Abstract
Accumulation of lipofuscin in the retinal pigmented epithelium (RPE) is observed in retinal degenerative diseases including Stargardt disease and age-related macular degeneration. Bis-retinoid N-retinyl-N-retinylidene ethanolamine (A2E) is a major component of lipofuscin. A2E has been implicated in RPE atrophy and retinal inflammation; however, mice with A2E accumulation display only a mild retinal phenotype. In the current study, human iPSC-RPE (hiPSC-RPE) cells were generated from healthy individuals to examine effects of A2E in human RPE cells. hiPSC-RPE cells displayed RPE-specific features, which include expression of RPE-specific genes, tight junction formation and ability to carry out phagocytosis. hiPSC-RPE cells demonstrated cell death and increased VEGF-A production in a time-dependent manner when they were cocultured with 10 μM of A2E. PCR array analyses revealed upregulation of 26 and 12 pro-inflammatory cytokines upon A2E and H2O2 exposure respectively, indicating that A2E and H2O2 can cause inflammation in human retinas. Notably, identified gene profiles were different between A2E- and H2O2-treated hiPSC-RPE cells. A2E caused inflammatory changes observed in retinal degenerative diseases more closely as compared to H2O2. Collectively, these data obtained with hiPSC-RPE cells provide evidence that A2E plays an important role in pathogenesis of retinal degenerative diseases in humans.
Keywords: iPSC-RPE, Lipofuscin, A2E, VEGF-A, Inflammation
1. Introduction
Bis-retinoid N-retinyl-N-retinylidene ethanolamine (A2E) accumulation in the retinal pigmented epithelial (RPE) cells is a hallmark of Stargardt disease, a juvenile form of macular degeneration, caused by ABCA4 mutations (Allikmets et al., 1997b, Moiseyev et al., 2010, Ahn et al., 2000, Sun et al., 1999). Age-related A2E accumulation and over-accumulation of A2E in patients with age-related macular degeneration (AMD) have also been reported (Moiseyev et al., 2010, Eldred and Lasky, 1993, Parish et al., 1998, Iriyama et al., 2008, Allikmets et al., 1997a). Roles of accumulation of A2E in RPE cells in these retinal degenerative diseases have been long under investigation. A2E is one byproduct of the visual cycle, which is a recycling mechanism of the visual chromophore 11-cis-retinal. A2E formation occurs through the condensation of two all-trans-retinal molecules and phosphatidylethanolamine (Travis et al., 2007, Kiser et al., 2014, Liu et al., 2000). In vertebrate vision, photoisomerization of 11-cis-retinal to all-trans-retinal activates photoreceptor G-protein coupled receptor, rhodopsin, and the light signal is converted to electrical signals which relay visual stimuli to the brain (Palczewski, 2012). Clearance of all-trans-retinal from photoreceptors is quickly performed under healthy conditions, but any delay of this reaction can lead to an increased concentration and condensation of all-trans-retinal to form byproducts including A2E and all-trans-retinal dimer (RALdi) in the RPE (Maeda et al., 2009, Chen et al., 2012). RPE cell death was observed by photooxidation of A2E due to generation of singlet oxygen and superoxide radicals (Sparrow et al., 2002). Other studies demonstrated that A2E can cause retinal inflammation which leads to retinal cell damage (Anderson et al., 2013, Radu et al., 2011, Zhou et al., 2006). A2E could play an important role in the pathogenesis of retinal degeneration; however, Abca4−/ − mice which accumulate high levels of A2E display a very mild phenotype suggesting a minor role of this molecule in mouse models of retinal degeneration (Weng et al., 1999).
Oxidative stress is another important contributing environmental factor to the development of retinal diseases including AMD (Jarrett and Boulton, 2012, Nita and Grzybowski, 2016). Anti-oxidative enzymes in RPE cells decrease with age, potentially allowing reactive oxygen species (ROS) generated to cause DNA damage and ultimately leading to the apoptosis of RPE cells (Samiec et al., 1998).
Although it has been accepted that the RPE is the major pathogenic target of macular degeneration, obtaining a sufficient number of RPE cells from suitable donors for disease modeling remains an obstacle. The ARPE19 cell line derived from human RPE cells, is the most widely employed in vitro model for studying RPE function; however, there has been a lack of consistency in the results generated in these cells as ARPE19 cells do not truly recapitulate the characteristics of human RPE cells. ARPE19 cells express low levels of several RPE markers, are unable to metabolize vitamin A and have a reduced capacity to express critical proteins which regulate and maintain tight junctions (Ablonczy et al., 2011, Dunn et al., 1996). Development of better RPE cell models for studying human diseases is in critical need.
Pluripotent stem cell (iPSC) technology has opened up a new era in disease modeling via providing the ability to differentiate adult somatic cells into any cell type in the body. iPSCs reprogrammed from adult somatic cells have an exciting potential in human disease modeling as well as cell sources for regenerative medicine. For example, iPSCs which are derived from skin or blood cells can be reprogrammed into beta islet cells to treat diabetes, blood cells to create new blood, or neurons to treat neurological disorders (Ye et al., 2013, Takahashi et al., 2007, Yu et al., 2007, Nakagawa et al., 2008). Several research groups have utilized iPSCs to differentiate into RPE-like cells with striking similarities to native RPE cells (Carr et al., 2009, Buchholz et al., 2009, Kokkinaki et al., 2011, Osakada et al., 2009a). RPE cells derived from iPSCs are analogous to human fetal RPE cells with respect to expression of key RPE markers and display RPE functionalities such as formation of tight junctions, protein secretion, phagocytosis and vitamin A metabolism (Chang et al., 2014). hiPSC-RPE cells have met standards for use in clinical trials and transplantation therapies have been conducted in patients with eye diseases (Schwartz et al., 2012, Mandai et al., 2017). hiPSCs provide access to physiologically relevant samples without the issues associated with paucity of adequate primary human RPE tissues and their limited proliferation potential.
In the current study, we isolated peripheral blood mononuclear cells from healthy donors, reprogrammed them to iPSCs followed by differentiation to RPE cells. hiPSC-RPE displayed all features akin to functionally normal RPE cells in vivo including morphology, monolayers and tight junction formation, secretory function and ability to carry out phagocytosis. Exposure to physiological stressors such as A2E and H2O2 mimicked distinct phenotypes of pathologic or aged RPE cells with inflammation and decrease in cell viability. Our study provides a unique experimental platform not only to understand distinct aspects of RPE function but also to dissect the complex cellular and molecular events in degenerative retinal diseases.
2. Materials and methods
2.1. Generation of human iPSCs
Blood samples were collected from healthy volunteers and peripheral blood mononuclear cells (PBMCs) were isolated using BD Vacutainer Cell Preparation Tubes containing sodium citrate. PBMCs were expanded and transduced with STEMCCA lentivirus vector using an earlier published protocol (Sommer et al., 2012). iPSC-like colonies were picked and maintained up to passage 10 on Matrigel (Corning Bioscience, USA) coated plates. After each passage differentiated cells were discarded and only iPSC-like colonies were propagated. After passage 10, iPSC-like colonies were tested for expression of pluripotency markers via quantitative RT-PCR and immunocytochemistry. All procedures were approved by the Institutional Review Boards (IRBs) at the Case Western Reserve University, Cleveland Ohio and adhered to the Declaration of Helsinki. All cell culture procedures were approved by Case Western Reserve University Institutional Biosafety Committee. All samples were obtained after patients had given informed consent.
2.2. Differentiation of human iPSCs to RPE cells
Fully characterized iPSC lines at passage 10 were used for differentiation. iPSCs were differentiated to functional RPE using a previously reported protocol (Osakada et al., 2009a, Osakada et al., 2009b). Briefly, cells were plated on gelatin coated dish with an inhibitor cocktail of CKI7 (Casein Kinase 1 Inhibitor) (Sigma, St. Lois, MO), SB431542 (Sigma) and ROCKi (Stemcell Technologies, Vancouver, Canada) in ReproCELL ReproStem Cell Culture medium (Stemgent Inc., MA) for one day. Culture medium was replaced by RPE differentiation medium with 20% KSR (ThermoFisher Scientific, MA) on day 1 and 3. On day 5, 7 and 9, KSR was reduced to 15% followed by 10% KSR from day 11 to day 18. Inhibitor cocktail was added up to day 18. Day 19 onwards cells were grown in 10% KSR until dark colonies appeared. Around day 30–35, when dark pigmented colonies appeared, cells were maintained in RPE maintenance medium. Around 10 days later, cells were detached and allowed to float as aggregates for 5 days to 2 weeks. Dark pigmented aggregates were then plated on CellStar coated plates (ThermoFisher Scientific, MA). After cells expanded, non-RPE cells were scrapped off manually and cells showing RPE morphology and pigmentation were passaged. RPE cells were allowed to mature for 30 days in RPE maintenance medium with bFGF (Stemcell Technologies) and SB431542 before being used for experiments.
2.3. RPE monolayers
RPE monolayers were established on 8 well chamber slides (Osakada et al., 2009a, Osakada et al., 2009b, Germany) or 96 well plates coated with CellStart for 30 days. 30,000–50,000 cells were seeded in each well from passage 4. Primary human RPE cells were obtained from Lonza (Walkersville, MD) and cultured in RPE maintenance medium. After the cells were attached medium was replaced every 2–3 days. At least 5 independent monolayers of cells were grown for each experiment.
2.3.1. Immunocytochemistry
Cells were washed once in PBS, and fixed in 4% paraformaldehyde solution (Electron Microscopy Sciences, Hatfield, PA) for 15 min. Cells were washed three times in PBS at room temperature and permeabilized by exposure to 1% Triton X-100 (Sigma, St. Lois, MO) for 1 min at room temperature. Samples were washed three times in PBS at room temperature and blocked with 1% goat serum for 45 min followed by incubation with primary antibodies (TRA-1-60, 1:50, SSEA4, 1:50, SSEA1, 1:50, OCT-4, 1; 50, EMD Millipore, MA), RPE65 antibody (1:250) (Golczak et al., 2010) and rabbit anti-ZO-1 antibody (1:200, Invitrogen) for 1 h at room temperature. The cells were washed three times in PBS, and incubated with secondary antibodies (Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 555 goat anti-mouse IgG (1:500; Invitrogen) for 30 min at room temperature followed by PBS wash three times for 5 min. DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes, Eugene, OR) was used to stain nuclei. Cells were visualized and imaged under an inverted fluorescence microscope.
2.3.2. RT-PCR and qRT-PCR
Total RNA was extracted with the RNeasy kit (Qiagen, Valencia, CA), treated with RNase-free DNAase I (Qiagen) and reverse transcribed using the cDNA synthesis kit (Qiagen) as previously described (Sahu et al., 2015, Parmar et al., 2016). PCR was performed using gene specific primer sequences designed using web tool Primer 3 and synthesized by Eurofins MWG Operon (Hunstville, Al). The PCR products were separated by electrophoresis on a 1% agarose gel and detected under UV illumination. Quantitative RT-PCR was performed with SYBR® Green Supermix (Bio-Rad, Hercules, CA) on the Light cycler 96 Real-Time PCR system (Roche Diagnostics, Risch-Rotkreuz, Switzerland). Relative expression of genes was normalized to the housekeeping gene Gapdh. Primers used for this study are listed in Table 1.
Table 1.
Human primer sequences used in the study.
| Gene | Primer sequence | Position | Size | Accession no |
|---|---|---|---|---|
| RPE65 | 5′-CAATGGGTTTCTGATTGTGGA-3′ | 208 | 150 | NM_000329 |
| 5′-CCAGTTCTCACGTAAATTGGCTA-3′ | 59 | |||
| MITF | 5′-AGAGTCTGAAGCAAGAGCACTG-3′ | 224 | 145 | NM_000248.3 |
| 5′-TGCGGTCATTTATGTTAAATCTTC-3′ | 80 | |||
| BEST1 | 5′-CATAGACACCAAAGACAAAAGC-3′ | 220 | 140 | NM_001139443 |
| 5′-GTGCTTCATCCCTGTTTTCC-3′ | 80 | |||
| MERTK | 5′-GTAAGGCTTGGCTTCTTCCC-3′ | 193 | 132 | NM_006343 |
| 5′-CCGTCCGGAGAGAAATTACA-3′ | 62 | |||
| PAX6 | 5′-TCCGTTGGAACTGATGGAGT-3′ | 206 | 146 | NM_000280 |
| 5′-TAAGGATGTTGAACGGGCAG-3′ | 61 | |||
| TYR1 | 5′-ACCATGTAGGATTCCCGGTT-3′ | 245 | 141 | NM_000372 |
| 5′-TGCCAACGATCCTATCTTCC-3′ | 105 | |||
| GAPDH | 5′-CCATCCACAGTCTTCTGGGT-3′ | 568 | 141 | NM_002046 |
| 5′-CCTCAAGATCATCAGCAAT-3′ | 427 | |||
| IL6 | 5′-CTGCAGCCACTGGTTCTGT-3′ | 239 | 143 | NM_000600 |
| 5′-CCAGAGCTGTGCAGATGAGT-3′ | 97 | |||
| CXCL1 | 5′-CTTCCTCCTCCCTTCTGGTC-3′ | 183 | 133 | NM_001511 |
| 5′-CCAAACCGAAGTCATAGCCA-3′ | 51 | |||
| CXCL12 | 5′-GCCCTTCCCTAACACTGGTT-3′ | 232 | 135 | NM_000609 |
| 5′-TTGACCCGAAGCTAAAGTGG-3′ | 98 | |||
| IL11 | 5′-GGGCGACAGCTGTATCTGG-3′ | 199 | 134 | NM_000882.3 |
| 5′-GGACAGGGAAGGGTTAAAGG-3′ | 66 | |||
| VEGFA | 5′-CACACAGGATGGCTTGAAGA-3′ | 265 | 136 | NM_003376 |
| 5′-AGGGCAGAATCATCACGAAG-3′ | 130 | |||
| C2 | 5′-GATTCGAGGAGCAGCGATAG-3′ | 248 | 136 | NM_000063 |
| 5′-ATGTGGGATGGAGAAACAGC-3′ | 113 | |||
| C5 | 5′-CCCTCCACAGCAGACATTTT-3′ | 178 | 148 | NM_001735 |
| 5′-TCAAGGCAAAGGTGTTCAAA-3′ | 31 | |||
| CFH | 5′-TTCGCTTTTTCTTTTAAGGCA-3′ | 234 | 133 | NM_000186 |
| 5′-CATGTATGGAGAATGGCTGG-3′ | 102 | |||
| TIMP3 | 5′-GTACTGCACATGGGGCATCT-3′ | 279 | 149 | NM_000362 |
| 5′-AGGACGCCTTCTGCAACTC-3′ | 131 | |||
| MMP9 | 5′-CTCAGGGCACTGCAGGAT-3′ | 262 | 133 | NM_004994 |
| 5′-CGACGTCTTCCAGTACCGA-3′ | 130 | |||
| SERPINA3 | 5′-ATGGGCACCATTACCCACT-3′ | 234 | 140 | NM_001085 |
| 5′-ATGACTCCTTTCGCAGCCT-3′ | 95 | |||
| TIMP1 | 5′-TTGACTTCTGGTGTCCCCAC-3′ | 197 | 137 | NM_003254 |
| 5′-GCTTCTGGCATCCTGTTGTT-3′ | 61 | |||
| IL23A | 5′-CCACACTGGATATGGGGAAC-3′ | 208 | 131 | NM_016584 |
| 5′-ACTCAGTGCCAGCAGCTTTC-3′ | 78 | |||
| TGFB2 | 5′-CTCCATTGCTGAGACGTCAA-3′ | 222 | 133 | NM_003238 |
| 5′-CGACGAAGAGTACTACGCCA-3′ | 90 | |||
| CCL2 | 5′-ATAACAGCAGGTGACTGGGG-3′ | 180 | 141 | NM_002982 |
| 5′-TCTCAAACTGAAGCTCGCAC-3′ | 40 | |||
| IL1B | 5′-GGAGATTCGTAGCTGGATGC-3′ | 209 | 144 | NM_000576 |
| 5′-GAGCTCGCCAGTGAAATGAT-3′ | 66 | |||
| CXCL8 | 5′-AGCACTCCTTGGCAAAACTG-3′ | 172 | 137 | NM_000584 |
| 5′-CAAGAGCCAGGAAGAAACCA-3′ | 36 | |||
| C3 | 5′-CCAGCTCTCTGGGAACTCAC-3′ | 217 | 144 | NM_000064 |
| 5′-TTCCTGGACTGCTGCAACTA-3′ | 74 | |||
| APOE | 5′-CAGGTAATCCCAAAAGCGAC-3′ | 247 | 144 | NM_000041 |
| 5′-TTGCTGGTCACATTCCTGG-3′ | 104 | |||
| TNFSF10 | 5′-CTCTCTTCGTCATTGGGGTC-3′ | 232 | 139 | NM_003810 |
| 5′-TGCAGTCTCTCTGTGTGGCT-3′ | 94 | |||
| LIPC | 5′-CATCTCATGCAGCGTTTTGT-3′ | 200 | 133 | NM_000236 |
| 5′-TCCCCTGTGTTTCTCCATTC-3′ | 68 | |||
| ABCA4 | 5′-TACTCGGCCGATGGATAAAC-3′ | 261 | 147 | NM_000350 |
| 5′-TGTATGCCAACGTGGACTTC-3′ | 115 | |||
| IL7 | 5′-CGAGCAGCACGGAATAAAAA-3′ | 259 | 138 | NM_000880 |
| 5′-TCTAATGGTCAGCATCGATCA-3′ | 122 | |||
| TNFSF11B | 5′-TGAGATGAGCAAAAGGCTGA-3′ | 267 | 139 | NM_033012 |
| 5′-TTTCAAGGAGCTGTGCAAAA-3′ | 129 | |||
| OCT4 | 5′-CTGGTTCGCTTTCTCTTTCG-3′ | 199 | 150 | NM_203289 |
| 5′-CTTTGAGGCTCTGCAGCTTA-3′ | 50 | |||
| SOX2 | 5′-GGAAAGTTGGGATCGAACAA-3′ | 2215 | 145 | NM_003106 |
| 5′-GCGAACCATCTCTGTGGTCT-3′ | 2071 | |||
| REX1 | 5′-TGCCTAGTGTGCTGGTGGT-3′ | 177 | 148 | NM_174900 |
| 5′-GGTGGCATTGGAAATAGCAG-3′ | 30 | |||
| NANOG | 5′-TTGGGACTGGTGGAAGAATC-3′ | 216 | 138 | NM_024865 |
| 5′-GATTTGTGGGCCTGAAGAAA-3′ | 79 | |||
| B-ACTIN | 5′-CCTTGCACATGCCGGAG-3′ | 127 | 127 | NM_001101 |
| 5′-GCACAGAGCCTCGCCTT-3′ | 16 |
2.3.3. Enzyme-Linked Immunosorbent Assay (ELISA)
Production of VEGF-A, PEDF and IL6 was quantified by ELISA kits (VEGF-A; VEGF-A Quantikine ELISA kit (R&D Systems, Minneapolis, USA), PEDF; PEDF613, BioProducts (MD, USA), IL6; IL6 Quantikine ELISA kit (R&D Systems). For VEGF-A and PEDF measurements 50 μl of culture supernatant was collected and spun down at 10,000 rpm for 10 min. The clear supernatant collected was used for protein quantification. Cell lysates were prepared with Nonidet P-40 lysis buffer containing 20 mM Tris, pH 8.0, 137 mM NaCl, and 1% Nonidet P-40. Protein concentration was measured with a BCA protein assay kit (Pierce, MA, USA). Triplicate wells were used for all conditions tested.
2.3.4. Phagocytosis assay
Cells were cultured at a density of 105 cells/cm2 in RPE maintenance medium. On the day of the assay cell medium was removed and 100 μl of pHrodo™ BioParticles® (Molecular Probes) conjugate was added to the 96 well plate. Cell density was scaled at 106 cells for 1 vial of 100 μl of the bead conjugate. The cells were then incubated at 37 °C for 2 h, 6 h and 24 h. After incubation, the cells were washed three times with PBS to remove the undigested beads. Cells were then fixed with 4% paraformaldehyde for 15 min followed by three washes with PBS. For imaging the cells were counterstained using DAPI and observed with an inverted fluorescence microscope (Leica, Wetzlar, Germany). The average fluorescent intensity per cell was measured on SpectaMax Plus 384 microplate reader (Molecular devices, Sunnyvale, CA) at 509/533 nm. The average fluorescence value of control wells with no beads is subtracted from wells containing beads at the end of the assay to yield a cell-specific, net phagocytosis signal. Each assay was repeated three times.
2.3.5. Cell death assay
Cell death was assessed using LDH-Cytotoxicity Colorimetric Assay Kit II (BioVision, Milpitas, CA) following manufacturer’s instructions. Cells were cultured in 96 well plates at a density of 104 cells/well at 37 °C. Activity of lactate dehydrogenase (LDH) released from dead cells into the culture supernatants was measured at 450 nm wavelength with a microplate reader (Multiscan FC Microplate Reader, Fisher Scientific Inc., Pittsburgh, PA). The percentage cytotoxicity was calculated as [(test sample – cell negative control)/(lysis control – cell negative control)] × 100.
2.3.6. A2E treatment
Cells were grown to 95–100% confluency in 96-well plates, at 37 °C with 5% O2 and 5% CO2. A2E was purified according to method published earlier (Parish et al., 1998). Stock A2E solution (40 mM in DMSO) was diluted to a final concentration of 1–100 μM in prewarmed RPE maintenance medium and added to the plates, as specified. Manipulations involving A2E (and vehicle) addition, and all medium changes and feedings were conducted under dim (60–65 lx) light. For the repeated A2E feeding procedure, media containing A2E (10 μM) was replaced for 4 days and culture supernatant was collected.
2.3.7. H2O2 treatment
Cells were grown to 95–100% confluency in 96-well plates, at 37 °C with 5% O2 and 5% CO2. Cells were treated with 100 μM H2O2 for 4 days. Fresh H2O2 was added and culture supernatant was collected every 24 h.
2.3.8. RT2 profiler PCR Array
Quantitative mRNA expression analysis of inflammatory chemokines and cytokines was performed using Pathway-Focused gene expression profiling using Light cycler@96 Real-time PCR system (Roche). Total RNA was isolated using RNeasy kit (Qiagen), reverse transcribed using the cDNA synthesis kit (Qiagen) as previously described. cDNA from each experimental condition was used to run PCR array analysis according to the manufacturer’s protocol with the RT2 SYBR® master mix (ThermoFisher scientific). mRNA expression of each gene was normalized using the expression of multiple housekeeping genes. Average normalization was applied to the data and the expression signals were converted to log2 scale for analysis. Data analysis was performed using Excel macro provided with the kit and Gene Set Enrichment Analysis (GSEA) software (Subramanian et al., 2005).
2.4. Statistical analysis
The results were presented as mean ± SD. Statistical analyses were performed using the t-test for comparing 2 groups, and one-way ANOVA was used to detect differences among 3 or more groups. The results were considered statistically significant at P < 0.05.
3. Results
3.1. Generation and characterization of iPSC from healthy donor blood
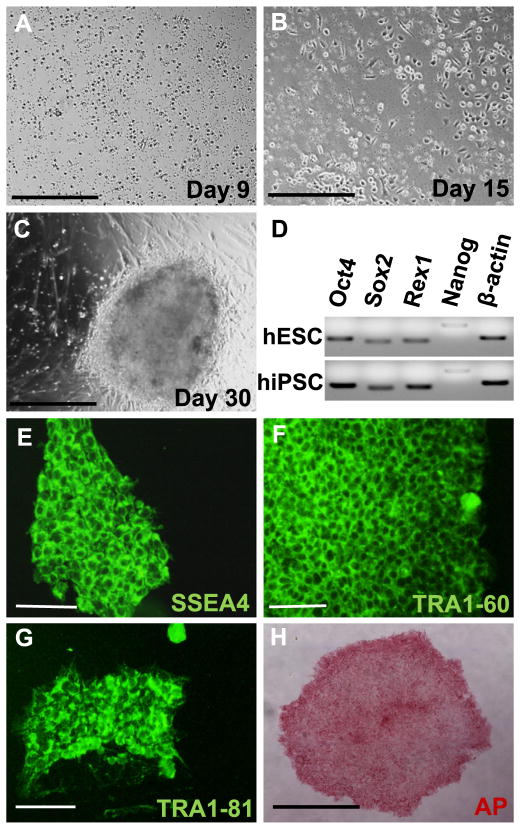
Peripheral blood mononuclear cells (PBMCs) isolated from healthy donors with no known history of retinal pathology were expanded for 9 days. On day 9, PBMCs were transduced with STEMCCA lentiviral vector encoding reprogramming factors Oct4, Klf4, Sox2 and c-Myc. Transduced PBMCs on day 15 showed change in morphology from round circular to more elongated shape. Around day 30, fully formed hiPSC-like colonies became visible (Fig. 1A–C). hiPSC-like colonies were picked and passaged manually for 10 times. With each passaging step, differentiated cells were excluded and only undifferentiated colonies were propagated. After passage 10, expression of pluripotency marker genes Oct4, Sox2, Rex1 and Nanog was detected (Fig. 1D). hiPSC-like colonies displayed positive immunostaining for stem cell markers including SSEA4, TRA1-60 and TRA1-81 (Fig. 1E–G). hiPSC colonies also stained positive for alkaline phosphatase (Fig. 1H). These results indicate that hiPSC lines generated in this study are pluripotent.
Fig. 1.
hiPSCs were generated from healthy donor blood. (A) PBMCs during expansion at day 9. (B) PBMCs after transduction with Lentiviral vector expressing Oct4, Klf4, Sox2 and c-Myc at day 15. (C) At around day 30, fully formed iPSC-like colonies appear. (D) Pluripotency specific gene expression for Oct4, Sox2, Rex1 and Nanog was checked by qRT-PCR. (E–H) Pluripotency is confirmed by immunocytochemistry with SSEA4, TRA-1-60, TRA1-81 (green) and alkaline phosphatase staining (red). Scale bar in A, B and H represents 500 μm and in E, F and G represents 100 μm.
3.2. Characterization of RPE cells differentiated from hiPSC
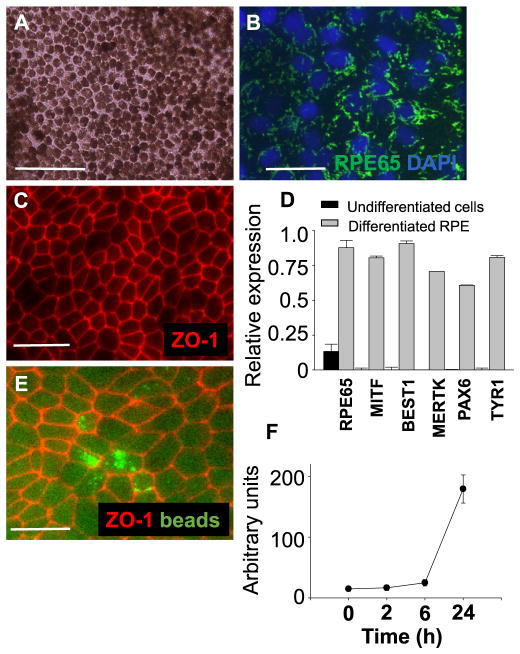
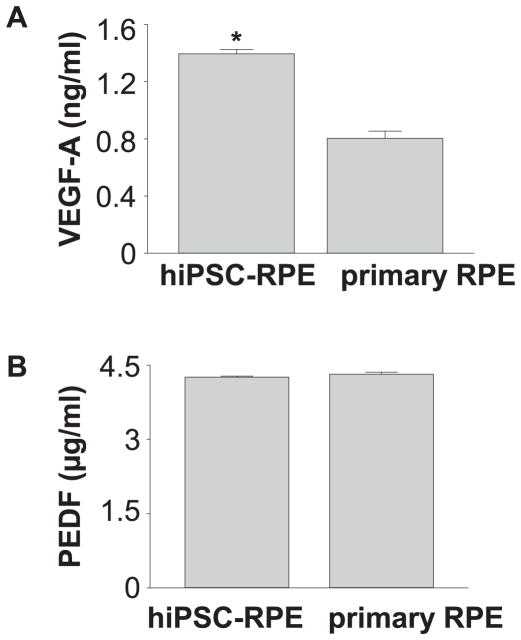
hiPSC lines from a healthy donor were used for RPE differentiation. Differentiated hiPSC-RPE cells displayed tightly packed pigmented and polygonal cells (Fig. 2A). RPE65, a crucial gene for RPE function was expressed abundantly in hiPSC-RPE cells (Fig. 2B). ZO-1 expression was detected by immunocytochemistry indicating that tight junctions are intact in the RPE monolayer (Fig. 2C). Expression of RPE-specific genes RPE65, MITF, BEST1, MERTK, PAX6, and TYR1 was detected by qRT-PCR (Fig. 2D). Expression of these markers was weakly or not detected in undifferentiated hiPSC lines. To confirm the functionality of hiPSC-RPE cells, their ability of phagocytosis and secretion of cytokines was examined. Phagocytosis ability of hiPSC-RPE cells was measured as a function of time by incubation with pHrodo® beads. Internalization of beads by hiPSC-RPE cells markedly increased at 24 h, indicating that hiPSC-RPE cells possess phagocytosis ability (Fig. 2E, F). Secretion of VEGF-A and PEDF from RPE cells was investigated by measuring amounts of these proteins in the culture supernatants when RPE cells were plated as a monolayer. Higher VEGF-A secretion in hiPSC-RPE cells than that of human primary RPE cells was documented and similar PEDF production was observed among hiPSC-RPE cells and human primary RPE cells (Fig. 3A, 3B). These data demonstrated successful differentiation of hiPSC lines into pigmented cells with typical RPE characteristics.
Fig. 2.
Differentiated RPE cells display RPE-specific features. (A) Morphology of RPE cells differentiated from hiPSC lines is shown by light microscopy. (B, C) RPE65 (green) and ZO-1 (red) immunocytochemistry shows localization in hiPSC-RPE cells. Nuclei were stained with DAPI (blue). (D) qRT-PCR gene expression analysis shows relative gene expression of RPE65, MITF, BEST1, MERTK, PAX6 and TYR1 normalized to GAPDH. (E) Phagocytosis ability of hiPSC-RPE cells is demonstrated by ingestion of beads (green) and immunostaining with ZO-1 (red). (F) Average fluorescence measured over time is shown. Bars indicate mean ± S.D. Scale bars represent 100 μm.
Fig. 3.
iPSC-RPE cells maintain RPE protein secretions. (A, B) Protein quantification by ELISA shows VEGF-A and PEDF levels in hiPSC-RPE and human RPE cells. Mean ± S.D. of individual wells is indicated.
3.3. A2E causes cell death in hiPSC-RPE cells
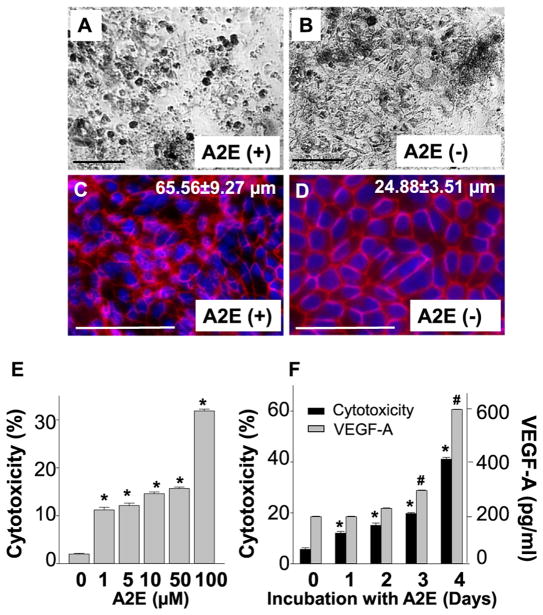
A2E accumulation in RPE cells is a hallmark of Stargardt disease and AMD (Sparrow and Boulton, 2005). To examine effects of A2E in RPE cells, hiPSC-RPE cells were co-incubated with 10 μM of A2E for 4 days, which can achieve a physiological concentration of A2E in human RPE cells (Sparrow et al., 1999). Fresh A2E was replaced in the culture medium every day for 4 days. Light microscopy revealed co-incubation with A2E increased the number of round-shaped cells as compared to the untreated cells (Fig. 4A, B). Upon immunostaining with a tight junction protein ZO-1, decreased numbers of regular polygonal shape of the RPE with enlarged cellular size (65.56 ± 9.27 μm vs 24.88 ± 3.51 μm, A2E-treated vs untreated) was observed, indicating that A2E led to disruption of tight junctions, loss of RPE monolayer integrity and decreased cell numbers as compared to untreated cells (Fig. 4C, D). Indeed, cell death assay by measuring lactate dehydrogenase (LDH) in the culture supernatants revealed a dose- and time-dependent increase in cell death when hiPSC-RPE cells were cultured with A2E (Fig. 4E, F, Supplemental Fig. 1). Increased IL6 and VEGF-A secretion was also observed in a dose- and time-dependent fashion (Fig. 4F, Supplemental Fig. 2).
Fig. 4.
A2E causes cell death and disruption of cell barrier integrity in iPSC-RPE cells. (A, B) Phase micrographs of A2E treated hiPSC-RPE cells compared to untreated cells. (C, D) Immunocytochemistry for ZO-1 (red) in A2E treated and untreated hiPSC-RPE cells. Cells are counterstained with DAPI (blue). Quantification of average cell diameters is presented here. (E) A2E induced cytotoxicity at different concentration was measured by LDH assay in A2E treated hiPSC-RPE cells after 24 h. (F) Cytotoxicity (in black) and VEGF-A (in grey) levels were measured by ELISA in hiPSC-RPE cells treated with A2E for four days. Bars indicate mean ± S.D. Scale bars indicate 100 μm.
3.4. A2E can cause inflammatory changes in hiPSC-RPE cells
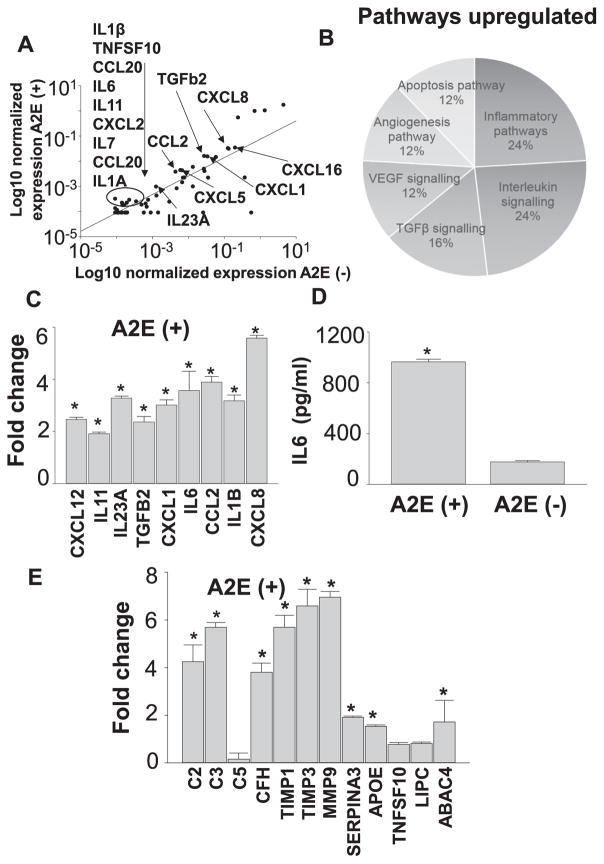
Previous studies have reported inflammatory changes caused by A2E (Radu et al., 2011, Anderson et al., 2013). Therefore, we examined if A2E can elicit inflammatory changes by employing RNA array profiling of 84 human chemokines and cytokines in hiPSC-RPE cells treated with 10 μM of A2E for 4 days. Chemokine and cytokine expression was compared between the treated and untreated cells. Applying a 2-fold cut-off, 26 cytokines were found upregulated as listed in Table 2. Scatter plot shows the changes of some of these upregulated genes (Fig. 5A). Genes found upregulated were uploaded in GSEA (Gene set enrichment analysis software) databa. Annotated gene sets contain genes classified according to their GO biological processes. Six major biological pathways were found upregulated besides inflammation including interleukin signaling, TGFβ signaling, VEGF signaling, Apoptosis and Angiogenesis pathways (Fig. 5B). Two-fold or more upregulation of CXCL12, IL23A, TGFB2, CXCL1, IL16, CCL2, IL1B and CXCL8 was validated by qRT-PCR (Fig. 5C). IL6 protein levels were found upregulated as measured by ELISA (Fig. 5D). These observations provide evidence that A2E can cause inflammatory changes which are observed in retinal degenerative diseases including AMD (Buschini et al., 2011, Ambati et al., 2013, Kauppinen et al., 2016). To investigate further if A2E can lead to any changes in the AMD-associated genes which were not included in the RNA array, qRT-PCR of C2, C3, C5, CFH, TIMP1, TIMP3, MMP9, SERPINA3, APOE, TNFSF10, LIPC and ABCA4 was performed with A2E-treated hiPSC-RPE cells. These genes have been reported as AMD-associated genes based on the genome-wide association studies (GWAS) with AMD subjects (Fritsche et al., 2013). Complement proteins C2, C3 and CFH were upregulated, while C5 was unchanged. Other genes TIMP1, TIMP3, MMP9, SERPINA3, APOE and ABCA4 were also upregulated, while TNFSF10 and LIPC were unchanged (Fig. 5E).
Table 2.
Upregulated genes (≥2.0 fold) after A2E treatment.
| Symbols | Gene names | Fold changesa |
|---|---|---|
| CX3CL1 | C-X-C Motif Chemokine Ligand 1 | 2.1 |
| BMP4 | Bone Morphogenetic Protein 4 | 2.2 |
| CXCL12 | C-X-C Motif Chemokine Ligand 12 | 2.2 |
| IL11 | Interleukin 11 | 2.4 |
| IL1A | Interleukin 1 Alpha | 2.5 |
| CNTF | Ciliary Neurotrophic Factor | 2.6 |
| IL23A | Interleukin 23 Subunit Alpha | 3.0 |
| TNFRSF11B | TNF Receptor Superfamily Member 11b | 3.1 |
| CXCL5 | C-X-C Motif Chemokine Ligand 5 | 3.1 |
| CXCL16 | C-X-C Motif Chemokine ligand 16 | 3.4 |
| GPI | Glucose-6-Phosphate Isomerase | 3.8 |
| LIF | Leukemia Inhibitory Factor | 3.9 |
| TGFB2 | Transforming Growth Factor Beta 2 | 4.2 |
| IL7 | Interleukin 7 | 4.5 |
| CXCL2 | C-X-C Motif Chemokine Ligand 2 | 4.6 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 5.4 |
| IL6 | Interleukin 6 | 5.6 |
| VEGFA | Vascular Endothelial Growth Factor A | 5.6 |
| CCL2 | C-C Motif Chemokine Ligand 2 | 5.7 |
| CCL20 | C-C Motif Chemokine Ligand 20 | 6.3 |
| SPP1 | Secreted Phosphoprotein 1 | 6.94 |
| IL1B | Interleukin 1 Beta | 9.8 |
| BMP2 | Bone Morphogenetic Protein 4 | 12.0 |
| CSF1 | Colony Stimulating Factor 1 | 13.5 |
| TNFSF10 | Tumor Necrosis Factor Superfamily Member 10 | 15.8 |
| CXCL8 | C-X-C Motif Chemokine Ligand 8 | 16.0 |
Fold changes were calculated against values from non-treated cells.
Fig. 5.
Inflammatory changes are induced by A2E in iPSC-RPE cells. (A) RNA array analyses show the most highly upregulated cytokines and chemokines. (B) Pathway analysis by GSEA shows the most enriched biological pathways. (C) qRT-PCR analyses show cytokines significantly upregulated upon treatment with A2E. Fold changes against values obtained with A2E-untreated cells are presented. (D) Quantification of IL6 protein was carried out by ELISA. (E) qRT-PCR of AMD-associated genes in A2E treated hiPSC-RPE cells. Fold changes against values obtained with A2E-untreated cells are presented. Data are presented as the means ± S.D., n = 6. *P < 0.05.
3.5. Oxidative stress with H2O2 causes significant upregulation of inflammatory cytokines in hiPSC-RPE cells
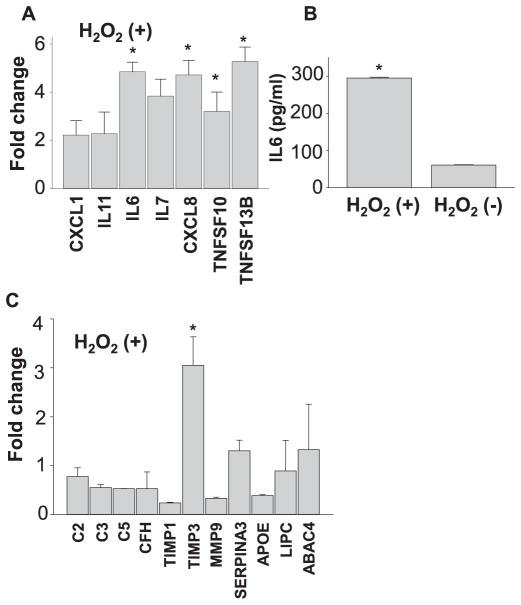
Photooxidation of A2E generates oxidative stress, which can be one of the major contributors to the pathogenesis of retinal diseases (Sparrow et al., 2003, Zhou et al., 2006, Chang et al., 2008, Anderson et al., 2013, Sparrow et al., 2002, Schutt et al., 2000, Rozanowska et al., 1995, Parish et al., 1998). To investigate if oxidative stress can recapitulate A2E-associated changes, hiPSC-RPE cells were exposed to 100 μM of H2O2 for 4 days. RNA array profiling of 84 human chemokines and cytokines using qRT-PCR was also carried out with treated and untreated cells. Applying a 2-fold cut-off, expression of 12 cytokines were found upregulated. qRT-PCR validation of CXCL1, IL11, IL6, IL7, CXCL8, TNFSF10 and TNFSF13B showed 2-fold or more upregulation (Fig. 6A and Table 3). Secreted levels of IL6 protein increased with H2O2 treatment as measured by ELISA (Fig. 6B). To investigate changes in AMD-associated genes with H2O2 treatment, qRT-PCR of C2, C3, C5, CFH, TIMP1, TIMP3, MMP9, SERPINA3, APOE, TNFSF10, LIPC and ABCA4 was performed. TIMP3 was upregulated 3-fold while no significant increase was observed in any of the other drusen/AMD-associated genes (Fig. 6C). Upon comparing the results from array analyses with A2E and H2O2 stressed hiPSC-RPE cells, 9 genes CXCL1, IL11, IL6, CXCL8, TNFSF10, IL7, TNFRSF11B and IL23A and CSF1 were found to be common while 3 genes were exclusive to H2O2 induced stress namely CSF3, CXCL11, and TNFSF13B (Fig. 7). Notably, A2E-treated hiPSC-RPE cells displayed similar inflammatory changes documented in AMD, whereas H2O2 treated hiPSC-RPE cells displayed minimal inflammatory changes.
Fig. 6.
Oxidative stress with H2O2 causes significant upregulation of inflammatory cytokines in iPSC-RPE cells. (A) Validation by qRT-PCR analyses of cytokines upregulated by oxidative stress. Fold changes against values obtained with H2O2-untreated cells are presented. (B) Quantification of IL6 protein was carried out by ELISA. (C) qRT-PCR of AMD-associated genes in H2O2 treated hiPSC-RPE cells. Fold changes against values obtained with H2O2-untreated cells are presented.
Table 3.
Upregulated genes (≥ 2.0 fold) after H2O2 treatment.
| Symbols | Gene names | Fold changesa |
|---|---|---|
| CSF1 | Colony Stimulating Factor 1 | 2.06 |
| CXCL11 | C-X-C Motif Chemokine Ligand 11 | 2.13 |
| CXCL1 | C-X-C motif chemokine ligand 1 | 2.50 |
| TNFSF10 | Tumor Necrosis Factor Superfamily Member 10 | 2.50 |
| IL23A | Interleukin 23 Subunit alpha | 2.98 |
| TNFSF13B | Tumor Necrosis Factor Superfamily Member 13B | 3.3 |
| IL7 | Interleukin 7 | 3.59 |
| IL11 | Interleukin 11 | 4.15 |
| CSF3 | Colony Stimulating Factor 1 | 4.18 |
| IL6 | Interleukin 6 | 4.45 |
| TNFRSF11B | Tumor Necrosis Factor Superfamily Member 11B | 5.68 |
| CXCL8 | C-X-C Motif Chemokine Ligand 8 | 7.9 |
Fold changes were calculated against values from non-treated cells
Fig. 7.
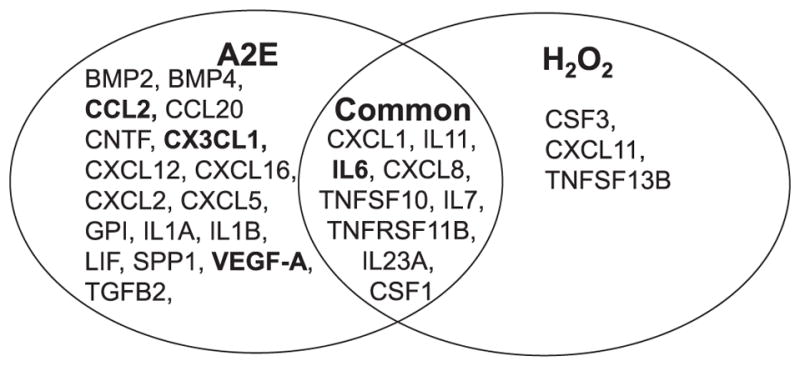
A2E treatment induces more inflammatory and AMD-related changes than H2O2 treated iPSC-RPE cells. Venn diagram representing the key up-regulated cytokines in A2E and H2O2 treated hiPSC-RPE cells. BOLD indicates AMD-related genes.
4. Discussion
Dysfunction or death of RPE cells underlies pathogenesis of retinal degenerative disorders including Stargardt disease and AMD. The complex functional alterations and phenotypes in diseased RPE cells are still not completely understood. hiPSCs have emerged as a powerful tool for human disease modeling to study pathophysiology as wells as for therapeutic development such as drug screening and cell-based transplantation therapy (Borooah et al., 2013, Hibaoui and Feki, 2012, Juopperi et al., 2011, Kim, 2015, Buchholz et al., 2009, Deleidi and Yu, 2016). RPE cells and other ocular cells are of particular interest because they are applicable for treating degenerative eye diseases (Mandai et al., 2017). We employed iPSC technology to differentiate hiPSCs from blood of healthy donors to RPE cells. hiPSCs derived from PBMCs were successfully differentiated into RPE cells. This successful RPE differentiation was confirmed by morphology, cytokine secretion, and ability of phagocytosis. First, hiPSC-RPE cells displayed a typical hexagonal/cobblestone morphology with pigmentation which is unique for human RPE cells. This unique feature of RPE cells can serve an important indicator for cell identity (Buchholz et al., 2009, Singh et al., 2013). One essential characteristic of RPE cells is the formation of tight junctions. Immunofluorescent staining for the tight junction protein, ZO-1, showed continuous junctions between hexagonal RPE cells. The tight junctions dynamically interact with numerous other proteins to regulate the paracellular permeability, the polar orientation of the membrane proteins, and protein expression (Cereijido et al., 2008, Zihni et al., 2016, Ablonczy et al., 2011). hiPSC-RPE cells in this study also successfully expressed RPE gene transcripts similar to human primary RPE cells.
Secretion properties which are characteristic for native RPE and hiPSC-RPE cells underlie the capacity of hiPSC-RPE cells to form functional, polarized monolayers (Brandl et al., 2014, Ablonczy et al., 2011, Singh et al., 2013). In accordance with the previous findings, we documented hiPSC-RPE cells to secrete high levels of VEGF-A in levels comparable to human primary RPE cells. VEGF-A is associated with formation of choroidal neovascularization and it is the principle cytokine responsible for neovascularization in the matured eye (Grossniklaus et al., 2010, Wang et al., 2016). hiPSC-RPE cells also secreted high levels of Pigment-epithelium-derived-factor (PEDF) comparable to human primary RPE cells. PEDF is known to be secreted in large quantities by the native RPE and is an important antagonist that limits the mitogenic activity of VEGF-A (Wang et al., 2016).
Phagocytosis is another key function of RPE cells (Strauss, 2005). Internalization of pHrodo® beads by immunofluorescence was observed in hiPSC-RPE cells verifying the phagocytotic properties of these cells. Collectively, all of these results suggest that the current methodologies for iPSC establishment and RPE differentiation can produce physiologically functional hiPSC-RPE cells in vitro.
hiPSC-RPE cells were then used to examine how A2E can affect these human cells. Previous research and clinical observation implicate noxious roles of A2E to RPE cells (Vives-Bauza et al., 2008, Perusek et al., 2015, Finnemann et al., 2002, Sparrow et al., 1999, Sparrow and Boulton, 2005); however, a mouse model with A2E accumulation only shows a mild phenotype (Wu et al., 2010, Weng et al., 1999). Difficulty in studying the formation and consequences of lipofuscin granules in RPE cell culture is compounded by the fact that these pigment granules do not normally occur in established RPE cell lines and pigment granules are rapidly lost in adult human primary culture (Boulton, 2014, Feeney, 1978, Hu and Bok, 2001). It is still not clear as to what cellular events are mediated by A2E, which ultimately lead to RPE damage and retinal degeneration in humans. Exposure to a physiological concentration of A2E (Sparrow et al., 1999) successfully mimicked distinct cellular disease phenotypes demonstrated by disrupted cell morphology, decreased epithelial integrity, increase in cell death and increased inflammation. A dose- and time-dependent A2E effect was also documented in hiPSC-RPE cells. Additionally, A2E treatment caused an increase in VEGF-A secretion, suggesting breakdown of barrier integrity. VEGF family of proteins are potent modulators of barrier function in both retinal endothelia and the RPE (Ablonczy and Crosson, 2007, Dahrouj et al., 2014). VEGF proteins can increase endothelial permeability and proliferation and are an important factor in retinal degeneration, particularly AMD pathogenesis (Gemenetzi and Patel, 2017, Amadio et al., 2016, Solomon et al., 2014). Treatments, including pegaptanib, ranibizumab, and bevacizumab, block the effects of VEGF-A and constitute the main pharmacological tools against neovascular AMD (or also called wet AMD) (Simunovic and Maberley, 2015, Iacono et al., 2010, Keane and Sadda, 2012). Few studies have used anti-VEGF therapies to reduce neovascularization in patients with Stargardt disease (Battaglia Parodi et al., 2015, Querques et al., 2010).
RNA array analyses of A2E-treated RPE cells revealed upregulation of 26 pro-inflammatory cytokines. IL-6 and IL-8 pro-inflammatory genotypes are reportedly associated to AMD pathobiology (Ricci et al., 2013, Seddon et al., 2005, Goverdhan et al., 2008). These interleukins are increased in the aqueous humor of patients with wet AMD and their concentrations have been correlated with retinal neovascular activity and the volume of macular edema (Smith et al., 2001, Anderson et al., 2013). Although progression from inflammation to pathogenesis of Stargardt disease has been experimentally suggested (Radu et al., 2011, Kohno et al., 2014) there is limited information available for changes in inflammatory genes in patients with Stargardt disease. Of note increased expression of several AMD-associated proteins by A2E was documented, implying the potential of hiPSC-RPE cells as a powerful in vitro model of AMD. Additionally, after identification that CFH Y402H variant is associated with AMD in 2005, complement regulatory protein is considered as a major risk factor for developing AMD (Klein et al., 2005, Edwards et al., 2005, Haines et al., 2005, Hageman et al., 2005, Zareparsi et al., 2005). This discovery has also suggested that inflammation can be an important contribution to AMD pathogenesis. Recent GWAS have also identified approximately 20 AMD-associated genetic variants, several of which have been localized to complement component genes, including CFH, CFI, CFB, and C3 (Tan et al., 2016). These studies have implicated that dysfunctional RPE cells lead to secretion of several proteins of the complement-associated pathway, including TIMP3, APOJ, annexin, crystallins, APOE, vitronectin, and amyloid β (Rabin et al., 2013, Johnson et al., 2011). In addition to AMD, the most recent study also revealed that complement modulation in RPE cells can reduce disease phenotypes in a mouse model of Stargardt disease, and accumulating information indicates complement roles in Stargardt disease as well as AMD (Lenis et al., 2017). Our current study provided evidence that A2E-treated hiPSC-RPE cells facilitate the secretion of these complement-associated proteins which are associated with AMD and Stargardt disease.
Oxidative stress could be an important factor in contributing to the development of retinal degeneration (Winkler et al., 1999, Harman, 1956, Beatty et al., 2000, Jomova et al., 2010). RPE cells are highly exposed to oxidative stress due to high rate of metabolism, exposure to scattered light, lipofuscin content, and hypoxia, all of which result in the generation of reactive oxygen species (ROS) (Juel et al., 2013, Kang et al., 2009, Reuter et al., 2010). Several studies have reported that lowered expression of anti-oxidative enzymes such as SOD1, catalase and heme oxygenase 1 in the RPE, correlating with age or with incident AMD (Gu et al., 2003, Kapphahn et al., 2006, Imamura et al., 2006). In the current study, hiPSC-RPE cells subjected to oxidative stress by H2O2 exposure showed an increase in inflammatory responses. RNA array analyses of H2O2-treated RPE cells identified an upregulation of 12 pro-inflammatory cytokines. Interestingly, only TIMP3 was found increased up to 3-fold while no significant increase was observed in any of the other AMD-associated genes, in contrast to that all of 11 genes except C5 were increased in A2E-treated hiPSC-RPE cells. Notably A2E treatment of hiPSC-RPE cells recapitulates the dynamic changes in disease progression better as compared to H2O2 treatment.
5. Conclusions
In summary, our results demonstrate the successful reprogramming of human blood cells to iPSCs which can be further differentiated to RPE cells with structural and functional capabilities characteristic for native RPE cells. Employment of these hiPSC-RPE cells provides clear evidence that A2E plays an important role in pathogenesis of retinal diseases by causing RPE cell death and activating immune systems.
Supplementary Material
Acknowledgments
Funding
This work was supported by grants from Research to Prevent Blindness (Catalyst Award and Unrestricted Grant) and the National Institutes of Health (R01EY022658 and P30EY011373). L.P. was supported by Prevent Blindness, Ohio Affiliate, young investigator student fellowship award for female scholars in vision research.
The authors thank Drs. Paul Tesar and Elizabeth Shick (iPS Core Facility, Department of Genetics, Case Western Reserve University) and Drs. Baseer Ahmad, Scott Howell, Tatiana Riedel and the Visual Sciences Research Center Core Facility (Department of Ophthalmology and Visual Sciences, Case Western Reserve University) for their comments and technical support.
Abbreviations
- A2E
N-retinyl-N-retinylidene ethanolamine
- AMD
age-related macular degeneration
- iPSC
induced pluripotent stem cells
- RPE
retinal pigmented epithelium
- KLF4
kruppel-like factor 4
- OCT4
octamer-binding transcription factor 4
- SOX2
sex determining region Y-box 2
- c-MYC
V-myc avian myelocytomatosis viral oncogene homolog
- CFH
Complement factor H
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- VEGF
vascular endothelial growth factor
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2018.01.014.
Footnotes
Disclosure of potential conflicts of interest
The authors of the manuscript have no potential conflicts of interest to disclose.
References
- Ablonczy Z, Crosson CE. VEGF modulation of retinal pigment epithelium resistance. Exp Eye Res. 2007;85:762–771. doi: 10.1016/j.exer.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablonczy Z, Dahrouj M, Tang PH, Liu Y, Sambamurti K, Marmorstein AD, Crosson CE. Human retinal pigment epithelium cells as functional models for the RPE in vivo. Invest Ophthalmol Vis Sci. 2011;52:8614–8620. doi: 10.1167/iovs.11-8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Wong JT, Molday RS. The effect of lipid environment and retinoids on the ATPase activity of ABCR, the photoreceptor ABC transporter responsible for Stargardt macular dystrophy. J Biol Chem. 2000;275:20399–20405. doi: 10.1074/jbc.M000555200. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997a;277:1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997b;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- Amadio M, Govoni S, Pascale A. Targeting VEGF in eye neovascularization: What’s new?: a comprehensive review on current therapies and oligonucleotide-based interventions under development. Pharmacol Res. 2016;103:253–269. doi: 10.1016/j.phrs.2015.11.027. [DOI] [PubMed] [Google Scholar]
- Ambati J, Atkinson JP, Gelfand BD. Immunology of age-related macular degeneration. Nat Rev Immunol. 2013;13:438–451. doi: 10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OA, Finkelstein A, Shima DT. A2E induces IL-1ss production in retinal pigment epithelial cells via the NLRP3 inflammasome. PLoS One. 2013;8:e67263. doi: 10.1371/journal.pone.0067263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia Parodi M, Munk MR, Iacono P, Bandello F. Ranibizumab for subfoveal choroidal neovascularisation associated with Stargardt disease. Br J Ophthalmol. 2015;99:1268–1270. doi: 10.1136/bjophthalmol-2014-305783. [DOI] [PubMed] [Google Scholar]
- Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- Borooah S, Phillips MJ, Bilican B, Wright AF, Wilmut I, Chandran S, Gamm D, Dhillon B. Using human induced pluripotent stem cells to treat retinal disease. Prog Retin Eye Res. 2013;37:163–181. doi: 10.1016/j.preteyeres.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton ME. Studying melanin and lipofuscin in RPE cell culture models. Exp Eye Res. 2014;126:61–67. doi: 10.1016/j.exer.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl C, Zimmermann SJ, Milenkovic VM, Rosendahl SM, Grassmann F, Milenkovic A, Hehr U, Federlin M, Wetzel CH, Helbig H, Weber BH. In-depth characterisation of Retinal Pigment Epithelium (RPE) cells derived from human induced pluripotent stem cells (hiPSC) NeuroMolecular Med. 2014;16:551–564. doi: 10.1007/s12017-014-8308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- Buschini E, Piras A, Nuzzi R, Vercelli A. Age related macular degeneration and drusen: neuroinflammation in the retina. Prog Neurobiol. 2011;95:14–25. doi: 10.1016/j.pneurobio.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Carr AJ, Vugler AA, Hikita ST, Lawrence JM, Gias C, Chen LL, Buchholz DE, Ahmado A, Semo M, Smart MJ, Hasan S, Da Cruz L, Johnson LV, Clegg DO, Coffey PJ. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4:e8152. doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereijido M, Contreras RG, Shoshani L, Flores-Benitez D, Larre I. Tight junction and polarity interaction in the transporting epithelial phenotype. Biochim Biophys Acta. 2008;1778:770–793. doi: 10.1016/j.bbamem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Chang JY, Bora PS, Bora NS. Prevention of oxidative stress-induced retinal pigment epithelial cell death by the PPARgamma agonists, 15-deoxy-delta 12, 14-prostaglandin J(2) PPAR Res. 2008;2008:720163. doi: 10.1155/2008/720163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Chang WC, Hung KH, Yang DM, Cheng YH, Liao YW, Woung LC, Tsai CY, Hsu CC, Lin TC, Liu JH, Chiou SH, Peng CH, Chen SJ. The generation of induced pluripotent stem cells for macular degeneration as a drug screening platform: identification of curcumin as a protective agent for retinal pigment epithelial cells against oxidative stress. Front Aging Neurosci. 2014;6:191. doi: 10.3389/fnagi.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Okano K, Maeda T, Chauhan V, Golczak M, Maeda A, Palczewski K. Mechanism of all-trans-retinal toxicity with implications for stargardt disease and age-related macular degeneration. J Biol Chem. 2012;287:5059–5069. doi: 10.1074/jbc.M111.315432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahrouj M, Alsarraf O, Mcmillin JC, Liu Y, Crosson CE, Ablonczy Z. Vascular endothelial growth factor modulates the function of the retinal pigment epithelium in vivo. Invest Ophthalmol Vis Sci. 2014;55:2269–2275. doi: 10.1167/iovs.13-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleidi M, Yu C. Genome editing in pluripotent stem cells: research and therapeutic applications. Biochem Biophys Res Commun. 2016;473:665–674. doi: 10.1016/j.bbrc.2016.02.113. [DOI] [PubMed] [Google Scholar]
- Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Eldred GE, Lasky MR. Retinal age pigments generated by self-assembling lysosomotropic detergents. Nature. 1993;361:724–726. doi: 10.1038/361724a0. [DOI] [PubMed] [Google Scholar]
- Feeney L. Lipofuscin and melanin of human retinal pigment epithelium. Fluorescence, enzyme cytochemical, and ultrastructural studies. Invest Ophthalmol Vis Sci. 1978;17:583–600. [PubMed] [Google Scholar]
- Finnemann SC, Leung LW, Rodriguez-Boulan E. The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2002;99:3842–3847. doi: 10.1073/pnas.052025899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Jr, Buitendijk GH, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht AM, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan CC, Cheng CY, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdottir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JP, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis NCNK, Khan JC, Kim IK, Kiyohara Y, Klein BE, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Said S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJ, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(433–9):439e1–2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemenetzi M, Patel PJ. A systematic review of the treat and extend treatment regimen with anti-VEGF agents for neovascular age-related macular degeneration. Ophthalmol Ther. 2017;6:79–92. doi: 10.1007/s40123-017-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczak M, Kiser PD, Lodowski DT, maeda A, Palczewski K. Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J Biol Chem. 2010;285:9667–9682. doi: 10.1074/jbc.M109.063941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverdhan SV, Ennis S, Hannan SR, Madhusudhana KC, Cree AJ, Luff AJ, Lotery AJ. Interleukin-8 promoter polymorphism-251A/T is a risk factor for age-related macular degeneration. Br J Ophthalmol. 2008;92:537–540. doi: 10.1136/bjo.2007.123190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus HE, Kang SJ, Berglin L. Animal models of choroidal and retinal neovascularization. Prog Retin Eye Res. 2010;29:500–519. doi: 10.1016/j.preteyeres.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Meer SG, Miyagi M, Rayborn ME, Hollyfield JG, Crabb JW, Salomon RG. Carboxyethylpyrrole protein adducts and autoantibodies, biomarkers for age-related macular degeneration. J Biol Chem. 2003;278:42027–42035. doi: 10.1074/jbc.M305460200. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, NOUREDDINE M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hibaoui Y, Feki A. Human pluripotent stem cells: applications and challenges in neurological diseases. Front Physiol. 2012;3:267. doi: 10.3389/fphys.2012.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Bok D. A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol Vis. 2001;7:14–19. [PubMed] [Google Scholar]
- Iacono P, Battaglia Parodi M, Bandello F. Antivascular endothelial growth factor in diabetic retinopathy. Dev Ophthalmol. 2010;46:39–53. doi: 10.1159/000320008. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Noda S, Hashizume K, Shinoda K, Yamaguchi M, Uchiyama S, Shimizu T, Mizushima Y, Shirasawa T, Tsubota K. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: a model of age-related macular degeneration. Proc Natl Acad Sci U S A. 2006;103:11282–11287. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriyama A, Fujiki R, Inoue Y, Takahashi H, Tamaki Y, Takezawa S, Takeyama K, Jang WD, Kato S, Yanagi Y. A2E, a pigment of the lipofuscin of retinal pigment epithelial cells, is an endogenous ligand for retinoic acid receptor. J Biol Chem. 2008;283:11947–11953. doi: 10.1074/jbc.M708989200. [DOI] [PubMed] [Google Scholar]
- Jarrett SG, Boulton ME. Consequences of oxidative stress in age-related macular degeneration. Mol Asp Med. 2012;33:399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LV, Forest DL, Banna CD, Radeke CM, Maloney MA, Hu J, Spencer CN, Walker AM, Tsie MS, Bok D, Radeke MJ, Anderson DH. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2011;108:18277–18282. doi: 10.1073/pnas.1109703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomova K, Vondrakova D, Lawson M, Valko M. Metals, oxidative stress and neurodegenerative disorders. Mol Cell Biochem. 2010;345:91–104. doi: 10.1007/s11010-010-0563-x. [DOI] [PubMed] [Google Scholar]
- Juel HB, Faber C, Svendsen SG, Vallejo AN, Nissen MH. Inflammatory cytokines protect retinal pigment epithelial cells from oxidative stress-induced death. PLoS One. 2013;8:e64619. doi: 10.1371/journal.pone.0064619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juopperi TA, Song H, Ming GL. Modeling neurological diseases using patient-derived induced pluripotent stem cells. Future Neurol. 2011;6:363–373. doi: 10.2217/FNL.11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang KH, Lemke G, Kim JW. The PI3K-PTEN tug-of-war, oxidative stress and retinal degeneration. Trends Mol Med. 2009;15:191–198. doi: 10.1016/j.molmed.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapphahn RJ, Giwa BM, Berg KM, Roehrich H, Feng X, Olsen TW, Ferrington DA. Retinal proteins modified by 4-hydroxynonenal: identification of molecular targets. Exp Eye Res. 2006;83:165–175. doi: 10.1016/j.exer.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Kauppinen A, Paterno JJ, Blasiak J, Salminen A, Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cell Mol Life Sci. 2016;73:1765–1786. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane PA, Sadda SR. Development of anti-VEGF therapies for intraocular use: a guide for clinicians. J Ophthalmol. 2012;2012:483034. doi: 10.1155/2012/483034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. iPSC technology—powerful hand for disease modeling and therapeutic screen. BMB Rep. 2015;48:256–265. doi: 10.5483/BMBRep.2015.48.5.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Palczewski K. Chemistry of the retinoid (visual) cycle. Chem Rev. 2014;114:194–232. doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, CHew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, Sangiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Maeda T, Perusek L, Pearlman E, Maeda A. CCL3 production by microglial cells modulates disease severity in murine models of retinal degeneration. J Immunol. 2014;192:3816–3827. doi: 10.4049/jimmunol.1301738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenis TL, Sarfare S, Jiang Z, Lloyd MB, Bok D, Radu RA. Complement modulation in the retinal pigment epithelium rescues photoreceptor degeneration in a mouse model of Stargardt disease. Proc Natl Acad Sci U S A. 2017;114:3987–3992. doi: 10.1073/pnas.1620299114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Itagaki Y, Ben-Shabat S, Nakanishi K, Sparrow JR. The biosynthesis of A2E, a fluorophore of aging retina, involves the formation of the precursor, A2-PE, in the photoreceptor outer segment membrane. J Biol Chem. 2000;275:29354–29360. doi: 10.1074/jbc.M910191199. [DOI] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Golczak M, Chou S, Desai A, Hoppel CL, Matsuyama S, Palczewski K. Involvement of all-trans-retinal in acute light-induced retinopathy of mice. J Biol Chem. 2009;284:15173–15183. doi: 10.1074/jbc.M900322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, Shibata Y, Terada M, Nomiya Y, Tanishima S, Nakamura M, Kamao H, Sugita S, Onishi A, Ito T, Fujita K, Kawamata S, Go MJ, Shinohara C, Hata KI, Sawada M, Yamamoto M, Ohta S, Ohara Y, Yoshida K, Kuwahara J, Kitano Y, Amano N, Umekage M, Kitaoka F, Tanaka A, Okada C, Takasu N, Ogawa S, Yamanaka S, Takahashi M. Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- Moiseyev G, Nikolaeva O, Chen Y, Farjo K, Takahashi Y, Ma JX. Inhibition of the visual cycle by A2E through direct interaction with RPE65 and implications in Stargardt disease. Proc Natl Acad Sci U S A. 2010;107:17551–17556. doi: 10.1073/pnas.1008769107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nita M, Grzybowski A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxidative Med Cell Longev. 2016;2016:3164734. doi: 10.1155/2016/3164734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Ikeda H, Sasai Y, Takahashi M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nat Protoc. 2009a;4:811–824. doi: 10.1038/nprot.2009.51. [DOI] [PubMed] [Google Scholar]
- Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y, Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009b;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CA, Hashimoto M, Nakanishi K, Dillon J, Sparrow J. Isolation and onestep preparation of A2E and iso-A2E, fluorophores from human retinal pigment epithelium. Proc Natl Acad Sci U S A. 1998;95:14609–14613. doi: 10.1073/pnas.95.25.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar T, Parmar VM, Arai E, Sahu B, Perusek L, Maeda A. Acute stress responses are early molecular events of retinal degeneration in Abca4−/−Rdh8−/−mice after light exposure. Invest Ophthalmol Vis Sci. 2016;57:3257–3267. doi: 10.1167/iovs.15-18993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perusek L, Sahu B, Parmar T, Maeno H, Arai E, Le YZ, Subauste CS, Chen Y, Palczewski K, Maeda A. Di-retinoid-pyridiniumethanolamine (A2E) accumulation and the maintenance of the visual cycle are independent of Atg7-mediated autophagy in the retinal pigmented epithelium. J Biol Chem. 2015;290:29035–29044. doi: 10.1074/jbc.M115.682310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querques G, Bux AV, Prascina F, Noci ND. Intravitreal avastin for choroidal neovascularization associated with stargardt-like retinal abnormalities in pseudoxanthoma elasticum. Middle East Afr J Ophthalmol. 2010;17:387–389. doi: 10.4103/0974-9233.71586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin DM, Rabin RL, Blenkinsop TA, Temple S, Stern JH. Chronic oxidative stress upregulates Drusen-related protein expression in adult human RPE stem cell-derived RPE cells: a novel culture model for dry AMD. Aging (Albany NY) 2013;5:51–66. doi: 10.18632/aging.100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu RA, Hu J, Yuan Q, Welch DL, Makshanoff J, Lloyd M, Mcmullen S, Travis GH, Bok D. Complement system dysregulation and inflammation in the retinal pigment epithelium of a mouse model for Stargardt macular degeneration. J Biol Chem. 2011;286:18593–18601. doi: 10.1074/jbc.M110.191866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci F, Staurenghi G, Lepre T, Missiroli F, Zampatti S, Cascella R, Borgiani P, Marsella LT, Eandi CM, Cusumano A, Novelli G, Giardina E. Haplotypes in IL-8 gene are associated to age-related macular degeneration: a case-control study. PLoS One. 2013;8:e66978. doi: 10.1371/journal.pone.0066978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanowska M, Jarvis-Evans J, Korytowski W, Boulton ME, Burke JM, Sarna T. Blue light-induced reactivity of retinal age pigment. In vitro generation of oxygen-reactive species. J Biol Chem. 1995;270:18825–18830. doi: 10.1074/jbc.270.32.18825. [DOI] [PubMed] [Google Scholar]
- Sahu B, Sun W, Perusek L, Parmar V, Le YZ, Griswold MD, Palczewski K, Maeda A. Conditional ablation of retinol dehydrogenase 10 in the retinal pigmented epithelium causes delayed dark adaption in mice. J Biol Chem. 2015;290:27239–27247. doi: 10.1074/jbc.M115.682096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samiec PS, Drews-Botsch C, Flagg EW, Kurtz JC, Sternberg P, Jr, Reed RL, Jones DP. Glutathione in human plasma: decline in association with aging, age-related macular degeneration, and diabetes. Free Radic Biol Med. 1998;24:699–704. doi: 10.1016/s0891-5849(97)00286-4. [DOI] [PubMed] [Google Scholar]
- Schutt F, Davies S, Kopitz J, Holz FG, Boulton ME. Photodamage to human RPE cells by A2-E, a retinoid component of lipofuscin. Invest Ophthalmol Vis Sci. 2000;41:2303–2308. [PubMed] [Google Scholar]
- Schwartz SD, Hubschman JP, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123:774–782. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- Simunovic MP, Maberley DA. Anti-vascular endothelial growth factor therapy for proliferative diabetic retinopathy: a systematic review and meta-analysis. Retina. 2015;35:1931–1942. doi: 10.1097/IAE.0000000000000723. [DOI] [PubMed] [Google Scholar]
- Singh R, Phillips MJ, Kuai D, MEYER J, Martin JM, Smith MA, Perez ET, Shen W, Wallace KA, Capowski EE, Wright LS, Gamm DM. Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest Ophthalmol Vis Sci. 2013;54:6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, Hofman A, Jensen S, Wang JJ, De Jong PT. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Solomon SD, Lindsley K, Vedula SS, Krzystolik MG, Hawkins BS. Anti-vascular endothelial growth factor for neovascular age-related macular degeneration. Cochrane Database Syst Rev. 2014:CD005139. doi: 10.1002/14651858.CD005139.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer AG, Rozelle SS, Sullivan S, Mills JA, Park SM, Smith BW, Iyer AM, French DL, Kotton DN, Gadue P, Murphy GJ, Mostoslavsky G. Generation of human induced pluripotent stem cells from peripheral blood using the STEMCCA lentiviral vector. J Vis Exp. 2012;68:4327. doi: 10.3791/4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp Eye Res. 2005;80:595–606. doi: 10.1016/j.exer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Sparrow JR, Parish CA, Hashimoto M, Nakanishi K. A2E, a lipofuscin fluorophore, in human retinal pigmented epithelial cells in culture. Invest Ophthalmol Vis Sci. 1999;40:2988–2995. [PubMed] [Google Scholar]
- Sparrow JR, Zhou J, Ben-Shabat S, Vollmer H, Itagaki Y, Nakanishi K. Involvement of oxidative mechanisms in blue-light-induced damage to A2E-laden RPE. Invest Ophthalmol Vis Sci. 2002;43:1222–1227. [PubMed] [Google Scholar]
- Sparrow JR, Vollmer-Snarr HR, Zhou J, Jang YP, Jockusch S, Itagaki Y, Nakanishi K. A2E-epoxides damage DNA in retinal pigment epithelial cells. Vitamin E and other antioxidants inhibit A2E-epoxide formation. J Biol Chem. 2003;278:18207–18213. doi: 10.1074/jbc.M300457200. [DOI] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Molday RS, Nathans J. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J Biol Chem. 1999;274:8269–8281. doi: 10.1074/jbc.274.12.8269. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tan PL, Bowes Rickman C, Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics. 2016;10:23. doi: 10.1186/s40246-016-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis GH, Golczak M, Moise AR, Palczewski K. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu Rev Pharmacol Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Anand M, Shiraz AK, Magrane J, Gao J, Vollmer-Snarr HR, Manfredi G, Finnemann SC. The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J Biol Chem. 2008;283:24770–24780. doi: 10.1074/jbc.M800706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Han X, Wittchen ES, Hartnett ME. TNF-alpha mediates choroidal neovascularization by upregulating VEGF expression in RPE through ROS-dependent beta-catenin activation. Mol Vis. 2016;22:116–128. [PMC free article] [PubMed] [Google Scholar]
- Weng J, Mata NL, Azarian SM, Tzekov RT, Birch DG, Travis GH. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in Abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Winkler BS, Boulton ME, Gottsch JD, Sternberg P. Oxidative damage and age-related macular degeneration. Mol Vis. 1999;5:32. [PMC free article] [PubMed] [Google Scholar]
- Wu L, Nagasaki T, Sparrow JR. Photoreceptor cell degeneration in Abcr (−/−) mice. Adv Exp Med Biol. 2010;664:533–539. doi: 10.1007/978-1-4419-1399-9_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Swingen C, Zhang J. Induced pluripotent stem cells and their potential for basic and clinical sciences. Curr Cardiol Rev. 2013;9:63–72. doi: 10.2174/157340313805076278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, Ott J, Hoh J, Abecasis GR, Swaroop A. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci U S A. 2006;103:16182–16187. doi: 10.1073/pnas.0604255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.