Abstract
Purpose of review
Disorders of sex development (DSD) are a diverse group of conditions affecting gonadal development, sexual differentiation or chromosomal sex. In this review, we will discuss recent literature on the genetic causes of DSD, with a focus on novel genetic sequencing technologies, new phenotypes associated with known DSD genes, and increasing recognition of the role of genetic regulatory elements in DSD.
Methods
We performed a comprehensive search of PubMed through August 2016 to identify important peer-reviewed publications from 2015–2016 on the topic of DSD genetics.
Summary of Recent Findings
Whole-exome sequencing was used to successfully identify genetic causes of DSD in 35% of a cohort of 46,XY subjects who had not previously received a genetic diagnosis.
A novel mutation in NR5A1 has been identified as a cause of 46,XX testicular and ovotesticular DSD, demonstrating a previously unappreciated role of NR5A1 in preventing testicular differentiation in 46,XX individuals.
Genetic regulatory elements of SOX9 have been identified as causes of 46,XX and 46,XY DSD.
Key terms: Gene, genetics, exome, disorder of sex development
Introduction
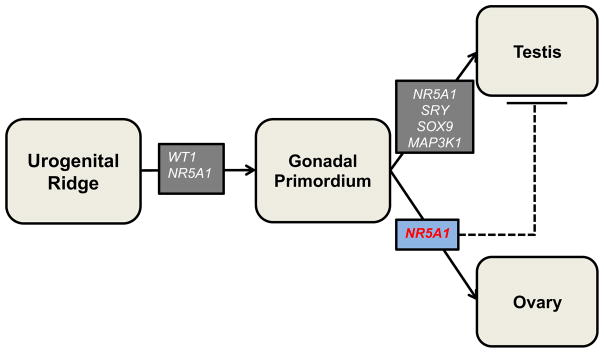
Disorders of sex development (DSD) represent a heterogeneous group of “congenital conditions in which development of chromosomal, gonadal, or anatomic sex is atypical” [1]. Over the past 30 years, we have learned much about the genetics of these disorders. By the late 1980s the human androgen receptor gene (AR) had been characterized [2] and by the early 1990s the critical testis-determining region on the Y chromosome, SRY, was described [3]. In the past two decades, the pace of genetic discovery has increased rapidly, with the identification of many new genetic causes of DSD. More than 50 genes have now been implicated in the pathogenesis of these disorders [4]. These genes encode essential factors in gonadal development or in sex-hormone synthesis or responsiveness (fig 1).
Fig 1.
Role of select DSD genes in typical genital differentiation
Dashed line: Proposed action of gene, solid line: demonstrated action of gene
This review focuses on recent insights into the genetic causes of disorders of sex development as well as the use of innovative sequencing technologies to facilitate understanding of the pathogenesis and phenotypic spectrum of DSD. Recent reports in the DSD field have been in three areas: 1) application of whole-exome sequencing to the identification of genetic causes of DSD, 2) expansion of the observed phenotypes associated with variants in known DSD-associated genes and 3) an examination of the role of genomic regulation in human disease.
Whole-exome sequencing in genetic diagnosis of DSD
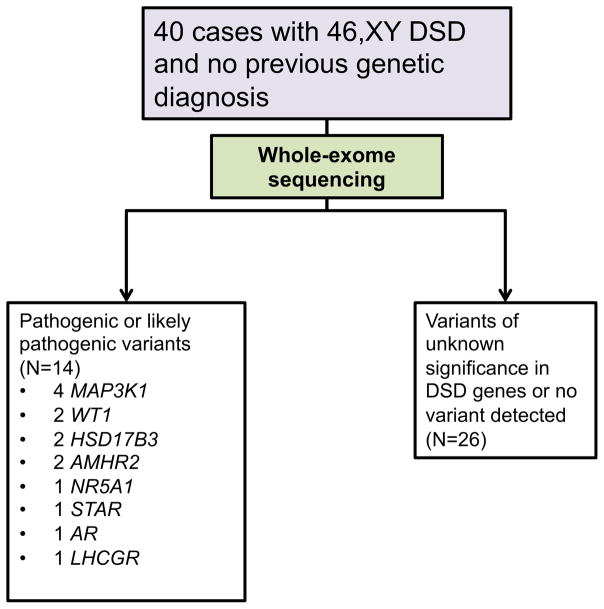
In the last decade, whole exome-sequencing (WES) has greatly improved the ability to identify disease-causing genetic variants. WES is a method for sequencing all of the protein-coding regions (exons) of an individual’s genome, which can then be compared to databases containing large numbers of control exomes to identify possible disease-causing variants [5]. Recently, this approach has been applied to individuals with DSD [6]. Baxter et al. performed whole-exome sequencing in 40 subjects with 46,XY DSD who had not previously received a genetic diagnosis and identified pathogenic or likely pathogenic variants in known DSD genes in 35% of these patients (fig 2).
Fig 2.
Study design and genetic sequencing results in Baxter et al [7].
NR5A1
Baxter et al. identified a pathogenic, heterozygous NR5A1 variant in one individual in the 46, XY DSD cohort who previously carried a clinical diagnosis of androgen insensitivity syndrome [7]*. NR5A1 (also known as steroidogenic factor 1, or SF-1), encodes a nuclear receptor expressed in the gonadal ridge as well as in Leydig cells later in gonadal development [8]. NR5A1 acts as a transcription factor involved in the formation of the bipotential gonad and is also essential for the subsequent differentiation of the testis via several downstream factors including SOX9 (see discussion of this gene below) [8,9]. Homozygous NR5A1 mutations are rare and cause complete gonadal dysgenesis as well as adrenal insufficiency, as NR5A1 also has a critical role in development of the adrenal cortex. In humans, heterozygous NR5A1 mutations are found in 10–15% of individuals with 46,XY DSD, but are not associated with adrenal insufficiency [10,11]. 46,XY individuals with pathogenic heterozygous NR5A1 mutations may have a broad range of phenotypes ranging from isolated male-factor infertility [12], to mild undervirilization with hypospadias and/or cryptorchidism, to severe undervirilization. Gonadal phenotypes range from normal to dysgenesis and anorchia [11]. Previous work has established that DSD gene mutations play a role in milder forms of undervirilization not typically classified as DSD, such as isolated hypospadias [10,13]. Our group recently used whole-exome sequencing to identify novel pathogenic NR5A1 mutations in two unrelated 46,XY individuals with bifid scrotum and penoscrotal hypospadias but not cryptorchidism [14]*. Additional novel phenotypes associated with NR5A1 mutations are discussed below.
WT1
Two subjects in the Baxter study had variants in WT1, which encodes a transcription factor involved in embryonic renal and gonadal development. [7] Alternative splicing of WT1 RNA transcripts can result in more than 20 isoforms of the protein [15].
Several phenotypes are associated with mutations in the WT1 gene. The most severe phenotype, “WAGR” syndrome, is characterized by Wilms tumor, aniridia, genitourinary malformations (specifically undervirilization in 46,XY children), and intellectual disability. WAGR syndrome is caused by a deletion of the WT1 gene. Frasier syndrome in 46,XY children is caused by splice-site mutations in the WT1 gene, which alter the ratio of protein isoforms. Frasier syndrome is characterized by varying degrees of gonadal dysgenesis (partial to complete), renal failure later in childhood due to focal segmental glomerulosclerosis, and increased risk for gonadoblastoma. Denys-Drash syndrome is caused by missense mutations affecting the zinc-finger DNA binding region of the WT1 gene, and is characterized by varying degrees of gonadal dysgenesis (partial to complete), renal failure early in childhood due to diffuse mesangial sclerosis, and increased risk for Wilms tumor and gonadoblastoma [16]. Isolated diffuse mesangial sclerosis has been attributed to WT1 mutations, in some cases the same missense mutations that have been described in association with Denys-Drash in other individuals [15,17–20].
In the Baxter study, one subject with WT1 variant presented with Müllerian structures and dysgenetic gonads bilaterally. On histology, the left gonad was consistent with an immature testis, and the right gonad contained fibro-fatty connective tissue with a possible vas deferens detected. This individual had end-stage renal disease and a previous history of bilateral nephrectomy, though no history of Wilms tumor. The second subject had cryptorchidism and a urogenital sinus, with bilateral testes, one with normal histology and one dysgenetic. There was no known renal disease in this patient, though medical records were incomplete [7].
A recent report investigating genotype-phenotype relationships in a cohort of pediatric patients with steroid-resistant nephrotic syndrome identified WT1 mutations in 21 of the 354 subjects (5.4%). Of these subjects, 12 had a 46,XY karyotype, and all of these subjects had a disorder of sex development, with eight having complete undervirilization of the external genitalia. The remaining four 46,XY subjects had partial undervirilization leading to hypospadias, with or without chordee and cryptorchidism. One 46,XY individual underwent gonadectomy at the age of nine years and was found to have gonadoblastoma. Of the nine 46,XX individuals with WT1 mutations, one individual was found to have absence of ovaries, but DSD was not observed in the remaining eight individuals; later ovarian function was not reported [21]*.
Baxter et al. also identified genetic variants in several additional DSD-associated genes, including DHH, MAP3K1, and MAMLD1 (which are involved in testicular development), LHCGR (which encodes the LH/hCG receptor), AMHR2 (which encodes the anti-mullerian hormone receptor) STAR and HSD17B3 (which are involved in testosterone synthesis), AR (which encodes the androgen receptor), and CHD7 (which is mutated in CHARGE syndrome, which includes genital anomalies among other features). These results highlight the utility of comprehensive genetic testing in the clinical setting to establish genetic diagnosis and in the research setting to expand our understanding of the frequency and phenotypic variability for specific genetic causes of DSD [7].
A Role for NR5A1 in ovarian fate specification
As discussed above, NR5A1 has been thought to be chiefly involved in early gonadal development and testis determination. The majority of NR5A1 mutations associated with DSD to date have been identified in 46,XY individuals with varying degrees of undervirilization. Phenotypic manifestations of NR5A1 mutations in 46,XX individuals can include ovarian hypoplasia, premature ovarian insufficiency, and/or primary or secondary amenorrhea with otherwise normal urogenital structures. In some cases, fertility is preserved [22].
Recently, two groups have described another phenotype associated with an NR5A1 variant in 46,XX individuals. Bashamboo et al. identified five cases of 46,XX testicular or ovotesticular DSD from four families due to identical mutations in NR5A1 resulting in an arginine-to-tryptophan change at position 92 (p.Arg92Trp), which lies in the DNA-binding domain of the NR5A1 protein. Testicular DSD is characterized by the presence of testicular tissue and virilized external genitalia in a 46,XX individual. Ovotesticular DSD is characterized by the coexistence of ovarian and testicular tissue in the gonads. Phenotypes ranged from ambiguous genitalia, to penoscrotal hypospadias, to micropenis noted at birth, to small testes and low testosterone noted in adolescence. Of note, one proband had a sibling with labial fusion, clitoral enlargement, and dysgenetic testes in the inguinal region who was found to have a 46,XY karyotype and the same NR5A1 mutation [23]*.
Baetens et al. reported the same mutation in three members of a cohort of eleven unrelated individuals with 46,XX testicular or ovotesticular DSD. Phenotypes ranged from mild clitoromegaly, to ambiguous genitalia, to micropenis and hypospadias [24]*.
Baetens et al. demonstrated expression of NR5A1 in the developing fetal human ovary and testis, in contrast to mouse studies that have shown expression in the early testis but minimal expression in the ovary [25], consistent with a role for NR5A1 in ovarian differentiation. The authors hypothesized that the variant identified in these cases disrupted NR5A1 activity in testis-opposing pathways in the developing ovary. Indeed, they specifically demonstrate that the Arg92Trp mutant NR5A1 protein exhibited less robust ability to synergize with β-catenin, a component of the Wnt signaling pathway, in promoting the expression of genes that inhibit testicular differentiation [24].
An additional phenotype for NR5A1 mutations in 46,XY individuals was explored by Ferlin et al. They found a prevalence of 1.8% for heterozygous missense NR5A1 mutations in infertile males with severe impairment in spermatogenesis, with a higher rate of mutations (2.7%) in men with a prior history of cryptorchidism, as compared to 1.4% of men without cryptorchidism [26]. This report was comparable to previous reports that identified an NR5A1 mutation prevalence of 4% in a cohort with idiopathic male infertility due to failure of spermatogenesis [27].
Mutations in the Regulatory Region of SOX9
Recent work in the field of DSD genetics has included an examination of the role of genetic regulatory elements in the pathogenesis of DSD, particularly involving the SOX9 gene. SOX9 is a target of SRY and is essential for testicular differentiation [28]. Loss-of-function mutations in SOX9 are a cause of 46,XY DSD, and large duplications near SOX9, hypothesized to be gain-of-function mutations, have been identified in patients with 46,XX DSD [29–31]. Studies in mice have identified specific enhancer regions required for the expression of SOX9 in early testis development [32]. Benko et al. had previously described five DSD cases associated with disruption of SOX9 regulatory regions. Three 46,XX individuals with virilization and ovotesticular DSD were found to have duplications in a regulatory region upstream of the SOX9 gene. Furthermore, two 46,XY individuals with gonadal dysgenesis and severe undervirilization were found to have deletions in the same region. Based on the location of the duplications and deletions discussed above, the authors hypothesize the existence of a 517–595 kilobase (kb) regulatory region upstream of SOX9 that when duplicated may drive a testicular fate for the bipotential gonad in 46,XX individuals, possibly through modification/relaxation of epigenetic repressors. When deleted in 46,XY individuals, SOX9 expression is restricted, leading to impaired testicular development [33].
Hyon et al. describe three males with phenotypically normal male genitalia who presented with infertility and were found to have low testicular volume and azospermia. On further evaluation, all three cases had 46,XX karyotypes and duplications upstream of the SOX9 gene overlapping with the region described by Benko et al. The authors were able to identify a smaller minimal critical regulatory region of 40–41.9 kb located ~600kb upstream of SOX9, which, when duplicated, may be sufficient to induce testicular differentiation in 46,XX individuals [34]*.
More recently, Kim et al. were able to further narrow the critical regulatory regions upstream of the SOX9 gene associated with 46,XX and 46,XY DSD. This group identified duplications that all shared a 68-kb region—located within the region upstream of SOX9 described by Benko et al.—in 3 unrelated individuals with 46,XX ovotesticular DSD. They similarly identified deletions upstream of SOX9 in one member of a six-generation kindred with autosomal dominant DSD in five 46,XY members, characterized by gonadal dysgenesis and severe undervirilization. Three additional families were described with undervirilization and gonadal dysgenesis in 46,XY family members and deletions in a region upstream of SOX9. All described deletions share a single 32.5 kb region which is not located within the region upstream of SOX9 described by Benko et al. It is near, though does not overlap, the 68kb region that they found to be associated with 46,XX DSD and overlaps with previously described deletions upstream of SOX9 causing 46,XY DSD [30,35]*.
The authors hypothesize the existence of two separate regulatory regions. The region that is deleted in cases of 46,XY DSD may serve as a SOX9 enhancer, and its absence may contribute to insufficient expression of the transcription factor, preventing normal testicular differentiation. Conversely, a separate SOX9 enhancer duplication may increase gene and protein product associated with Sertoli cell differentiation, which may “push” the undifferentiated gonad into a testicular fate [35].
These findings narrow the “critical regions” for regulation of SOX9 in the developing gonad and demonstrate the importance of non-coding regions of the genome in normal gene expression and their role in human disease.
Conclusion
Facilitated by ongoing developments in genetic sequencing technologies, advances continue to be made in understanding the genetics of DSD and, in turn, the biology of gonadal development. Recent findings have mostly focused on previously identified DSD genes; future work will undoubtedly use whole-exome sequencing and other emerging technologies to identify new DSD genes.
Whole-exome sequencing is becoming increasingly available in the clinical setting, and the ability to provide genetic diagnoses for a large fraction of DSD promises to change the way we advise and treat patients with these conditions. For example, establishing a specific genetic diagnosis may allow for improved guidance on oncologic risk and early identification of associated issues such as renal insufficiency. As the pace of innovation in genetic diagnosis continues to accelerate, the impact of whole-exome sequencing and related methods on patient care will be an essential area of future inquiry.
Key Points.
Whole-exome sequencing has proven to be an effective modality by which to identify genetic causes of DSD, though its use has thus far been limited to a research setting.
An expanded understanding of the genetic variants associated with DSD phenotypes has brought about a deeper understanding of the biology of gonadal development.
Acknowledgments
Financial support and sponsorship:
JK is supported by National Institutes of Health (NIH) Grant T32 DK007699. JS received support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
Footnotes
Conflicts of Interest:
The authors have no relevant conflicts of interest.
References and Recommended Reading
Papers of particular interest, published within the annual period of review, (2015–2016) have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hughes IA, Houk C, Ahmed SF, Lee P Lawson Wilkins Pediatric Endocrine Society/European Society for Paediatric Endocrinology Consensus Group. Consensus statement on management of intersex disorders. J Pediatr Urol. 2006;2:148–62. doi: 10.1016/j.jpurol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Tilley WD, Marcelli M, Wilson JD, McPhaul MJ. Characterization and expression of a cDNA encoding the human androgen receptor. Proc Natl Acad Sci U S A. 1989;86:327–331. doi: 10.1073/pnas.86.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair aH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf aM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 4.Hiort O, Birnbaum W, Marshall L, Wünsch L, Werner R, Schröder T, Döhnert U, Holterhus P-M. Management of disorders of sex development. [Internet] Nat Rev Endocrinol. 2014;10:520–9. doi: 10.1038/nrendo.2014.108. [DOI] [PubMed] [Google Scholar]
- 5.Singleton AB. Exome sequencing: A transformative technology [Internet] Lancet Neurol. 2011;10:942–946. doi: 10.1016/S1474-4422(11)70196-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggers S, Smith KR, Bahlo M, Looijenga LH, Drop SL, Juniarto ZA, Harley VR, Koopman P, Faradz SM, Sinclair AH. Whole exome sequencing combined with linkage analysis identifies a novel 3 bp deletion in NR5A1. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2014.130. doi:10.1038/ejhg. 2014. 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••7.Baxter RM, Arboleda VA, Lee H, Barseghyan H, Adam MP, Fechner PY, Bargman R, Keegan C, Travers S, Schelley S, et al. Exome sequencing for the diagnosis of 46, XY disorders of sex development. J Clin Endocrinol Metab. 2015;100:E333–E344. doi: 10.1210/jc.2014-2605. Application of whole-exome sequencing to 46,XY subjects with DSD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramayya MS, Zhou J, Kino T, Segars JH, Bondy CA, Chrousos GP. Steroidogenic Factor 1 Messenger Ribonucleic Acid Expression in Steroidogenic and Nonsteroidogenic Human Tissues: Northern Blot and in Situ Hybridization Studies. J Clin Endocrinol Metab. 1997;82:1799–1806. doi: 10.1210/jcem.82.6.3967. [DOI] [PubMed] [Google Scholar]
- 9.Lucas-Herald AK, Bashamboo A. Gonadal development. [Internet] Endocr Dev. 2014;27:1–16. doi: 10.1159/000363608. [DOI] [PubMed] [Google Scholar]
- 10.Suntharalingham JP, Buonocore F, Duncan AJ, Achermann JC. DAX-1 (NR0B1) and steroidogenic factor-1 (SF-1, NR5A1) in human disease. [Internet] Best Pract Res Clin Endocrinol Metab. 2015;29:607–619. doi: 10.1016/j.beem.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philibert P, Zenaty D, Lin L, Soskin S, Audran F, Léger J, Achermann JC, Sultan C. Mutational analysis of steroidogenic factor 1 (NR5a1) in 24 boys with bilateral anorchia: A French collaborative study. Hum Reprod. 2007;22:3255–3261. doi: 10.1093/humrep/dem278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Röpke A, Tewes A-C, Gromoll J, Kliesch S, Wieacker P, Tüttelmann F. Comprehensive sequence analysis of the NR5A1 gene encoding steroidogenic factor 1 in a large group of infertile males. [Internet] Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2012.290. doi:10.1038/ejhg. 2012. 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camats N, Pandey AV, Fernandez-Cancio M, Andaluz P, Janner M, Toran N, Moreno F, Bereket A, Akcay T, Garcia-Garcia E, et al. Ten novel mutations in the NR5A1 gene cause disordered sex development in 46,XY and ovarian insufficiency in 46,XX individuals. J Clin Endocrinol Metab. 2012:97. doi: 10.1210/jc.2011-3169. [DOI] [PubMed] [Google Scholar]
- •14.Swartz JM, Ciarlo R, Guo MH, Abrha A, Diamond DA, Chan Y-M, Hirschhorn JN. Two Unrelated Undervirilized 46,XY Males with Inherited NR5A1 Variants Identified by Whole-Exome Sequencing [Internet] Horm Res Paediatr. 2016:02115. doi: 10.1159/000448754. A case series describing NR5A1 variants in 46,XY DSD, identified using whole-exome sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler B, Biebermann H, Friedsam V, Gellermann J, Maier RF, Pohl M, Wieacker P, Hiort O, Gruters A, Krude H. Analysis of the Wilms’ tumor suppressor gene (WT1) in patients 46,XY disorders of sex development. J Clin Endocrinol Metab. 2011;96:1131–1136. doi: 10.1210/jc.2010-2804. [DOI] [PubMed] [Google Scholar]
- 16.Coppes MJ, Liefers GJ, Higuchi M, Zinn AB, Balfe JW, Williams BRG. Inherited WT1 mutation in Denys-Drash syndrome. Cancer Res. 1992;52:6125–6128. [PubMed] [Google Scholar]
- 17.Kadandale JS, Wachtel SS, Tunca Y, Sid Wilroy R, Martens PR, Tharapel AT. Localization of SRY by primed in situ labeling in XX and XY sex reversal. Am J Med Genet. 2000;95:71–74. doi: 10.1002/1096-8628(20001106)95:1<71::aid-ajmg14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 18.Miller-Hodges E, Hohenstein P. WT1 in disease: shifting the epithelial - mesenchymal balance [Internet] J Pathol. 2011;226 doi: 10.1002/path.2977. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 19.Hahn H, Cho YM, Park YS, You MW, Cheong H. Two cases of isolated diffuse mesangial sclerosis with WT1 mutations. J Korean Med Sci. 2006;21:160–164. doi: 10.3346/jkms.2006.21.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeanpierre C, Denamur E, Henry I, Cabanis MO, Luce S, Cécille A, Elion J, Peuchmaur M, Loirat C, Niaudet P, et al. Identification of constitutional WT1 mutations, in patients with isolated diffuse mesangial sclerosis, and analysis of genotype/phenotype correlations by use of a computerized mutation database. [Internet] Am J Hum Genet. 1998;62:824–33. doi: 10.1086/301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •21.Ahn YH, Park EJ, Kang HG, Kim SH. Genotype – phenotype analysis of pediatric patients with WT1 glomerulopathy. Pediatr Nephrol. 2016 doi: 10.1007/s00467-016-3395-4. An analysis of WT1 mutations in subjects with steroid-resistant nephropathy. [DOI] [PubMed] [Google Scholar]
- 22.Lourenço D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, et al. Mutations in NR5A1 associated with ovarian insufficiency. [Internet] N Engl J Med. 2009;360:1200–10. doi: 10.1056/NEJMoa0806228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••23.Bashamboo A, Donohoue PA, Vilain E, Rojo S, Calvel P, Seneviratne SN, Buonocore F, Barseghyan H, Bingham N, Rosenfeld JA, et al. A recurrent p. Arg92Trp variant in steroidogenic factor-1 (NR5A1) can act as a molecular switch in human sex development [Internet] Hum Mol Genet. 2016:ddw186. doi: 10.1093/hmg/ddw186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••24.Baetens D, Stoop H, Peelman F, Todeschini A-L, Rosseel T, Coppieters F, Veitia RA, Looijenga LHJ, De Baere E, Cools M. NR5A1 is a novel disease gene for 46,XX testicular and ovotesticular disorders of sex development [Internet] Genet Med. 2016 doi: 10.1038/gim.2016.118. doi:10.1038/gim. 2016. 118. Two case series describing novel 46,XX testicular and ovotesticular DSD phenotypes associated with a variant in NR5A1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen WH, Moore CCD, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: A link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 26.Ferlin A, Rocca MS, Vinanzi C, Ghezzi M, Di Nisio A, Foresta C. Mutational screening of NR5A1 gene encoding steroidogenic factor 1 in cryptorchidism and male factor infertility and functional analysis of seven undescribed mutations [Internet] Fertil Steril. 2015;104:163–169e1. doi: 10.1016/j.fertnstert.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 27.Bashamboo A, Ferraz-De-Souza B, Loureno D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi JP, Christin-Maitre S, et al. Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet. 2010;87:505–512. doi: 10.1016/j.ajhg.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai Y, Hiramatsu R, Matoba S, Kidokoro T. From SRY to SOX9: Mammalian testis differentiation. J Biochem. 2005;138:13–19. doi: 10.1093/jb/mvi098. [DOI] [PubMed] [Google Scholar]
- 29.Xiao B, Ji X, Xing Y, Chen Y, wei Tao J. A rare case of 46, XX SRY-negative male with a ~74-kb duplication in a region upstream of SOX9 [Internet] Eur J Med Genet. 2013;56:695–698. doi: 10.1016/j.ejmg.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Bhagavath B, Layman LC, Ullmann R, Shen Y, Ha K, Rehman K, Looney S, McDonough PG, Kim HG, Carr BR. Familial 46,XY sex reversal without campomelic dysplasia caused by a deletion upstream of the SOX9 gene [Internet] Mol Cell Endocrinol. 2014;393:1–7. doi: 10.1016/j.mce.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. Am J Med Genet. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 32.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 33.Benko S, Gordon CT, Mallet D, Sreenivasan R, Thauvin-Robinet C, Brendehaug A, Thomas S, Bruland O, David M, Nicolino M, et al. Disruption of a long distance regulatory region upstream of SOX9 in isolated disorders of sex development. [Internet] J Med Genet. 2011;48:825–30. doi: 10.1136/jmedgenet-2011-100255. [DOI] [PubMed] [Google Scholar]
- •34.Hyon C, Chantot-Bastaraud S, Harbuz R, Bhouri R, Perrot N, Peycelon M, Sibony M, Rojo S, Piguel X, Bilan F, et al. Refining the regulatory region upstream of SOX9 associated with 46,XX testicular Disorders of Sex Development (DSD) Am J Med Genet Part A. 2015;167:1851–1858. doi: 10.1002/ajmg.a.37101. [DOI] [PubMed] [Google Scholar]
- •35.Kim G-J, Sock E, Buchberger A, Just W, Denzer F, Hoepffner W, German J, Cole T, Mann J, Seguin JH, et al. Copy number variation of two separate regulatory regions upstream of SOX9 causes isolated 46,XY or 46,XX disorder of sex development [Internet] J Med Genet. 2015;52:240–247. doi: 10.1136/jmedgenet-2014-102864. These two reports analyze the role of variants in a known regulatory of the SOX9 gene in human disease. [DOI] [PubMed] [Google Scholar]