCP members on the BLAST(Basic Local Alignment Search Tool) program of the NCBI (National Center for Biotechnology Information) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Full-length nucleotide and protein sequences of the TCP family in Chinese cabbage were also searched and obtained from BRAD (http://brassicadb.org/brad/). The TCP family sequences of A. thaliana and O. sativa were downloaded from TAIR (http://www.arabidopsis.org/) and the O. sativa Genome Annotation Project (http://rice.plantbiology.msu.edu/) and were adjusted with the plant TFDB (Plant Transcription Factor Databas)database (http://planttfdb.cbi.edu.cn/) [36].
4.2. Chromosomal Location and Gene Structure
The chromosomal location and gene structures of BrTCP genes were acquired from BRAD and were assessed and enhanced using MapInspect (Mike Lischke, Berlin, Germany) and Photoshop software CS (San Jose, CA, USA).
4.3. Phylogenetic Analysis and Analysis of Conserved Domains and Motif
The protein sequences of identified BrTCPs and A. thaliana and O. sativa TCPswere downloaded and compared to generate the phylogenetic tree. Multiple TCP sequence alignments were performed using Clustal X2.0 (University College Dublin, Dublin, Ireland) [37], and MEGA6 (Tokyo Metropolitan University, Tokyo, Japan) was used to construct the phylogenetic trees with the NJ (neighbor joining) method [38] by using 1000 replicates.
To identify the conserved motif of BrTCPs, all protein sequences were analyzed using the online tool MEME with the following settings: repetition number, any; maximum motif number, 5; motif width, between 6 and 50; and minimum motif width, 6. In addition, InterPro (http://www.ebi.ac.uk/interpro/scan.html) and Pfam (http://pfam.xfam.org/search#tabview=tab1) were used for further confirmation and annotation [29].
4.4. Plant Materials
“A160” were sown in the greenhouse until the cotyledon were unfolding, and then seedlings were vernalized for 20 days at 4 °C. The tissues of lateral root, stem, rosette leaf and flower were harvested when cabbages were flowering at 28 °C under a 16 h light/8 h dark cycle.
Seeds of selfing lines A77-1-1, A120-1, and A160 (fold heading, straight cabbage, and overlapping heading cabbage, respectively) were sown in the field on 20 July. A77 inbred line, A160 inbred line and A120 inbred line were fold heading type with the involute leaves less than the axle wire, overlapping heading type with the involute leaves exceeding the axle wire, and straight heading type with revolute leaves, respectively. The fifth leaf were harvested at five leaf stage for seedling leaves, and rosette leaves were obtained at 10 leaf stage. The green lobus cardiacus were selected when the leaves incurved to form headings. All plant materials were for RNA isolation.
4.5. Total RNA Isolation and qRT-PCR Analysis
Total RNA of each tissue was extracted with the EasyPure Plant RNA Kit (TransGen, Beijing, China), and 2 mg of total RNA was reversed transcribed with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). For RT-PCR, 15 mL total volume with 7.5 mL of SYBR Green Real-Time PCR Master Mix (TOYOBO, Osaka, Japan), 0.25 mL of specific primers, and 5 mL of cDNA samples with 30–50-fold dilution were used. For each RT-PCR, 15 mL of total volume with gene-specific primers was used. The amplification was performed on a Bio-Rad (Berkeley, CA, USA) IQ5 Multi-Color Real-Time PCR Detection System. The reactions of tubulin were considered as internal references. All primers were listed in Supplementary Table S3.
Acknowledgments
This work was supported by the Foundation for the National key Research and Development Program of China (2016YFD0101701), by Ministry of Science and Technology of the People’s Republic of China.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/3/847/s1.
Author Contributions
Yan Liu and Yaowei Zhang planned and designed the study and wrote the manuscript. Xiaoyu Guan and Shengnan Liu performed the experiments. Meng Yang implemented the software. Junhui Ren. and Meng Guo provided supplementary experimental results. Zhihui Huang collected all of the plant materials in this work. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Doebley J., Stec A., Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 2.Kosugi S., Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cubas P., Lauter N., Doebley J., Coen E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. Cell Mol. Biol. 1999;18:215–222. doi: 10.1046/j.1365-313X.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 4.Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/S0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 5.Cooper A. A Case of Aneurism of the Carotid Artery. Med. Chir. Trans. 1809;1:1–12. doi: 10.1177/095952870900100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosugi S., Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. Cell Mol. Biol. 2002;30:337–348. doi: 10.1046/j.1365-313X.2002.01294.x. [DOI] [PubMed] [Google Scholar]
- 7.Takeda T., Suwa Y., Suzuki M., Kitano H., Ueguchi-Tanaka M., Ashikari M., Matsuoka M., Ueguchi C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. Cell Mol. Biol. 2003;33:513–520. doi: 10.1046/j.1365-313X.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 8.Vadde B.V.L., Challa K.R., Nath U. The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2017 doi: 10.1111/tpj.13772. [DOI] [PubMed] [Google Scholar]
- 9.Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014;164:710–720. doi: 10.1104/pp.113.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S., Luo Y., Cai Z., Cao X., Hu X., Yang J., Luo D. Functional diversity of CYCLOIDEA-like TCP genes in the control of zygomorphic flower development in Lotus japonicus. J. Integr. Plant Biol. 2013;55:221–231. doi: 10.1111/j.1744-7909.2012.01169.x. [DOI] [PubMed] [Google Scholar]
- 11.Juntheikki-Palovaara I., Tahtiharju S., Lan T., Broholm S.K., Rijpkema A.S., Ruonala R., Kale L., Albert V.A., Teeri T.H., Elomaa P. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae) Plant J. Cell Mol. Biol. 2014;79:783–796. doi: 10.1111/tpj.12583. [DOI] [PubMed] [Google Scholar]
- 12.Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2008;53:42–52. doi: 10.1111/j.1365-313X.2007.03308.x. [DOI] [PubMed] [Google Scholar]
- 13.Balsemao-Pires E., Andrade L.R., Sachetto-Martins G. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 2013;67:120–125. doi: 10.1016/j.plaphy.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C.P., Van Aken O., Millar A.H., Murcha M., Whelan J. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell. 2010;22:3921–3934. doi: 10.1105/tpc.110.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22:1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viola I.L., Uberti Manassero N.G., Ripoll R., Gonzalez D.H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011;435:143–155. doi: 10.1042/BJ20101019. [DOI] [PubMed] [Google Scholar]
- 17.Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 18.Kieffer M., Master V., Waites R., Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. Cell Mol. Biol. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukhtar M.S., Carvunis A.R., Dreze M., Epple P., Steinbrenner J., Moore J., Tasan M., Galli M., Hao T., Nishimura M.T., et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida D.M., Gregorio G.B., Oliveira M.M., Saibo N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017;93:61–77. doi: 10.1007/s11103-016-0547-7. [DOI] [PubMed] [Google Scholar]
- 21.Xiao B.Z., Chen X., Xiang C.B., Tang N., Zhang Q.F., Xiong L.Z. Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant. 2009;2:73–83. doi: 10.1093/mp/ssn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukhopadhyay P., Tyagi A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015;5:9998. doi: 10.1038/srep09998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J.G., Lee G.H., Kim J.S., Shim E.J., Park Y.D. An insertional mutagenesis system for analyzing the Chinese cabbage genome using Agrobacterium T-DNA. Mol. Cells. 2010;29:267–275. doi: 10.1007/s10059-010-0013-3. [DOI] [PubMed] [Google Scholar]
- 24.Ito H., Kato T. Studies on the head formation of Chinese cabbage. II. Relation between auxin and head formation. Agric. Hortic. 1958;26:771. (In Japanese) [Google Scholar]
- 25.Gao L.W., Lyu S.W., Tang J., Zhou D.Y., Bonnema G., Xiao D., Hou X.L., Zhang C.W. Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci. Rep. 2017;7:42229. doi: 10.1038/srep42229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukushima K., Hasebe M. Adaxial-abaxial polarity: The developmental basis of leaf shape diversity. Genesis. 2014;52:1–18. doi: 10.1002/dvg.22728. [DOI] [PubMed] [Google Scholar]
- 27.Liang J., Liu B., Wu J., Cheng F., Wang X. Genetic Variation and Divergence of Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Brassica rapa. Front. Plant Sci. 2016;7:94. doi: 10.3389/fpls.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun J.H., Bancroft I., Cheng F., et al. Brassica rapa Genome Sequencing Project, C. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 29.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 30.Du J., Hu S., Yu Q., Wang C., Yang Y., Sun H., Yang Y., Sun X. Genome-Wide Identification and Characterization of BrrTCP Transcription Factors in Brassica rapa ssp. rapa. Front. Plant Sci. 2017;8:1588. doi: 10.3389/fpls.2017.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danisman S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016;7:1930. doi: 10.3389/fpls.2016.01930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015;10:e1044192. doi: 10.1080/15592324.2015.1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyama T., Sato F., Ohme-Takagi M. A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 2010;74:2145–2147. doi: 10.1271/bbb.100442. [DOI] [PubMed] [Google Scholar]
- 34.Bresso E.G., Chorostecki U., Rodriguez R.E., Palatnik J.F., Schommer C. Spatial Control of Gene Expression by miR319-Regulated TCP Transcription Factors in Leaf Development. Plant Physiol. 2018;176:1694–1708. doi: 10.1104/pp.17.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., Mao Y., Yang J., He Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 2015;6:436. doi: 10.3389/fpls.2015.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin J., Zhang H., Kong L., Gao G., Luo J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42:D1182–D1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.