Abstract
Avian adenovirus has been reported in many countries and is an infectious agent related with inclusion body hepatitis, hepatitis-hydropericardium syndrome (HHS), and respiratory and enteric conditions in chickens worldwide. The objective of this study was to detect and establish the molecular sequences of the hexon gene from the avian adenovirus strains of group I (FAdV-I) isolated from birds with hepatitis-hydropericardium syndrome (HHS), malabsorption syndrome and runting-stunting syndrome, to characterize the serotype of virus affecting commercial flocks in Brazil. Molecular characterization was performed by polymerase chain reaction (PCR), using specific primers to amplify the Loop 1 (L1) variable region of the hexon gene in the FAdV-I genome and subsequent sequencing of the PCR product for each positive sample. The results have revealed the presence of the FAdV-8a, FAdV-8b, and FAdV-11 serotypes circulating in Brazilian chicken flocks. Phylogenetic analysis grouped these sequences into three (3) distinct groups, 14 samples were aligned with the FAdV-11 group, three (3) samples in the FAdV-8b group and one (1) sample in the FAdV-8a group. The serotypes FAdV-8a, FAdV-8b, and FAdV-11 are circulating in Brazilian chicken flocks. Therefore, these results are very important for improvement biosecurity measurements and vaccine production.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0430-z) contains supplementary material, which is available to authorized users.
Keywords: Avian adenovirus, IBH, HHS, RSS, Serotypes, PCR
Introduction
Avian adenovirus group I (FAdV-I) mainly infects birds and is classified within the genus Aviadenovirus in the family Adenoviridae [10]. Avian adenovirus is composed by 3 groups (I–III) [11], and was characterizer into 5 species (A–E) and 12 serotypes (FAdV-1, FAdV-2, FAdV-3, FAdV-4, FAdV-5, FAdV-6, FAdV-7, FAdV-8a, FAdV-8b, FAdV-9, FAdV-10, FAdV-11) [12]. The viral particles are non-enveloped and icosahedral in geometry. They are 70–90 nm in diameter, with a linear double-stranded DNA genome between 35 and 45 kb in size [13]. The capsid has 252 capsomeres, of which 240 are formed by the hexon protein and 12 by the penton protein, which forms the vertices from which fibres that have antigenic properties become detached [12].
Inclusion body hepatitis (IBH) associated with FAdV-I occurs worldwide [1]. It is an acute disease that mainly affects young chickens between 3 and 7 weeks of age [29], and is caused by several avian adenovirus serotypes [12]. FAdV-I can be transmitted vertically and horizontally through progeny and faeces, respectively [1]. There are studies demonstrating infections caused by this virus in Brazil, which is associated with enteric diseases, liver problems, and hydropericardium [18]. Furthermore, there are reports of adenovirus infections resulting in increased feed conversion rate and increased mortality [11, 27, 30]. Therefore, the effects of avian adenovirus infections are of great economic importance in poultry farming [20] and associated with enteric virus, such as ANV [8]. In addition to the liver, the virus can be found in the upper respiratory tract, digestive tract, pancreas, kidneys, spleen and heart [23]. Studies with experimentally inoculated birds have demonstrated that avian adenovirus infection can result in weight loss, apathy, and mortality [2].
The goal of this work was to detect and establish the molecular characterization of FAdV-1 obtained from broilers with HHS, MAS, RSS, in Brazil.
Materials and methods
Virus samples
The samples were stored during the period of 2011–2015, where 18 strains identified as avian adenovirus group I obtained from chickens at eight farms in different Brazilian states were identified by PCR. The birds presented clinical signs related to enteric problems and low zootechnical performance. After necropsy, all birds presented hydropericardium. Viral strains were identified from the following organs: liver, bursa, heart, intestine, pancreas, trachea, and kidneys [17], as shown in Table 1.
Table 1.
Description of samples obtained from chickens with poor development and malabsorption syndrome in different poultry companies
| Sample designation | Bird and origin of samples | Main case history or necropsy findings | Serotype identified based on molecular characterization | GenBank accession number |
|---|---|---|---|---|
| USP-BR-418.1 | Broiler—Liver | Hydropericardium—hepatitis | FAdV-8b | KY229168 |
| USP-BR-418.14 | Broiler—Bursa | Hydropericardium—hepatitis | FAdV-11 | KY229169 |
| USP-BR-420.12 | Broiler—Heart | Malabsorption—undigested feed | FAdV-8b | KY229185 |
| USP-BR-420.16 | Broiler—bursa | Malabsorption—undigested feed | FAdV-8b | KY229170 |
| USP-BR-420.17 | Broiler—intestine | Malabsorption—undigested feed | FAdV-11 | KY229171 |
| USP-BR-420.18 | Broiler—pancreas | Malabsorption—undigested feed | FAdV-11 | KY229172 |
| USP-BR-420.26 | Broiler—bursa | Malabsorption—undigested feed | FAdV-11 | KY229173 |
| USP-BR-420.27 | Broiler—intestine | Malabsorption—undigested feed | FAdV-11 | KY229174 |
| USP-BR-420.28 | Broiler—pancreas | Malabsorption—undigested feed | FAdV-11 | KY229175 |
| USP-BR-424.4 | Broiler—pancreas- | Malabsorption—undigested feed | FAdV-11 | KY229176 |
| USP-BR-453.2 | Broiler—trachea | NI | FAdV-8a | KY229177 |
| USP-BR-471.14 | Broiler—trachea | NI | FAdV-11 | KY229184 |
| USP-BR-475.1 | Broiler—kidney | Runting-stunting syndrome | FAdV-11 | KY229178 |
| USP-BR-475.3 | Broiler—pancreas | Runting-stunting syndrome | FAdV-11 | KY229179 |
| USP-BR-475.4 | Broiler—pancreas | Runting-stunting syndrome | FAdV-11 | KY229180 |
| USP-BR-475.11 | Broiler—kidney | Runting-stunting syndrome | FAdV-11 | KY229182 |
| USP-BR-102.8D | NI—liver | NI | FAdV-11 | KY229181 |
| USP-BR-G21.B | NI—liver | NI | FAdV-11 | KY229183 |
FAdV fowl adenovirus group I; USP University of São Paulo; BR Brazil; NI not informed
DNA extraction
For isolation of DNA, macerated tissue of each organ and 0.1 M PBS (Phosphate Buffered Saline), pH 7.4 was added to 1.5 mL tubes at a 1:1 ratio. The mixture was vortexed for 10 s and subjected to three freeze–thaw cycles. Extraction was performed using the phenol/chloroform method, as described previously [3].
Polymerase chain reaction (PCR) for detection of FAdV-I
The PCR reaction was carried out as described with modifications [19]. The reaction mix contained 0.5 μmol each of the sense and antisense primers, 3.5 μL of 10X buffer, 4 μL of 1.25 mM dNTP, 0.75 μL of 50 mM MgCl2, 1 U of DNA polymerase (Taq Platinum, Invitrogen™), 2.5 μL of extracted DNA and ultrapure water to bring the total volume to 25 μL. The amplification reaction occurred under the following conditions: a thermal cycle of 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 60 s, annealing at 52 °C for 45 s, and extension at 72 °C for 60 s. The final extension cycle was 72 °C for 10 min. The reaction was stored at -20 °C. The primers used in the PCR reaction correspond to the conserved segments of the Pedestal 1 (P1) regions adjacent to the Loop 1 (L1) variable segment of the hexon gene (hexon A: CAARTTCAGRCAGACGGT and hexon B: TAGTGATGMCGSGACATCAT). This resulted in amplification of an 897 bp segment [19].
Purification and sequencing
The product amplified by the FAdV-I hexon gene PCR was purified using the GPX™ PCR DNA and Gel Band Purification kit (GE Healthcare, Piscataway, New Jersey, USA) according to the manufacturer’s instructions. Each purified product was sequenced in sense and antisense directions using the BigDye® Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems by Life Technologies, Carlsbad, California, USA). Sequencing reactions were performed on an ABI 3730 DNA Analyser (Applied Biosystems by Life Technologies).
Phylogenetic analysis
The nucleotide sequences were edited using the CLC Main Work Bench 7.7.1 software and aligned with reference sequences representative of the FAdV-I serotypes [10] present in GenBank with the following accession numbers: FAdV-1 Strain CELO (AC_000014) from Germany; FAdV-2 strain P7-A (AF339915) and FAdV-3 strain 75 (AF508949), both from Belgium; FAdV-4 strain ON1 (NC_015323) from Canada; FAdV-5 strain 340 (AF508952), FAdV-6 strain CR119 (AF508954) and FAdV-7 strain YR36 (AF508955), all from Belgium; FAdV-8a strain TR59 (KT862810) from Japan; FAdV-8b strain 764 (KT862811) from the United Kingdom; FAdV-9 strain A-2A (AF083975.2) from Canada; FAdV-10 strain C-2B (KT717889) from the USA and FAdV-11 strain 380 (AF339925) from Belgium. Alignment was performed using the CLUSTAL W method available in Clustal X 2.0 software. Phylogenetic analysis of nucleotides was inferred using the maximum likelihood model [30] integrated into MEGA software 7.0.18. The nucleotide and amino acid sequence similarity matrix was generated in BioEdit Sequence Alignment Editor v. 7.2.5.
Results and discussion
Sequencing analysis and phylogenetic tree
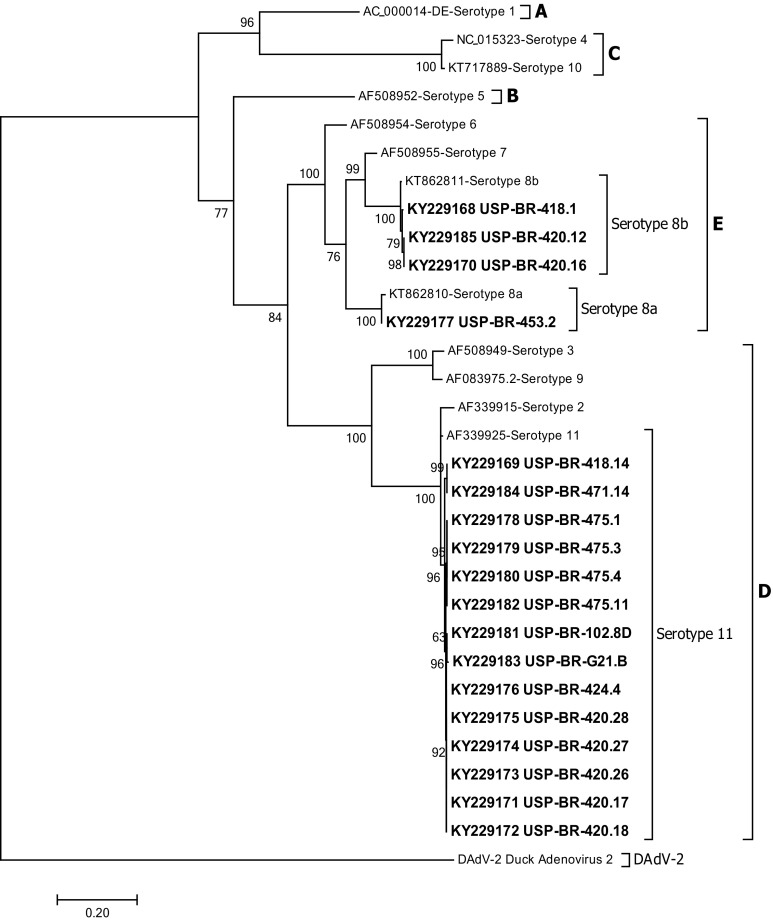
Avian adenovirus has been reported in many countries and is a virus associated with diseases such as inclusion body hepatitis (IBH), hydropericardium syndrome (HS), and respiratory and enteric conditions [2, 4, 6, 14, 20, 24]. FAdV-I has also been described in South American countries like Ecuador, Chile and Peru [16, 26, 31], including Brazil [22]. In this study, FAdV-I was detected in its target tissues where replication occurs [7, 29], based on PCR amplification of the 897 bp fragment of the hexon gene [19]. Sequencing and alignment of sequences in GenBank showed that 14/18 viral strains (77.7%) corresponded to the FAdV-11 serotype, 3/18 strains (16.6%) corresponded to the FAdV-8b serotype, and 1/18 strains (5.5%) corresponded to the FADV-8a serotype (Table 2 in ESM). The sequences were deposited in GenBank under the accession numbers described below: USP-BR-418.1 (KY229168), USP-BR-418.14 (KY229169), USP-BR-420.12 (KY229185), USP-BR-420.16 (KY229170), USP-BR-420.17 (KY229171), USP-BR-420.18 (KY229172), USP-BR-420.26 (KY229173), USP-BR-420.27 (KY229174), USP-BR-420.28 (KY229175), USP-BR-424.4 (KY229176), USP-BR-453.2 (KY229177), USP-BR-471.14 (KY229184), USP-BR-475.1 (KY229178), USP-BR-475.3 (KY229179), USP-BR-475.4 (KY229180), USP-BR-475.11 (KY229182), USP-BR-102.8D (KY229181), and USP-BR-G21.B (KY229183). The phylogenetic tree was inferred using phylogenetic reconstruction analysis with the Maximum Likelihood statistical model with 1000 replicates (Bootstrap), the Tamura-Nei substitution model and a Gamma distribution with Invariant sites (TN93 + G + I). This model was suggested by previous analysis with the Maximum Likelihood statistical method using MEGA 7 Software. Phylogenetic analysis revealed that of the 18 viral strains, fourteen (14/18) belonged to serotype 11 of the D species, three (3/18) belonged to serotype 8b and one (1/18) to serotype 8a, both of species E (Fig. 1). In addition, phylogenetic analysis was carried out based on the nucleotide sequence and its resulting amino acid sequence. Sequencing of this fragment and its subsequent analysis allowed characterization of different serotypes among the samples. The results obtained in this study corroborate those found by other authors and highlight the importance of this method for the detection and characterization of FAdV-I serotypes [19–21]. Although infections caused by FAdV-I are commonly related to primary immunosuppressive infections such as Infectious Chicken Anemia (CAV) or Infection Bursa Disease (IBD) [5], there are also reports of infection by FAdV-I without immunological changes [4].
Fig. 1.
Phylogenetic relationships of Brazilian FAdV-I sequences from chickens presented enteric problems, impairment, malabsorption diseases, hydropericardium and hepatitis in comparison to isolates from different countries. The tree was inferred based on alignments of partial hexon gene segment L1 and adjacent P1, using the Maximum Likelihood statistical method included in the MEGA 5 software. Numbers along the branches refer to bootstrap value for 1000 replicates. The scale bar represents the number of substitutions per site. Highlighted names represent the sequences used in this study, and letters A to E represent fowl adenovirus groups. DAdV-2 Duck adenovirus 2 sequence was employed as the out-group
Similarity matrix
The similarity matrix of the USP-BR-418.1 sample with serotype 8b showed nucleotide (n) and amino acid (aa) similarities of 99.3 and 99.6% respectively. The USP-BR-418.14 sample had 97.5% (n) and 96.8% (aa) similarity to serotype 11. Both samples USP-BR-420.12 and USP-BR-420.16 had 99.0% (n) and 99.6% (aa) similarity to serotype 8b. The sequences USP-BR-420.17, USP-BR-420.18, USP-BR-420.26, USP-BR-420.27, USP-BR-420.28 and USP-BR-424.4 showed 97.6% (n) and 96.8% (aa) similarity to serotype 11, and the USP-BR-453.2 sample showed 99.2% (n) and 99.6% (aa) similarity to serotype 8a. The samples USP-BR-471.14, USP-BR-475.1, USP-BR-475.3, USP-BR-475.4, USP-BR-475.11 and USP-BR-102.8D showed 97.5% (n) and 96.8% (aa) similarity to serotype 11, while the USP-BR-G21.B sample had similarities of 97.2% (n) and 96.0% (aa) for serotype 11 (Table 2 in ESM).
Determination of groups
In addition, the FAdV-4 serotype has been associated with hydropericardium syndrome [15]. Besides, all the FAdV-I serotypes are associated with inclusion body hepatitis corpuscles [25], whereas the FAdV-1 serotype is associated with gizzard erosions [28]. This study showed the presence of group I adenoviruses and their serotypes with the associated enteric disorders described in Table 1. The molecular characterization classified the samples as serotypes 8a, 8b and 11, making this the first report of the presence of these serotypes as probable causes of enteric infections in Brazilian poultry. These samples belonged to birds with signs of enteric disease and feed malabsorption, so these clinical signs could be related to these serotypes. The samples were from eight different chicken farms, and in seven of them (87.5%), the presence of serotype 11 occurred in different organs such as the liver, intestine, bursa, kidneys, pancreas, and trachea. This result suggests that serotype 11 could be predominant in Brazil. The trachea is an important viral replication organ as well as the pharynx, heart, and kidneys [23]. In this study, serotypes 11 and 8a were found in trachea, corroborating the results of [6]. The bursa is also an important organ where FAdV-I is found in natural infections [29], which is confirmed in this report, which shows that serotypes 11 and 8b were present in the bursa. In the samples obtained from farms 418 and 420, serotypes 8b and 11 were found, demonstrating the presence of mixed infections that have also been described [19]. However, it is unknown whether mixed FAdV-I infections influence the severity of the disease or whether there is synergism between the serotypes [20]. Phylogenetic analysis based on the nucleotide sequence of the variable region L1 of the hexon gene classified the nucleotide sequences into 3 distinct groups and confirmed the presence of the serotype 11 of the D species and the serotypes 8a and 8b of the E species in Brazilian poultry flocks. These results emphasize the worldwide distribution of FAdV-I, since serotype 8a aligned with a reference strain from Japan, serotype 8b with a UK reference strain, and serotype 11 with a reference strain from Belgium. Other studies will be necessary to associate the pathogenicity of these serotypes with the clinical changes described. It is necessary to provide more epidemiological data to establish the prevalence of these viruses and justify the development of vaccine programmes in breeders, broiler and laying hens to prevent vertical and horizontal virus infections and prevent the spread of this virus [5, 9].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Grants from the FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo—Grants #2013/08560-5 and 2015/09348-5) and CNPq (Conselho Nacional de Desenvolvimento Cientifico e Tecnológico—Grant #453920/2014-4). Also, the authors would like to thank the poultry companies in Brazil that generously sent the samples for the development of this study and for the diagnosis of enteric viruses.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0430-z) contains supplementary material, which is available to authorized users.
References
- 1.Alemnesh W, Hair-Bejo M, Aini I, Omar AR. Pathogenicity of fowl adenovirus in specific pathogen free chicken embryos. J Comp Pathol. 2010;146:223–229. doi: 10.1016/j.jcpa.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Alvarado IR, Villegas P, El-Attrache J, Jensen E, Rosales G, Perozo F, Purvis LB. Genetic characterization, pathogenicity, and protection studies with an avian adenovirus isolate associated with inclusion body hepatitis. Avian Dis. 2007;51:27–32. doi: 10.1637/0005-2086(2007)051[0027:GCPAPS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P. A reagent for the single-step simultaneus isolation of RNA, DNA and protein for the cell and tissues samples. Biotechniques. 1993;15:532–536. [PubMed] [Google Scholar]
- 4.Christensen NH, Saifuddin M. A primary epidemic of inclusion body hepatitis in broilers. Avian Dis. 1989;33:622–630. doi: 10.2307/1591135. [DOI] [PubMed] [Google Scholar]
- 5.Gomis S, Goodhope R, Ojkic D, Willson P. Inclusion body hepatitis as a primary disease in broilers in Saskatchewan, Canada. Avian Dis. 2006;2006(50):550–555. doi: 10.1637/7577-040106R.1. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin MA. Adenovirus inclusion body ventriculitis in chickens and captive bobwhite quail (Colinus virginianus) Avian Dis. 1993;37:568–571. doi: 10.2307/1591688. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin MA, Hill DL, Dekich MA, Putnam MR. Multisystemic adenovirus infection in broiler chicks with hypoglycemia and spiking mortality. Avian Dis. 1993;37:625–627. doi: 10.2307/1591701. [DOI] [PubMed] [Google Scholar]
- 8.Gowthaman V, Singh S, Dhama K, Barathidasan R, Srinivasan P, Saravanan S, Gopalakrishnamurthy T, Deb R, Mathapati B, Ramakrishnan M. Detection and partial genetic characterisation of a novel variant of Avian nephritis virus in Indian poultry flocks showing diverse clinical signs. Acta Vet Hung. 2015;63:499–507. doi: 10.1556/004.2015.046. [DOI] [PubMed] [Google Scholar]
- 9.Grgić H, Philippe C, Ojkić D, Nagy É. Study of vertical transmission of fowl adenoviruses. Can J Vet Res. 2006;70:230–233. [PMC free article] [PubMed] [Google Scholar]
- 10.Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarria M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G. Family Adenoviridae. In: King A, Adams M, Carstens E, Lefkowitz E, editors. Virus taxonomy classification and nomenclature of viruses. London: Elsevier Press Inc.; 2012. pp. 125–141. [Google Scholar]
- 11.Hess M. Detection and differentiation of avian adenoviruses: a review. Avian Pathol. 2000;29:195–206. doi: 10.1080/03079450050045440. [DOI] [PubMed] [Google Scholar]
- 12.Hess M. Aviadenovirus infection. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V, editors. Diseases of poultry. Ames: Wiley; 2013. pp. 290–300. [Google Scholar]
- 13.Jiang P, Ojkic D, Tuboly T, Huber P, Nagy E. Application of the polymerase chain reaction to detect fowl adenoviruses. Can J Vet Res. 1999;1999(63):124–128. [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Luo L, Luo Q, Zhang T, Zhao K, Wang H, Zhang R, Lu Q, Pan Z, Shao H, Zhang W, Wen G. Genome sequence of a fowl adenovirus serotype 4 strain lethal to chickens, isolated from China. Genome Announc. 2016;4(2):e00140-16. doi: 10.1128/genomeA.00140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim TH, Lee HJ, Lee DH, Lee YN, Park JK, Youn HN, Kim MS, Youn HS, Lee JB, Park SY, Choi IS, Song CS. Identification and virulence characterization of fowl adenoviruses in Korea. Avian Dis. 2011;55:554–560. doi: 10.1637/9730-032011-Reg.1. [DOI] [PubMed] [Google Scholar]
- 16.Mazaheri A, Prusas C, Voss M, Hess M. Some strains of serotype 4 fowl adenoviruses cause inclusion body hepatitis and hydropericardium syndrome in chickens. Avian Pathol. 1998;1998(27):269–276. doi: 10.1080/03079459808419335. [DOI] [PubMed] [Google Scholar]
- 17.Mettifogo E, Nuñez LF, Chacón JL, Santander Parra SH, Astolfi-Ferreira CS, Jerez JA, Jones RC, Piantino Ferreira AJ. Emergence of enteric viruses in production chickens is a concern for avian health. Sci World J. 2014;450423. 10.1155/2014/450423. [DOI] [PMC free article] [PubMed]
- 18.Mettifogo E, Nuñez LFN, Santander Parra SH, Astolfi-Ferreira CS, Piantino Ferreira AJ. Fowl adenovirus group I as a causal agent of inclusion body hepatitis/hydropericardium syndrome (IBH/HPS) outbreak in brazilian broiler flocks. Pesq Vet Bras. 2014;34:733–737. doi: 10.1590/S0100-736X2014000800004. [DOI] [Google Scholar]
- 19.Meulemans G, Boschmans M, Berg TP, Decaesstecker M. Polymerase chain reaction combined with restriction enzyme analysis for detection and differentiation of fowl adenoviruses. Avian Pathol. 2001;30:655–660. doi: 10.1080/03079450120092143. [DOI] [PubMed] [Google Scholar]
- 20.Mittal D, Jindal N, Tiwari AK, Khokhar RS. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virus Dis. 2014;25:114–119. doi: 10.1007/s13337-013-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niczyporuk JS. Phylogenetic and geographic analysis of fowl adenovirus field strains isolated from poultry in Poland. Arch Virol. 2016;161:33–42. doi: 10.1007/s00705-015-2635-4. [DOI] [PubMed] [Google Scholar]
- 22.Nuñez LFN, Parra SHS, Astolfi-Ferreira CS, Carranza C, Torre D, Pedroso AC, Ferreira AJP. Detection of enteric viruses in pancreas and spleen of broilers with runting-stunting syndrome (RSS) Pesq Vet Bras. 2016;36:595–599. doi: 10.1590/S0100-736X2016000700006. [DOI] [Google Scholar]
- 23.Nuñez LFN, Piantino Ferreira AJ. Viral agents related to enteric disease in commercial chicken flocks, with special reference to Latin America. World’s Poult Sci J. 2013;69:853–864. doi: 10.1017/S0043933913000858. [DOI] [Google Scholar]
- 24.Ojkić D, Krell PJ, Tuboly T, Nagy É. Characterization of fowl adenoviruses isolated in Ontario and Quebec, Canada. Can J Vet Res. 2008;72:236–241. [PMC free article] [PubMed] [Google Scholar]
- 25.Ojkic D, Martin E, Swinton J, Vaillancourt J-P, Boulianne M, Gomis S. Genotyping of Canadian isolates of fowl adenoviruses. Avian Pathol. 2008;37:95–100. doi: 10.1080/03079450701805324. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez J, Koga Y, Alvarado A, Tinoco R. Molecular characterization of Peruvian Fowl Adenovirus (FAdV) isolates. Adv Microbiol. 2014;4:595–603. doi: 10.4236/aim.2014.410065. [DOI] [Google Scholar]
- 27.Sentíes-Cué CG, Wills RW, Stayer PA, Burleson MA, Magee DL. Epidemiology and effect on production parameters of an outbreak of inclusion body hepatitis in broilers. Avian Dis. 2010;54:74–78. doi: 10.1637/8958-061109-Reg.1. [DOI] [PubMed] [Google Scholar]
- 28.Seuberlich T, Doherr MG, Botteron C, Nicolier A, Heim D, Zurbriggen A. Comparison of the fibers of Fowl adenovirus A serotype 1 isolates from chickens with gizzard erosions in Europe and apathogenic reference strains. J Vet Diagn Invest. 2010;2010(22):97–101. doi: 10.1177/104063871002200613. [DOI] [PubMed] [Google Scholar]
- 29.Steer PA, Sandy JR, O’Rourke D, Scott PC, Browning GF, Noormohammadi AH. Chronological analysis of gross and histological lesions induced by field strains of fowl adenovirus serotypes 1, 8b and 11 in one-day-old chickens. Avian Pathol. 2015;44:106–113. doi: 10.1080/03079457.2015.1007919. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toro H, Prusas C, Raue R, Cerda L, Geisse C, González C, Hess M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999;43:262–270. doi: 10.2307/1592616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.