Abstract
Trimethylamine N-oxide (TMAO) respiration is carried out mainly by the Tor system in Escherichia coli. This system is encoded by the torCAD operon and comprises a periplasmic TMAO reductase (TorA) and a c-type cytochrome (TorC), which shuttles electrons to TorA. Expression of the tor operon is positively controlled by the TorS/TorR phosphorelay system in response to TMAO availability and negatively regulated by apocytochrome TorC. Interaction studies showed that, when immature, TorC can no longer bind TorA efficiently but can bind the periplasmic detector region of sensor TorS. ApoTorC negative autoregulation and TMAO induction are thus mediated by the same sensor protein. As apocytochromes related to TorC could not down-regulate the tor operon, we concluded that this negative control is highly specific. Moreover, the N-terminal half of apoTorC played no role in this control but the immature C-terminal domain of TorC strongly down-regulated the tor operon and interacted with the TorS detector region. This sophisticated autoregulatory pathway thus involves the C-terminal domain of apoTorC and allows optimal TorC biogenesis by preventing from saturation the c-type cytochrome maturation machinery.
Without oxygen, bacteria often use a variety of small exogenous compounds as electron acceptors for respiration (1, 2). For example, several bacteria can reduce trimethylamine N-oxide (TMAO) into volatile trimethylamine to provide cells with energy (3). TMAO is a widespread organic compound found at a high concentration in the tissues of marine animals, where it plays the role of a powerful chemical chaperone protecting proteins against denaturation by urea or hydrostatic pressure (4). TMAO respiration has been widely studied in Escherichia coli, Rhodobacter species, and, more recently, in Shewanella and Vibrio species (5, 6). Although these bacteria are phylogenetically distant from each other, the TMAO respiratory systems are often related. Generally, reduction of TMAO involves a pentahemic c-type cytochrome and a periplasmic molybdoenzyme as a terminal reductase. In E. coli, these two conserved structural proteins are encoded by the torCAD operon (7). torC, torA, and torD encode the c-type cytochrome TorC, the periplasmic molybdoenzyme TorA, and the TorA-specific chaperone TorD, respectively.
TorC is anchored to the inner membrane by a hydrophobic N-terminal extremity but its main body, which comprises two domains of equivalent size (about 20 kDa), is in the periplasm (8). The N-terminal domain of TorC (TorCN) contains four heme-binding motifs CXXCH and is highly homologous to the members of the large NirT/NapC family of c-type cytochromes (9). The TorC C-terminal domain (TorCC) is found mainly in DMSO/TMAO respiratory systems and carries a fifth heme-binding site close to its C terminus. We recently showed that TorC is an essential component of the TMAO respiratory chain and that it shuttles electrons from the membranous menaquinones to TorA. Surprisingly, the TorCN domain not only transfers electrons to the TorCC domain but also binds to TorA; in contrast, TorCC contains the electron donor site to TorA but is not involved in TorA binding (8). Like other c-type cytochromes, TorC is synthesized first as an immature cytochrome (apocytochrome) that must cross the inner membrane via the Sec pathway before maturation occurs in the periplasm (10). Cytochrome maturation starts with disulfide bond formation between the two conserved cysteines of the heme-binding site, and this oxidation pathway is driven by the DsbA/DsbB system (11). The covalent attachment of hemes to apocytochrome is then catalyzed by the Ccm proteins (12–14). Cytochrome maturation is thus a complex process, and overproduction of various cytochromes, including TorC, leads to the accumulation of immature proteins. Fortunately, overexpression of the ccm genes improves the maturation level of many c-type cytochromes (15).
When we looked for proteins that modulate torCAD operon expression, we noticed that overproduction of TorC as well as miniTn10 insertions within genes involved in the maturation pathway both strongly down-regulated torA–lacZ fusion (10). As this negative regulation was relieved by overexpression of the ccm genes and abolished in a torC mutant strain, we suspected an unknown regulatory pathway involving the immature TorC cytochrome.
Expression of the tor operon is strictly controlled by the TorS/TorR system in response to TMAO availability. TMAO is probably detected by the periplasmic N-terminal region of the transmembrane TorS sensor (16). Once activated, TorS transphosphorylates the TorR response regulator by a four-step phosphorelay. In turn, TorR-P binds to and induces the tor operon promoter (17). When TMAO is exhausted, TorR-P is rapidly dephosphorylated by TorS through a truncated reverse phosphorelay (18). On the basis of genetic evidence, we supposed that the TorS/TorR phosphorelay system also mediates the apoTorC negative control (10). In this study, we show that this supposition is true and that, in contrast to holoTorC, apoTorC can bind to the detector region of TorS. In addition, we give evidence that this sophisticated regulatory pathway is not triggered by immature c-type cytochromes different from TorC and that it specifically requires the apoform of the C-terminal domain of TorC. Thus the TorC bacterial cytochrome has both structural and regulatory functions according to its maturation state.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions.
Bacterial strains and plasmids are listed in Table 1. For β-galactosidase activity determination, strains were grown overnight anaerobically in Luria broth medium at 37°C. Values represent the average of at least three determinations with a variation of no more than 15% from the mean. When necessary, ampicillin (100 μg/ml), TMAO (10 mM), arabinose (0.01–0.1‰), or isopropyl β-d-thiogalactoside (IPTG) (1 mM) was added.
Table 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotypes and/or characteristics | Ref. |
|---|---|---|
| Strain | ||
| MC4100 | araD139 Δ(lacIPOZYA–argF)U169 rpsL thi | 7 |
| LCB620 | MC4100 torA8∷MudII 1734 (torA–lacZ) | 7 |
| LCB512 | ΔtorC derivative of LCB620 | This work |
| Plasmid | ||
| pBC | torC cloned into pBAD24 | 8 |
| pBCN | First half of torC cloned into pBAD24 | 8 |
| pBCC | Second half of torC cloned into pBAD24 | 8 |
| pBCcyto | Sequence encoding soluble TorC cloned into pBAD24 | This work |
pBC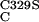
|
pBCC with mutation C329S | This work |
| pET-C | Sequence encoding soluble TorC cloned into pET21 | This work |
| pET-CN | Sequence encoding soluble TorCN cloned into pET21 | This work |
| pET-CC | Sequence encoding soluble TorCC cloned into pET21 | This work |
| ptorY | torY cloned into pJF119EH | 19 |
| pJ800 | D. vulgaris cyc cloned into pUC8 | 20 |
| pBCso | Sequence encoding periplasmic TorC from Shewanella oneidensis cloned into pBAD24 | This work |
| pET-Sp | Sequence encoding periplasmic TorS region cloned into pET21 | This work |
Analytical Procedure.
Proteins were analyzed by SDS/PAGE. After electrophoresis, the presence of heme-containing proteins was revealed by staining the gel with 3,3′,5,5′-tetramethylbenzidine (TMBZ) as described by Thomas et al. (21). For Western blotting, proteins were transferred to a Hybond ECL nitrocellulose membrane; the ECL-Western blotting system (Amersham Pharmacia) was used for detection.
Construction of a torC Deletion Mutant (LCB512).
To delete the chromosomal torC gene of strain LCB620 (torA–lacZ), we used the method described by Datsenko and Wanner (22). First, the torC gene was replaced by a chloramphenicol-resistance gene generated by PCR. The primers were 60 nucleotides long and included 40-nucleotide homology extensions, corresponding to the 5′ region upstream of the start codon of torC gene or to the 3′ extremity of the torC gene, and 20-nucleotide priming sequences that hybridized to the resistance gene flanking homologies of plasmid pKD3. The PCR mixture was treated with DpnI and used to transform strain LCB620 carrying the λ Red helper plasmid pKD46. The resistance cassette was then eliminated by using plasmid pCP20 expressing the FLP recombinase, which acts on the repeated sites flanking the resistance gene.
Construction of Plasmids.
To synthesize apoforms of TorC (apoTorC, from position 39 to 390 relative to TorC), TorCN (apoTorCN, from position 39 to 194), and TorCC (apoTorCC, from position 198 to 390), the corresponding torC coding sequence was amplified by PCR from MC4100 chromosomal DNA. We used primers that contain a SmaI site followed by a Shine-Dalgarno motif and the appropriate torC sequence and primers that correspond to a SmaI site followed by a sequence encoding a C-terminal His6-tag and the appropriate torC sequence. The purified PCR products were digested by SmaI and ligated into the same site of pET21 to give pET-C, pET-CN, and pET-CC. To synthesize the His-tagged periplasmic N-terminal domain of TorS (TorSN, from position 17 to 319), the corresponding torS coding sequence was amplified by using appropriate oligonucleotides and cloned into pET21, to give pET-Sp. To create plasmid pBCcyto, a DNA fragment encoding cytoplasmic TorC from position 39 to 390 and a C-terminal His6-tag was cloned into pBAD24 (23) downstream of the arabinose-inducible PBAD promoter. A mutation of the heme-binding site C329XXCH was introduced by PCR mutagenesis on pBCC, leading to plasmid pBCCC329S. The mutation TGT to TCT changed a Cys into Ser at position 329 of TorC. To create plasmid pBCso, a DNA fragment encoding the soluble TorCso from position 39 to 392 was amplified by PCR with Shewanella oneidensis chromosomal DNA as template, using an oligonucleotide that contains an EcoRI site followed by the TorT signal peptide coding sequence and the appropriate torC sequence, and a primer that corresponds to a SmaI site followed by a His6-tag coding sequence and the complementary sequence of the 3′ end of torC. The digested PCR product was cloned into the corresponding cloning sites of pBAD24. All PCR products and fusion sites were confirmed by sequencing. Transformations were performed according to the method of Chung and Miller (24).
Purification of Recombinant Proteins.
TorA, TorC, TorCC, and TorCN were produced and purified as described by Gon et al. (8). TorSN, apoTorC, apoTorCN, and apoTorCC were produced from BL21DE3 cells harboring plasmid pET-Sp, pET-C, pET-CN, and pET-CC, respectively. The recombinant strains were grown in Luria broth medium until the optical density at 600 nm reached 0.8 unit. Overproduction of proteins was then induced with 1 mM IPTG for 4 h at 37°C. Overproduced soluble proteins TorSN and apoTorCC were found in the cytoplasm. French press extracts were equilibrated with 20 mM sodium phosphate buffer (pH 7.4) and loaded onto a HiTrap chelating Ni2+ column (Amersham Pharmacia Biotech). The proteins were eluted with a step gradient of imidazole from 20 to 500 mM. TorSN and apoTorCC were eluted with 200 mM and 500 mM imidazole, respectively. Overproduced proteins apoTorC and apoTorCN were found in inclusion bodies. The proteins were solubilized by a treatment with 6 M urea and then refolded by dilution of urea by dialysis (25).
Chemical Cross-Linking Studies.
These experiments were carried out by using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) as a cross-linker. Proteins (5 μM) were incubated 30 min at room temperature in 10 mM cacodylate buffer (pH 6.0) with EDC (1 mM). The interactions were analyzed by SDS/PAGE. Western blot detections were performed with anti-His antibodies (Invitrogen) or anti-TorA antibodies.
Surface Plasmon Resonance Experiments.
The surface plasmon resonance (BIAcore apparatus, Amersham Pharmacia Biosensor) was used to analyze the binding between TorA or TorSN and TorC derivatives. All experiments were carried out at 25°C. TorA in 10 mM sodium acetate buffer (pH 5) and TorSN in 50 mM sodium formate (pH 4.5) were immobilized on a sensor chip CM5 (BIAcore) through amine coupling. The TorC derivatives were prepared in 20 mM sodium phosphate buffer (pH 7.4) and injected over the test and control (no protein immobilized) surfaces at a flow rate of 10 μl/min. Analysis of interactions was carried out by using the BIA evaluation 3.0 software.
Results
Negative Regulation of the tor Operon Specifically Involves Apocytochrome TorC.
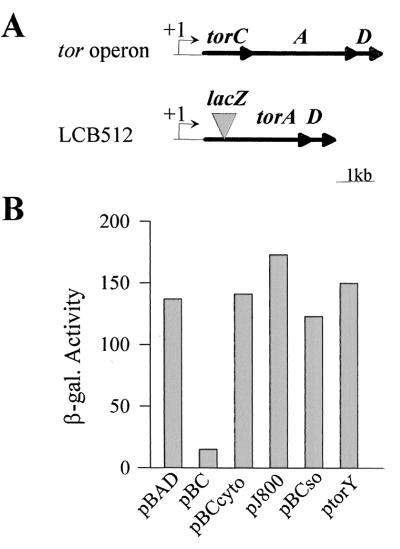
We had previously shown that the immature form of the TorC c-type cytochrome (apoTorC) down-regulates the transcription of the torCAD operon in E. coli (10). To better understand the specificity and the mechanism of this puzzling control, we constructed a mutant strain (LCB512) carrying a chromosomal deletion of the entire torC gene. The strain also contains a torA–lacZ fusion to follow the expression of the tor operon promoter (Fig. 1A). When apoTorC was produced in trans from plasmid pBC, the β-galactosidase activity of the torA–lacZ fusion decreased to about 1/10 (Fig. 1B). This decrease confirms that, unless matured, TorC strongly represses the expression of the tor operon. As c-type cytochrome maturation takes place in the periplasm, the negative effect could be mediated by the TorC polypeptide either before or after its translocation through the inner membrane. To settle this point, we cloned a 5′ deleted torC gene into plasmid pBAD24. The resulting plasmid (pBCcyto) allows production of a TorC polypeptide devoid of its N-terminal hydrophobic segment and, as a result, the truncated protein is found in the cytoplasm (data not shown). As shown in Fig. 1B, production of the cytoplasmic form of TorC from pBCcyto did not reduce the activity of the torA–lacZ fusion, indicating that apoTorC acts as a negative regulator after translocation to the periplasm.
Figure 1.
Effect of apocytochromes on the torA–lacZ fusion activity. (A) Physical map of the tor operon in MC4100 and LCB512 (ΔtorC, torA–lacZ). The +1 arrow indicates the transcription start site. (B) The β-galactosidase activities of torA–lacZ fusion of strain LCB512 producing no protein (pBAD), apocytochrome TorC (pBC), cytoplasmic apoTorC (pBCcyto), apocytochrome c3 from Desulfovibrio vulgaris Hildenborough (pJ800), apoTorC from Shewanella oneidensis (pBCso), or apoTorY (ptorY) are expressed in Miller units. The strains were grown anaerobically with TMAO (10 mM). The PBAD promoter was induced by addition of 0.05‰ arabinose; 1 mM IPTG was added to the strain containing pJ800 or ptorY.
To determine whether this negative control involves any immature c-type cytochrome, cytochrome c3 of Desulfovibrio vulgaris Hildenborough was overproduced from plasmid pJ800 in strain LCB512. As expected (20), the Ccm maturation machinery was overloaded and only apocytochrome c3 was observed in the periplasm. However, expression of the torA–lacZ fusion was not affected by the presence of large amount of apocytochrome c3. The negative regulatory process might thus be mediated by apoTorC only. To confirm this hypothesis, we tested the effect of plasmids encoding TorC-related proteins. Plasmids pBCso and ptorY encode cytochrome TorC of Shewanella oneidensis (TorCso) and TorY, a TorC homologue of E. coli, respectively. TorCso and TorY are pentahemic cytochromes that display 38% and 34% identity to TorC from E. coli, respectively. Production of each of the two apoproteins did not significantly modify the activity of the tor fusion (Fig. 1B), indicating that the negative control is not observed even in the presence of apocytochromes related to TorC. This control is thus an autoregulatory process specifically involving apoTorC of E. coli.
Negative Autoregulation Involves the C-Terminal Domain of ApoTorC.
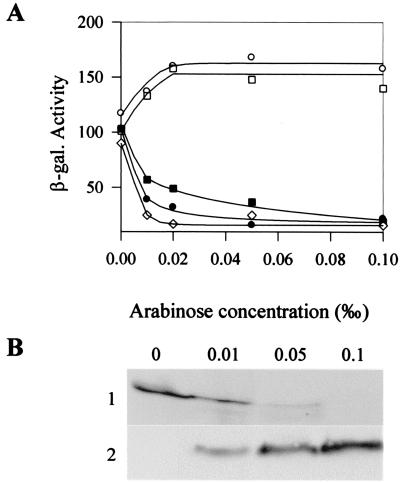
TorC is made up of two domains of similar size. The N-terminal domain (TorCN) contains a membrane anchor and four heme-binding sites, whereas the C-terminal domain (TorCC) possesses only one heme-binding motif (8). As each TorC domain contains heme-binding sites, negative regulation, which depends on the absence of heme insertion, might require only one of the two domains of TorC. To test this hypothesis, we used plasmids pBCN and pBCC, which respectively encode the membrane-anchored N-terminal domain of TorC (TorCN) and the periplasmic C-terminal domain of TorC (TorCC). The cloned torC fragments are under the control of the PBAD promoter, and different concentrations of arabinose were used to follow the effect of increasing amounts of the immature forms of TorCN and TorCC on the expression of the tor fusion. As exemplified for pBCC, when the arabinose concentration rose in the medium, maturation of the cytochromes was rapidly stopped, but the amount of apocytochromes increased (Fig. 2B and data not shown). Interestingly, production of increasing amounts of apoTorCN resulted in a slightly increased activity of the torA–lacZ fusion, but production of apoTorCC led to a rapid drop of the fusion activity (Fig. 2A). As the expression levels of the fusion were not significantly different with or without apoTorCN, we conclude that the N-terminal domain of TorC plays no role in the regulatory mechanism. In contrast, that production of apoTorCC strongly decreased the activity of the fusion and that the pattern of repression by apoTorCC was close to that of the entire apoTorC protein strongly suggest the C-terminal domain of TorC is responsible for the tor down-regulation. To confirm that tor repression requires apoTorCC, we mutated Cys-329 of the heme-binding site of TorCC to prevent heme insertion. As shown on Fig. 2, very low amounts of the mutated TorCC domain were sufficient to down-regulate the tor fusion; the negative effect was maximum even at the lowest arabinose concentration, whereas equivalent repression by the wild-type TorCC domain necessitated higher concentrations of arabinose. Together these data show that the C-terminal domain of TorC acts as a strong negative regulator of the tor operon when it is immature.
Figure 2.
Effect of increasing amounts of apoTorC, apoTorCC, and apoTorCN on the torA–lacZ fusion activity. (A) β-Galactosidase activities of the torA–lacZ fusion of strain LCB512 (ΔtorC) producing either no apocytochrome (pBAD, ○), apoTorC (pBC, ●), apoTorCN (pBCN, □), apoTorCC (pBCC, ■), or TorCCC329S (pBCCC329S, ⋄) are expressed in Miller units. Strains were grown anaerobically with TMAO (10 mM); induction of the PBAD promoter was performed with arabinose at the indicated concentrations. (B) Maturation state of TorCC. Periplasmic fractions (60 μg) of LCB512 containing plasmid pBCC grown as in A were loaded on an SDS/15% polyacrylamide gel. After electrophoresis, the gel was stained for heme with 3,3′,5,5′-tetramethylbenzidine (TMBZ) (1) or revealed by Western blotting with anti-His antibodies (2).
Direct Interaction Between ApoTorC and the Periplasmic Detector Region of Sensor TorS.
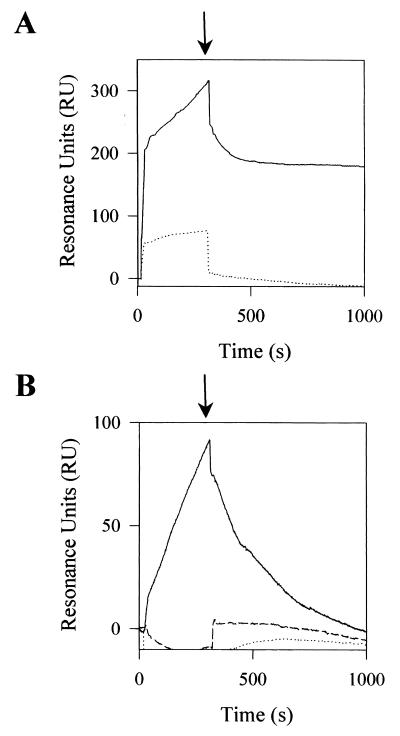
tor operon expression is under the positive control of the TorS/TorR two-component system. TorS is a transmembrane sensor that detects the presence of TMAO in the medium and, in response, transphosphorylates TorR (17). Epistatic experiments have previously shown that the TorC autoregulatory pathway could implicate the TorS/TorR system (10). To confirm this result, we introduced plasmid pBCC into strain LCB726, which carries a TorS constitutive mutation and a torA–lacZ fusion. As expected, overproduction of apoTorCC from pBCC did not decrease the activity of the fusion (616 and 647 Miller units with 0 and 0.1‰ arabinose, respectively). Because the negative effect is abolished by a TorS constitutive mutation, apoTorC seems to act upstream of TorS. Our simplest hypothesis is that apoTorC can directly bind to TorS so that the TorS kinase activity is inhibited. As apoTorC plays the role of a negative effector from the periplasmic space, to fit our model it should contact a periplasmically located TorS region. To test the model in vitro, we overproduced the large N-terminal region of TorS (from position 17 to 319) putatively oriented to the periplasm, and the soluble forms of apoTorC, apoTorCN, and apoTorCC. Each polypeptide contains a C-terminal His-tag and was overproduced in the cytoplasm. The His-tagged proteins were then purified to near homogeneity by using Ni2+ columns (Fig. 3). To characterize any binding between apoTorC and the N-terminal region of TorS (hereafter called TorSN), we first carried out BIAcore experiments. Purified TorSN was coupled to the dextran matrix of a sensor chip and apoTorC was injected into the TorSN-containing sensor chip. The sensorgram clearly reflected an association between two proteins, indicating apoTorC directly binds to TorSN (Fig. 4A). Moreover, the association was stable, since the amount of resonance units slowly decreased after the injection step. As a control, we injected purified mature TorC. After injection the amount of recovered resonance units was very low, showing that, in contrast to apoTorC, mature TorC cannot interact with TorSN.
Figure 3.
Coomassie blue-stained SDS/20% PAGE of Ni2+ column-purified extracts. Twenty micrograms of TorSN (lane 1) and 10 μg of apoTorC (lane 2), apoTorCC (lane 3), and apoTorCN (lane 4) were loaded on the gel.
Figure 4.
BIAcore analysis of the interactions between TorSN and the TorC derivatives. The sensorgrams of interactions between immobilized TorSN and (A) apoTorC (solid line) and holoTorC (dotted line) or (B) apoTorCC (solid line), TorCC (dashed line), and apoTorCN (dotted line) are expressed in resonance units (RU). Proteins (50 μl at 1 μM) were injected into a sensor chip with dextran matrix coupled either to TorSN or to no protein as a control. The control flow cell (no protein immobilized) was subtracted from the experimental flow cells. Arrows indicate the end of the injection.
The same approach was used for each domain of TorC. No interaction between immobilized TorSN and mature TorCC or apoTorCN was observed; in contrast, injection of apoTorCC gave a positive signal indicating binding between TorSN and apoTorCC (Fig. 4B). The TorSN–apoTorCC complex, however, appeared less stable than that of TorSN–apoTorC because the resonance units decreased more rapidly in the former case after the injection step. To confirm that apoTorCC binds to TorSN, we performed in vitro cross-linking experiments. TorSN and apoTorCC alone or together were incubated with EDC and the mixtures were loaded onto an SDS/polyacrylamide gel. After electrophoresis, Western blotting was carried out with anti-His antibodies. As indicated by the arrow on Fig. 5, an additional band was detected in the track containing the two proteins, compared with the tracks with TorSN and apoTorCC alone. Moreover, on the basis of its electrophoretic mobility, this band could correspond to a protein complex of about 54 kDa, which is consistent with the molecular mass of a TorSN (34 kDa)–apoTorCC (21 kDa) heterodimer complex. As a last control, the same experiment was carried out with TorSN and mature TorCC. No additional band was revealed in the track containing TorSN and TorCC, indicating no TorSN–TorCC complex was formed. In conclusion, we have shown by using two distinct methods that the immature C-terminal domain of TorC can bind to the N-terminal detector region of sensor TorS.
Figure 5.
Cross-link of TorSN and apoTorCC or holoTorCC. TorSN (5 μM) was incubated with the cross-linker EDC (1 mM) with either apoTorCC (5 μM) or TorCC (5 μM) for 30 min at room temperature. Samples were loaded on an SDS/15% polyacrylamide gel. After electrophoresis, proteins were visualized by Western blotting with anti-His antibodies. The arrow indicates a complex formed between apoTorCC and TorSN.
In Contrast to HoloTorC, Immature TorC Weakly Binds to TorA.
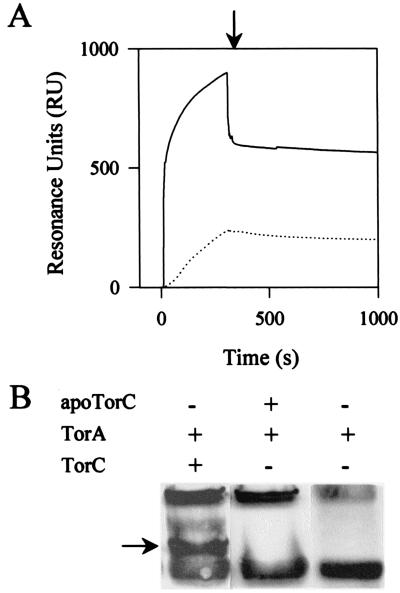
Once matured, TorC tightly binds periplasmic TorA and shuttles electrons from the menaquinones to TorA (8). To better understand the dual role of TorC according to its maturation state, we investigated whether the immature form of TorC could still bind the TorA enzyme. The same two approaches as earlier were used—that is, BIAcore and cross-link techniques. Purified TorA was attached to the dextran matrix and the same amount of mature TorC or apoTorC was injected into the sensor chip. As expected, the sensorgram with TorC confirmed that it strongly binds to TorA (Fig. 6A). Injection of apoTorC led to a sensorgram different from that of TorC; the amount of resonance units was clearly lower during the whole experiment. This difference suggests that apoTorC does not bind to TorA as strongly as TorC does. In the second set of experiments, TorA and TorC or apoTorC were mixed, incubated with EDC, and then subjected to SDS/PAGE; a Western blot was performed with antibodies against TorA (Fig. 6B). When TorA and apoTorC were incubated together, no additional band appeared in comparison to the experiment with TorA alone. In contrast, two additional bands were detected when TorA was incubated with holoTorC. Moreover, the molecular mass of the darker band could correspond to that of a TorA–TorC heterodimer; a Western blot with anti-His antibodies confirmed the presence of His-tagged TorC in the two additional bands (data not shown). That no cross-link could be detected between TorA and apoTorC agrees with the BIAcore experiments and confirms that apoTorC cannot strongly bind to TorA.
Figure 6.
Analysis of the interactions between TorA and TorC or apoTorC proteins. (A) the sensorgrams of interactions between immobilized TorA and TorC (solid line) or apoTorC (dotted line) are expressed in resonance units (RU). Proteins (50 μl at 1 μM) were injected into a sensor chip with dextran matrix coupled either to TorA or to no protein as a control. The control flow cell (no protein immobilized) was subtracted from the experimental flow cells. The arrow indicates the end of the injection. (B) Cross-link of TorA and TorC or apoTorC. TorA (5 μM) was incubated with EDC (1 mM) in the presence of either TorC (5 μM) or apoTorC (5 μM) for 30 min at room temperature. Samples were loaded on an SDS/12.5% polyacrylamide gel. After electrophoresis, proteins were visualized by Western blotting with anti-TorA antibodies. The arrow indicates a complex between TorC and TorA.
Discussion
Cytochromes of the c type are recognized as essential structural intermediates that shuttle electrons between components of respiratory chains (12). To date, no bacterial c-type cytochrome except TorC from E. coli has been shown to have both regulatory and structural functions (10). Another interesting example of a respiratory protein that can be either a negative autoregulator or a respiratory enzyme is the PutA protein from Salmonella typhimurium (26). Indeed, in the absence of proline, PutA behaves as a repressor by binding to the putA promoter, yet it reaches the inner membrane and becomes a proline dehydrogenase upon interaction with proline (27). We have shown that TorC is also a down-regulator of its own synthesis in the absence of heme insertion, but the autoregulatory process is indirect because apoTorC acts from the periplasmic compartment of the cell. Consequently, the effect of apoTorC must be mediated by a transmembrane protein: the TorS sensor proved to be this regulatory intermediate. TorS is involved in induction of torCAD operon in the presence of TMAO. The N-terminal part of TorS comprises a large periplasmic region of about 300 aa flanked by two transmembrane segments, and TMAO detection most probably involves this periplasmic detector region, because a small deletion in it (mutant TorS726) mimics the presence of TMAO and renders TorS constitutively active (16). Our results indicate that apoTorC can also bind to the TorS detector region but the effect of apoTorC binding is opposite to that of TMAO, since apoTorC inhibits kinase TorS and TMAO activates TorS. The mechanism of TorS kinase inhibition is still unknown, but apoTorC either prevents TMAO binding by hiding the TMAO-binding site or counteracts the TorS conformational change induced by TMAO. To answer this question, the precise location of apoTorC and TMAO binding sites should be determined. In any case, this study shows that a certain sensor could detect both positive and negative signals. Moreover, the versatility of sensory detection is illustrated in this work, as the detector region of TorS appears to interact with effectors as different as the small organic compound TMAO or the apoTorC protein. Sensors have been reported to detect a wide range of signals, including proteins and small molecules, and even environmental changes or stresses. However, the direct interaction of an effector with its dedicated sensor has been demonstrated only in a restricted number of cases, and the precise nature of effectors is often unknown (28, 29).
An autoregulatory pathway based on the maturation state of the TorC cytochrome exists most probably because the maturation process is complex and rapidly overloaded. Strikingly, when TorCC was slightly overproduced, the amount of holoTorCC dropped, showing that the maturation machinery can be easily blocked in E. coli (Fig. 2B). Fine-tuning of torCAD operon expression by apoTorC autoregulation is thus essential to allow optimal biogenesis of TorC and to prevent maturation from jamming.
TorC is made up of two domains, and this work reveals that autoregulation does not depend on the entire apocytochrome but only on its C-terminal domain. This domain, which contains one heme-binding site, is homologous to the C-terminal portion of many c-type cytochromes, including TorY from E. coli and TorC from Shewanella oneidensis. However, production of apoTorY or apoTorCso did not down-regulate the tor operon. This observation highlights the specificity of the regulatory process and strongly suggests that TorS does not recognize immature cytochrome domains except apoTorCC from E. coli. The mature TorCC domain is the direct electron donor to TorA, and the good positioning of TorCC on TorA seems to result from the strong interaction between TorA and the TorCN domain (Fig. 7) (8). If apoTorC could still bind to TorA, then apoTorCC could not be available to interact with TorS. In fact, our experiments showed that apoTorC binds much more weakly to TorA than does TorC, suggesting that apoTorCC is neither held nor hidden by TorA. This finding also suggests that the folding of apoTorC clearly differs from that of holoTorC. In conclusion, TorC has dual function according to its maturation state and acts as a direct TorS inhibitor when immature or as an electron donor to TorA once mature (Fig. 7).
Figure 7.
Model for the dual role of TorC in E. coli. Immature TorC cytochrome inhibits (− arrow) TorS kinase activity from the periplasm by binding to the detector region of TorS. This interaction depends on the C-terminal domain of apoTorC. After heme insertion, holoTorC no longer binds to TorS, and the N-terminal domain of TorC then strongly interacts with TorA, allowing formation of a functional TorC–TorA respiratory complex.
Acknowledgments
We thank C. Iobbi-Nivol and G. Burkhart for reviewing the manuscript and A. Dolla and B. L. Wanner for the kind gift of plasmids. This work was supported by grants from the Centre National de la Recherche Scientifique, the Université de la Méditerranée, and a Ministère de l'Education Nationale, de la Recherche et de la Technologie fellowship to S.G.
Abbreviations
- TMAO
trimethylamine N-oxide
- IPTG
isopropyl β-d-thiogalactoside
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Richardson D J. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 2.Berks B C, Ferguson S J, Moir J W, Richardson D J. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 3.Barrett E L, Kwan H S. Annu Rev Microbiol. 1985;39:131–149. doi: 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- 4.Yancey P H, Fyfe-Johnson A L, Kelly R H, Walker V P, Aunon M T. J Exp Zool. 2001;289:172–176. doi: 10.1002/1097-010x(20010215)289:3<172::aid-jez3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 5.Dos Santos J P, Iobbi-Nivol C, Couillault C, Giordano G, Méjean V. J Mol Biol. 1998;284:421–433. doi: 10.1006/jmbi.1998.2155. [DOI] [PubMed] [Google Scholar]
- 6.Proctor L M, Gunsalus R P. Environ Microbiol. 2000;2:399–406. doi: 10.1046/j.1462-2920.2000.00121.x. [DOI] [PubMed] [Google Scholar]
- 7.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M C. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 8.Gon S, Giudici-Orticoni M T, Méjean V, Iobbi-Nivol C. J Biol Chem. 2001;276:11545–11551. doi: 10.1074/jbc.M008875200. [DOI] [PubMed] [Google Scholar]
- 9.Roldan M D, Sears H J, Cheesman M R, Ferguson S J, Thomson A J, Berks B C, Richardson D J. J Biol Chem. 1998;273:28785–28790. doi: 10.1074/jbc.273.44.28785. [DOI] [PubMed] [Google Scholar]
- 10.Ansaldi M, Bordi C, Lepelletier M, Méjean V. Mol Microbiol. 1999;33:284–295. doi: 10.1046/j.1365-2958.1999.01468.x. [DOI] [PubMed] [Google Scholar]
- 11.Fabianek R A, Hennecke H, Thöny-Meyer L. FEMS Microbiol Rev. 2000;24:303–316. doi: 10.1111/j.1574-6976.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 12.Thöny-Meyer L. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barker P D, Ferguson S J. Structure Fold Des. 1999;7:R281–R290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 14.Tanapongpipat S, Reid E, Cole J A, Crooke H. Biochem J. 1998;334:355–365. doi: 10.1042/bj3340355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arslan E, Schulz H, Zufferey R, Kunzler P, Thöny-Meyer L. Biochem Biophys Res Commun. 1998;251:744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 16.Jourlin C, Bengrine A, Chippaux M, Méjean V. Mol Microbiol. 1996;20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 17.Jourlin C, Ansaldi M, Méjean V. J Mol Biol. 1997;267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- 18.Ansaldi M, Jourlin-Castelli C, Lepelletier M, Théraulaz L, Méjean V. J Bacteriol. 2001;183:2691–2695. doi: 10.1128/JB.183.8.2691-2695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gon S, Patte J H, Méjean V, Iobbi-Nivol C. J Bacteriol. 2000;182:5779–5786. doi: 10.1128/jb.182.20.5779-5786.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock W B, Chemerika P J, Forrest M E, Beatty J T, Voordouw G. J Gen Microbiol. 1989;135:2319–2328. doi: 10.1099/00221287-135-8-2319. [DOI] [PubMed] [Google Scholar]
- 21.Thomas P E, Ryan D, Levin W. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 22.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. . (First Published May 30, 2000; 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman L M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung C T, Miller R H. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Plamann L. J Bacteriol. 1996;178:289–292. doi: 10.1128/jb.178.1.289-292.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrovsky P, Maloy S. Proc Natl Acad Sci USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muro-Pastor A M, Ostrovsky P, Maloy S. J Bacteriol. 1997;179:2788–2791. doi: 10.1128/jb.179.8.2788-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 29.Grebe T W, Stock J B. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]