Significance
Fluorination has become a key design strategy for the development of a broad range of small-molecule therapeutics. As a result, there is significant interest in understanding how fluorine substituents affect interactions between ligands and their protein targets. In this context, we elucidate the molecular details of fluorine recognition in the fluoroacetyl–CoA thioesterase, which has evolved a 106-fold selectivity for a single fluorine atom as part of its native function in organofluorine biosynthesis. Using a combination of thermodynamic, kinetic, and biophysical experiments, we show that fluorine recognition is entropically driven by interaction with a hydrophobic residue in the binding site. These findings highlight the importance of hydrophobic and solvation effects in mediating fluorine-binding selectivity driven by natural selection.
Keywords: enzyme selectivity, fluorine, organofluorine, molecular recognition, metabolism
Abstract
Fluorinated small molecules play an important role in the design of bioactive compounds for a broad range of applications. As such, there is strong interest in developing a deeper understanding of how fluorine affects the interaction of these ligands with their targets. Given the small number of fluorinated metabolites identified to date, insights into fluorine recognition have been provided almost entirely by synthetic systems. The fluoroacetyl–CoA thioesterase (FlK) from Streptomyces cattleya thus provides a unique opportunity to study an enzyme–ligand pair that has been evolutionarily optimized for a surprisingly high 106 selectivity for a single fluorine substituent. In these studies, we synthesize a series of analogs of fluoroacetyl–CoA and acetyl–CoA to generate nonhydrolyzable ester, amide, and ketone congeners of the thioester substrate to isolate the role of fluorine molecular recognition in FlK selectivity. Using a combination of thermodynamic, kinetic, and protein NMR experiments, we show that fluorine recognition is entropically driven by the interaction of the fluorine substituent with a key residue, Phe-36, on the lid structure that covers the active site, resulting in an ∼5- to 20-fold difference in binding (KD). Although the magnitude of discrimination is similar to that found in designed synthetic ligand–protein complexes where dipolar interactions control fluorine recognition, these studies show that hydrophobic and solvation effects serve as the major determinant of naturally evolved fluorine selectivity.
Site-selective fluorination has become an important strategy in the design of structurally diverse bioactive small molecules with a broad range of functions ranging from human therapeutics and imaging agents to biocides used for plants, fungi, and insects (1–4). The success of fluorine in the development of these compounds has mainly relied on the small size of fluorine to conserve binding of the fluorinated ligand to its macromolecular target while tuning a breadth of pharmacological characteristics. As such, studies on the impact of fluorination on metabolic stability, distribution, and other pharmacokinetic properties of small molecules have provided important information on how fluorine substituents contribute to their behavior in vivo (5, 6). In contrast, the role of fluorine in tuning binding affinity and selectivity of small molecules for their protein targets is relatively less well understood given the exceptional and context-dependent elemental traits of fluorine (4).
Fluorine’s high electronegativity and “polar hydrophobicity,” as well as the highly polarized nature of the carbon–fluorine bond can contribute in different ways to protein–ligand interactions (4, 7). In some cases, nonspecific interactions arising from the increase in hydrophobic surface area introduced by fluorine can exert an energetically significant effect on molecular recognition (8–11). In others, the increased steric bulk of fluorine can cause changes in compound conformation that lead to increased selectivity as observed in structure–activity relationship studies on fluoxetine (Prozac) (12, 13). In terms of specific recognition of the fluorine substituent itself, the high polarization of the C-F bond creates an opportunity for dipolar interactions to occur with protein-based functional groups (14, 15), which appears to play a key role in the increased potency of ciprofloxacin (Cipro) compared with analogs lacking the fluorine substituent (16, 17).
Despite the strong interest in fluorine–protein interactions, there are few systems in which evolutionarily optimized interactions between ligand and receptor can be studied given the limited existence of naturally occurring fluorinated metabolites and macromolecules that interact with them. As one of the few characterized organisms that produces fluorinated natural products, Streptomyces cattleya provides an interesting system for exploring native fluorine recognition by protein targets that have been subject to an unusual evolutionary pressure to distinguish between fluorine and other similar substituents. One of the organofluorines produced by S. cattleya is fluoroacetate, a potent poison that relies on the inability of the target host’s primary metabolic enzymes to sufficiently discriminate against the conservative replacement of hydrogen with fluorine (18, 19). S. cattleya itself produces a resistance protein, the fluoroacetyl CoA thioesterase (FlK) (20–22), which displays a remarkably high 106-fold preference for the fluorinated substrate compared with acetyl–CoA, a cellularly abundant competitor (21). This selectivity is largely derived from the increased kcat (104-fold increase) due to change in the hydrolysis mechanism for the fluorinated substrate (23). Nevertheless, the remaining 100-fold difference attributed to the decreased KM suggests that molecular recognition of the fluorine substituent also plays a role in substrate discrimination (21, 24). Given that the magnitude of the expected binding selectivity is similar to that observed for synthetic organofluorine inhibitors (4, 14, 15), elucidation of the factors that control fluorine recognition in FlK can provide insight into optimizing the design of fluorinated ligands.
Using a set of nonhydrolyzable substrate analogs to separate the contributions of fluorine recognition from catalytic selectivity, we show that FlK does indeed bind with increased affinity to ligands containing a fluorine substituent (21). We further show that a mobile residue on a unique hydrophobic lid structure (21), Phe-36, is involved in providing an entropic driving force for fluorinated substrate binding and controlling the off-rate of the substrate. NMR studies were then carried out to examine FlK dynamics, which demonstrated that both fluorine and Phe-36 are required to lower the off-rate of the substrate. Taken together, these results support a model in which a conformationally dynamic residue (Phe-36) plays an important role in fluorine-based substrate discrimination. In contrast to synthetic systems, in which enhanced ligand affinity is usually mediated by dipolar interactions that confer an enthalpic binding advantage (4), fluoroacetyl–CoA discrimination in FlK is achieved by creating an entropic advantage for fluorinated substrate binding and by increasing the residence time of the fluorinated substrate in the active site. This mechanism appears to be distinct to natural fluorine selectivity and provides an equal if not greater fluorine selectivity to enthalpically driven fluorine recognition.
Results and Discussion
Phe-36, a Conformationally Dynamic Residue at the FlK Active Site, Is Important for Substrate Binding and Specificity.
FlK exhibits a number of distinctive features compared with other members of the hotdog-fold thioesterase family that may contribute to its high substrate selectivity. Most of the characterized members of this family are quite promiscuous and accept a variety of medium-chain, long-chain, and aromatic acyl–CoAs with similar catalytic efficiency (25–28). In comparison, FlK is strongly selective for fluoroacetyl–CoA compared with acetyl–CoA based on the conservative exchange of fluorine for hydrogen at the α-carbon. While a large component of this discrimination occurs as a result of fluorine-induced changes in the chemical and kinetic hydrolysis mechanism, structural studies have shown that FlK contains an additional “lid” structure near the acyl group binding site that may play a role in fluorine recognition. Mutagenesis of key lid residues suggest that this structure plays a role in FlK selectivity (21). The lid is not found in other characterized hotdog-fold thioesterases (21, 29), although a number of proteins in the National Center for Biotechnology Information database also appear to contain the lid based on sequence conservation and homology modeling.
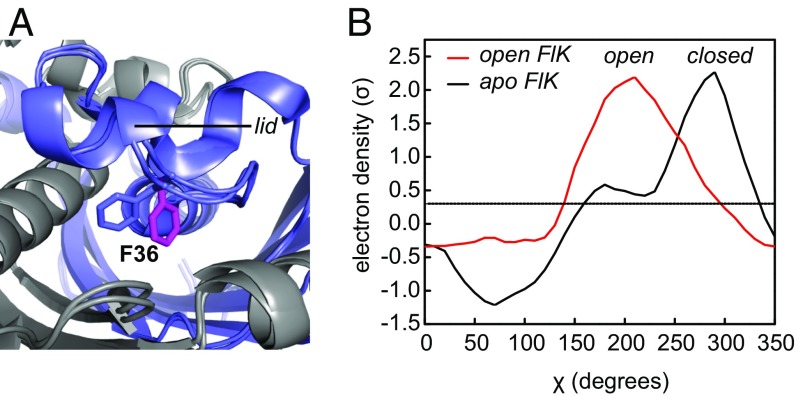
A residue within the lid, Phe-36, undergoes a conformational change upon soaking FlK crystals with substrate or products (Fig. 1A) (21). This conformational change opens up a channel between the active site and exterior solvent that is normally occluded by Phe-36. It is interesting to note that Phe-36 is unique to FlK from Streptomyces cattleya and an ortholog from Streptomyces xinghaiensis, whose genome also encodes a putative fluorinase enzyme (SI Appendix, Fig. S1). To further explore changes in Phe-36 conformation, we analyzed the FlK electron-density maps using Ringer, a program designed to detect molecular motions through electron-density sampling (30). The resulting plots of electron density (σ) versus the χ angle of Phe-36 reveal that apo–FlK appears to sample this alternative rotamer conformation as well, with about 10% occupancy under cryocrystallography conditions (Fig. 1B).
Fig. 1.
Conformational change observed in FlK upon soaking with substrate or products. (A) Structural alignment of FlK in the presence and absence of products. Phe-36 is shown in blue for apo–FlK and in magenta for substrate-soaked FlK. (B) Ringer plot showing the Phe-36 χ-angle for FlK crystals in the presence (open) and absence (closed) of substrate. Figures are based on PDB entry 3P2Q for apo FlK and 3P2S for open FlK.
While biochemical characterization of the F36A mutant had shown that kcat/KM for the fluoroacetyl–CoA was decreased by 140-fold without affecting acetyl–CoA parameters (21), the increase in KM for the fluorinated substrate cannot directly be attributed to a change in KD because of the multistep chemical and kinetic mechanism for FlK-catalyzed acyl–CoA hydrolysis (23). To further probe the role of Phe-36 in substrate affinity, we examined the presteady-state kinetics of the F36A mutant under saturating conditions with both acetyl–CoA and fluoroacetyl–CoA (SI Appendix, Fig. S2). In these experiments, we observed burst-phase kinetic behavior with acetyl–CoA similar to wild-type enzyme, with one equivalent of CoA released rapidly during the first enzyme turnover followed by a slower steady-state release of CoA (SI Appendix, Fig. S2A). The rate constants determined for each of these steps by nonlinear curve fitting were identical, within error, to the wild-type enzyme (23). Based on mechanistic studies of the wild-type enzyme, we conclude that the F36A mutant also follows a two-step chemical mechanism for acetyl–CoA hydrolysis, with rapid formation of an acyl–enzyme intermediate followed by slower intermediate breakdown. For fluoroacetyl–CoA, we observed single-phase presteady-state kinetics, consistent with a change in rate-limiting step from the breakdown of the acyl–enzyme intermediate to its formation as occurs in the wild-type enzyme (SI Appendix, Fig. S2B). As with acetyl–CoA, the kcat for hydrolysis of the fluorinated substrate by the F36A mutant is identical, within error, to that measured for the wild-type enzyme (23). From these experiments, we conclude that the chemical steps involved in substrate hydrolysis are the same between wild-type FlK and F36A FlK and that mutation of Phe-36 results in a change in KM by affecting substrate binding.
Fluorinated and Nonfluorinated Ligands Bind FlK with Different Affinities.
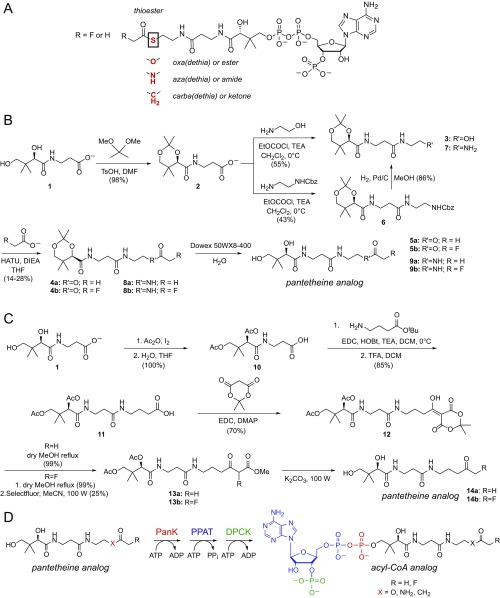
To isolate the role of molecular recognition of fluorine in FlK, we designed a series of substrate analogs intended to be nonhydrolyzable so that the role of catalysis in fluorine selectivity could be excluded. Toward this goal, we synthesized acyl–CoA analogs with either an acetyl or fluoroacetyl group in which the thioester sulfur atom was replaced by oxygen, nitrogen, or carbon to produce the less labile or nonhydrolyzable ester, amide, or ketone congener (Scheme 1A). We used a synthetic strategy in which the desired acylated pantetheine analogs [oxa(dethia): 5a, 5b; aza(dethia): 9a, 9b; carba(dethia): 14a, 14b] (Scheme 1 B and C) were first prepared chemically (31, 32) and then converted enzymatically to the corresponding CoA analogs using purified pantothenate kinase, phosphopantetheine adenylyltransferase, and dephospho–CoA kinase from Escherichia coli (31–33) (Scheme 1D and SI Appendix, Fig. S3).
Scheme 1.
Design and synthesis of ester, amide, and ketone substrate analogs of fluoroacetyl– and acetyl– CoA. (A) Design of nonhydrolyzable substrate analogs. Replacement of S with either O, NH, or CH2 generates the oxa(dethia), aza(dethia), and carba(dethia) congeners, respectively. (B) Synthesis of acetyl– and fluoroacetyl– oxa(dethia)-pantetheine (5a, 5b) and acetyl– and fluoroacetyl –aza(dethia)-pantetheine (9a, 9b). (C) Synthesis of acetyl– and fluoroacetyl– carba(dethia)-pantetheine (14a, 14b). (D) Enzymatic conversion of pantetheine analogs to their corresponding acyl–CoA analog (DPCK, dephosphocoenzyme A kinase; PanK, pantetheine kinase; PPAT, phosphopantetheine adenylyltransferase).
The fluorinated and nonfluorinated ester [FAcOCoA, fluoroacetyl–oxa(dethia) –CoA; AcOCoA, acetyl–oxa(dethia)–CoA], amide [FAcNCoA, fluoroacetyl–aza(dethia)–CoA; AcNCoA, acetyl–aza(dethia)–CoA], and ketone [FAcCCoA, fluoroacetyl–carba(dethia)–CoA; AcCCoA, acetyl–carba(dethia)–CoA] substrate analogs were then characterized to assess if they could be utilized for binding studies. To extrapolate the behavior of the analogs to the thioester substrate, the analog both needs to exist in the same form in solution as well as be unreactive toward hydrolysis by FlK on the timescale of the binding experiment. Since the carbonyl polarization of these derivatives is different from the corresponding thioester, they could potentially exist preferentially in solution as the hydrate or an enolate form and consequently bind in a different mode from the substrate. Based on NMR spectroscopy studies, there was no evidence of any other forms besides the carbonyl species for any of the six compounds under these conditions within the limit of detection (SI Appendix, Figs. S4–S6). We also tested whether FlK could catalyze hydrolysis of the ester or amide substrate analogs because esterases and amidases are found in the larger α/β-hydrolase superfamily even if they have yet to be identified in the hotdog-fold superfamily (34). In biochemical experiments with FlK, we found that the ester analog was hydrolyzed very inefficiently, at a rate constant more than 500-fold lower (kcat = 0.01 min−1) than measured for the thioester (SI Appendix, Fig. S7A) (21, 23), while no detectable hydrolysis was observed for the amide analog after 18 h in the presence of 100 µM FlK (SI Appendix, Fig. S7B). From these experiments, all three classes of substrate analogs can be used to study fluorine molecular recognition by FlK.
Using isothermal titration calorimetry (ITC), we measured the dissociation constant and the contributions made by entropy (ΔS) and enthalpy (ΔH) for each of the substrate analogs (Table 1 and SI Appendix, Fig. S8). Based on the measured KM for fluoroacetyl–CoA (8 ± 1 μM) (21), it appears that the fluorinated inhibitors bind with an apparent defect of one to two orders of magnitude compared with the native substrate, with the ester analog binding with the highest affinity and the amide analog binding with the lowest affinity. Within each fluorine/hydrogen pair, we measured an ∼5- to 20-fold lower affinity for the acetyl–CoA analogs compared with the fluoroacetyl–CoA analogs (Table 1 and SI Appendix, Fig. S8), indicating that fluorine recognition contributes to FlK selectivity. Interestingly, the magnitude of discrimination rivals the highest reported changes in affinity for binding of fluorinated synthetic inhibitors to their targets in cases where inhibitor conformation or the pKas of nearby groups are not the major contributors (∼10-fold) (4, 14, 15). However, despite the strong fluorine-based discrimination observed in the analog pairs, the 5- to 20-fold larger difference observed between the KMs for acetyl–CoA and fluoroacetyl–CoA suggests that other factors that are not completely recapitulated in the analog pairs may contribute to selectivity in the KM parameter.
Table 1.
Thermodynamic parameters for binding of substrate analog to wild-type and F36A FlK
| Wild-type FlK | F36A FlK | |||||
| Analog | KD (μM) | ΔH (kcal/mol) | TΔS (kcal/mol) | KD (μM) | ΔH (kcal/mol) | TΔS (kcal/mol) |
| FAcOCoA | 90 ± 2 | −12.1 ± 0.1 | −6.5 ± 0.1 | 220 ± 30 | −13.0 ± 0.1 | −8.0 ± 0.1 |
| AcOCoA | 565 ± 45 | −13.5 ± 0.3 | −9.0 ± 0.1 | 500 ± 70 | −9.9 ± 0.6 | −6.3 ± 0.1 |
| FAcNCoA | 1,700 ± 100 | −7.4 ± 0.1 | −3.7 ± 0.1 | 7,500 ± 200 | −7.3 ± 0.1 | −4.4 ± 0.1 |
| AcNCoA | 9,300 ± 500 | −8.4 ± 0.3 | −5.3 ± 0.1 | 6,200 ± 300 | −8.5 ± 0.1 | −5.5 ± 0.1 |
| FAcCCoA | 440 ± 90 | −5.6 ± 0.2 | −1.0 ± 0.3 | 2,700 ± 100 | −5.6 ± 0.1 | −2.1 ± 0.1 |
| AcCCoA | 7,700 ± 230 | −9.4 ± 0.2 | −6.6 ± 0.1 | n.d. | n.d. | n.d. |
ITC experiments of wild-type and F36A FlK with the ester [oxa(dethia): FAcOCoA, AcOCoA], amide [aza(dethia): FAcNCoA, AcNCoA], and ketone [carba(dethia: FAcCCoA, AcCCoA] analogs of fluoroacetyl– and acetyl–CoA were carried out. Inhibition constants, ΔH, and TΔS, were obtained from fitting. Errors are derived from nonlinear curve fitting. Data are mean ± SE (n = 3). n.d., not determined.
The observed differences in KD between fluorinated and nonfluorinated compounds correspond to a ΔΔG of ∼1 kcal/mol for the single fluorine substitution. Our ITC measurements demonstrate that the fluorinated and nonfluorinated compounds have similar enthalpies of binding (ΔH), whereas the fluorinated analogs demonstrate more favorable entropies of binding (ΔS), suggesting that the energetic contribution of fluorine to binding (KD) is mainly entropic in nature (Table 1). In comparison, binding affinity differences in synthetic systems have been mainly attributed to enthalpic changes related to dipolar interactions between the C-F bond and protein amides, carboxamides, or guanidinium groups, which typically contribute 0.2–0.3 kcal/mol per interaction to binding affinity (4).
Fluorinated Substrate Binding Is Entropically Coupled to Phe-36.
Because the origin of the binding selectivity for the fluorinated substrate is related to entropy and thus potentially the increase in hydrophobic surface area presented by the C-F group, we next set out to probe the role of the lid structure of FlK in substrate binding. The interior surface of the lid is lined with hydrophobic side chains (21, 29), which appear to serve the dual roles of excluding water from the active site as well as creating a hydrophobic environment. Previous mutagenesis studies of the lid revealed that replacement of any of these residues with alanine led to an increase in KM for the fluorinated substrate by one to two orders of magnitude while leaving the KM for acetyl–CoA relatively unchanged (21). Since Phe-36 seems to both be particularly important in fluorine-selective binding and shows no significant catalytic defect when mutated, the F36A mutant was used to further probe the molecular basis of the entropic advantage on substrate binding provided by the fluorine atom.
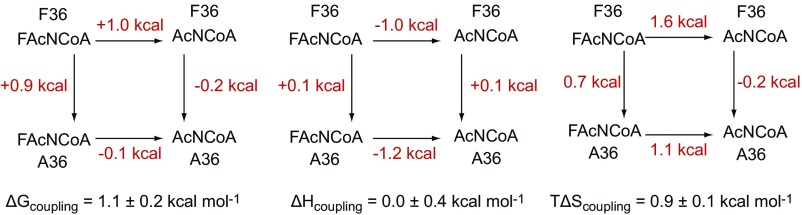
Based on ITC measurements, the fluorine/hydrogen pair in each analog series bound to the F36A FlK mutant with similar affinity, demonstrating that fluorine selectivity is lost in the absence of the Phe-36 sidechain (Table 1 and SI Appendix, Fig. S9). This behavior is caused by a loss of binding affinity for the fluorinated ligand rather than an increase in affinity for the nonfluorinated ligand. Further analysis of the data for the amide inhibitors by a chemical-enzymatic double-mutant cycle (35) shows that enhanced binding affinity of the fluorinated compound (ΔG) is energetically coupled to the Phe-36 side chain and relies on entropy (Fig. 2). From this analysis, we conclude that Phe-36 participates directly in providing an entropic driving force for fluorinated substrate recognition.
Fig. 2.
Analysis of binding of aza(dethia)–CoA substrate analogs to wild-type and F36A FlK. Double-mutant cycles were generated for mutation of F36 (wild type) to A36 as well as the switch between the fluorinated (FAc) and nonfluorinated (Ac) substrates. Errors for coupling energies were propagated from the errors in the individual thermodynamic terms.
Phe-36 Controls Fluorine Selectivity Through the Substrate Off-Rate as Suggested by ITC and NMR Experiments.
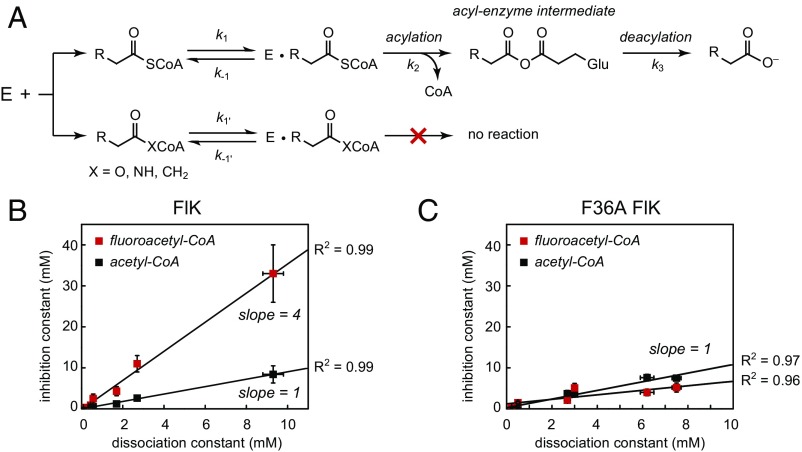
We also examined the ability of our substrate analogs to act as competitive inhibitors of fluoroacetyl–CoA and acetyl–CoA hydrolysis by FlK (Fig. 3A and SI Appendix, Table S1). For FlK-catalyzed acetyl–CoA hydrolysis, all of the substrate analogs exhibited competitive behavior with inhibition constants (KIs) within error of the dissociation constants measured by ITC (Fig. 3B). With fluoroacetyl–CoA, all of the substrate analogs also behaved as competitive inhibitors, but the measured inhibition constants for FlK were significantly higher than the dissociation constants measured by ITC (Fig. 3B). However, the approximation that KD ∼ KI relies on the assumption that k−1 > k2, which does not always hold for substrates for which the rates of catalytic steps are rapid. Therefore, the inequivalence of KI and KD that we observe for FlK with fluoroacetyl–CoA indicates that binding of fluoroacetyl–CoA to FlK is not in rapid equilibrium and further suggests that the first chemical step (k2) is faster than the substrate off-rate (k−1). A plot of KI versus KD reveals that for both fluoroacetyl–CoA and acetyl–CoA, these two parameters are linearly related with a slope of (k2/k−1 + 1) derived from the kinetic model for competitive inhibition (SI Appendix, Fig. S10). For acetyl–CoA, k2 is small relative to the off-rate, so k2/k−1 approaches zero and the observed slope is equal to 1 (Fig. 3B). For fluoroacetyl–CoA, the slope of this plot is equal to 4 and indicates that the enzyme acylation rate constant for this substrate (k2) is three times faster than the substrate dissociation rate constant (k−1), which explains why the substrate analogs are less effective at competing with the fluoroacetyl–CoA substrate.
Fig. 3.
FlK inhibition studies with substrate analogs. (A) Kinetic scheme for competitive inhibition of FlK by substrate analogs. R = H, F. (B) Plots of inhibition constant versus dissociation constant for substrate analogs for wild-type and F36A FlK. (KI measured with respect to fluoroacetyl–CoA, red; KI measured with respect to acetyl–CoA, black.) The data are fit to the equation: KI = (k2/k−1 + 1) × KD. Data are mean ± SE (n = 3).
The observed change in behavior with respect to the fluorine substituent could be caused either by slower acylation of the enzyme by acetyl–CoA (k2 = 3 s−1) (23) compared with fluoroacetyl–CoA (k2 = 270 s−1), or a slower off-rate related to the higher affinity of the fluorinated substrate (KD = k−1/k1), or to a combination of both factors. To probe the role of enhanced binding of fluoroacetyl–CoA, we examined the ability of the nonhydrolyzable substrate analogs to competitively inhibit the F36A mutant (SI Appendix, Table S1), which is compromised only in binding the fluorinated ligand. In this mutant, we again observed competitive behavior by all of the inhibitors with respect to both the fluoroacetyl–CoA and acetyl–CoA substrates. Interestingly, the KI was within error of KD and slopes of the plots of KI versus KD were equal to one regardless of the substrate used. Thus, k2/k−1 approaches zero for both acetyl–CoA and fluoroacetyl–CoA, showing that rapid equilibrium is restored for fluoroacetyl–CoA in this mutant. Since the rate constant measured by rapid chemical quench for enzyme acylation by fluoroacetyl–CoA is unchanged, we can conclude that mutation of Phe-36 increases the substrate off-rate (koff, k−1) such that it becomes faster than the first chemical step (k2). We thus conclude that Phe-36 is directly involved in controlling the off-rate for fluoroacetyl–CoA, allowing for kinetic discrimination of the fluorine substituent in substrate binding. Because acetyl–CoA is in rapid equilibrium for both wild-type and F36A FlK, we are unable to detect any potential changes in the off-rate for this substrate based on the competitive inhibition studies. However, ITC studies demonstrated that the acetyl–CoA analogs have identical dissociation constants for both wild-type and F36A FlK. Therefore, if the off-rate is increased in the mutant, the on-rate (kon, k1) must increase by an equal amount to maintain the observed KD (k−1/k1). The simplest explanation for the ITC data in combination with competitive inhibition studies is that mutation of Phe-36 increases the off-rate for fluoroacetyl–CoA while leaving the off-rate for acetyl–CoA unchanged.
The hypothesis that both Phe-36 and the fluorine substituent modulate substrate binding through decreasing the off-rate is further supported by 1H-NMR experiments with the carba(dethia) substrate analogs, FAcCCoA and AcCCoA. For protons close to the protein active site, their chemical shift in 1H-NMR spectra will become altered upon protein binding, as the chemical environment of these protons will be significantly different. When the substrate analog is not fully bound, an equilibrium exists between the free and protein-bound substrate analog, where the rate of exchange between the two states is defined as kex. As a result, the peak in the NMR spectrum from a proton in this equilibrium represents a combination of these two states. When the exchange rate is in the fast exchange regime, the chemical shift of the peak will change compared with free ligand, but the linewidth of the peak will not be altered. Alternatively, if the exchange rate is in the intermediate exchange regime, the resulting peak will be significantly broader as well as showing a change in chemical shift. An exchange rate in the slow exchange regime will manifest as two distinct peaks, one corresponding to free ligand and one corresponding to the ligand–protein complex.
When excess FAcCCoA (500 µM) was complexed with wild-type FlK (100 µM), the 1D 1H-spectrum of FAcCCoA in regions unique to the ligand showed significant line-broadening, reflective of an intermediate exchange rate for the ligand–protein-binding interaction (SI Appendix, Fig. S11) (36). However, when the same experiment is carried out with wild-type FlK and AcCCoA or F36A FlK and FAcCCoA, only a slight change in chemical shift is observed without significant alteration of the linewidths, indicating fast exchange. Therefore, the exchange rate (kex) of FAcCCoA bound to wild-type FlK is slower than that of both FAcCCoA bound to F36A FlK and AcCCoA bound to wild-type FlK.
In these experiments, ligand concentration [P], rather than protein concentration, is kept constant and in excess, hence the exchange rate can be represented as kex = k1[P] + k−1 (36). With [P] kept constant in all experiments, an increase in exchange rate as a result of either the F36A mutation or loss of the fluorine substituent would suggest either a higher on-rate (k1) or off-rate (k−1). However, an increase in off-rate instead of on-rate is most consistent with the results of the competitive inhibition studies, which suggested an increase in the fluorinated substrate off-rate in the F36A mutant, and with KD measurements for the substrate analogs, which showed that the nonfluorinated congeners maintained the same affinity for wild-type and F36A FlK. Taken together, these results suggest that both the fluorine moiety and Phe-36 are required for fluorinated substrate selectivity facilitated by a slower off-rate.
Protein NMR Confirms Phe-36 Plays a Fluorine-Specific Role in Substrate Binding.
To elucidate the role of Phe-36 in greater detail, we utilized NMR spectroscopy of FlK labeled with 13C at the δ-position of the aromatic rings of Phe and Tyr via feeding with [1-13C]-glucose (37). This feeding scheme results in a single peak for each Phe or Tyr residue in the aromatic region of 1H/13C heteronuclear single quantum correlation (HSQC) spectra, simplifying the interpretation of chemical shift perturbation. Transverse relaxation-optimized spectroscopy (TROSY) optimized for aromatic spin systems (38, 39) was employed for 2D 1H/13C experiments with the 13C-labeled wild-type and F36A FlK proteins (Fig. 4 A and B). The 1H/13C TROSY-HSQC spectrum of wild-type FlK in the aromatic region of Phe and Tyr exhibits at least nine peaks, which could correspond to the Cδ1-Hδ1 bond vectors of seven Phe residues and two Tyr residues in an FlK monomer. Considering the intensities of the peaks vary greatly, it is possible that there are additional Cδ1-Hδ1 signals that are too weak to observe at the present contour level. At least two Cδ1-Hδ1 peaks were overlapped (δ7.1 ppm in the 1H region) and could not be well defined. The two peaks at δ 133.5 and 134 ppm in the 13C dimension are attributed to the two Tyr residues, as confirmed by a separate 1H/13C TROSY-HSQC spectrum where 13C-Phe was incorporated into FlK (SI Appendix, Fig. S12). The resonance assignments of Phe-36 and Phe-40 were determined by generating point mutations from Phe to Ala (F36A) or Leu (F40L) and obtaining the corresponding 1H/13C TROSY-HSQC spectra (SI Appendix, Fig. S13). Additionally, the Phe-36 assignment was confirmed by 1H/15N Nuclear Overhauser effect spectroscopy (NOESY) (SI Appendix, Fig. S14). The assignments of both residues are within the three-peak cluster region between δ7.5 and 7.6 ppm in the 1H dimension (Fig. 4A). The third peak may derive from an alternative conformation for Phe-36 since similar secondary peaks have been observed in other proteins (40). Alternatively, it could correspond to Phe-33 but we were not able to obtain enough soluble F33A FlK protein for an HSQC spectrum to confirm either assignment.
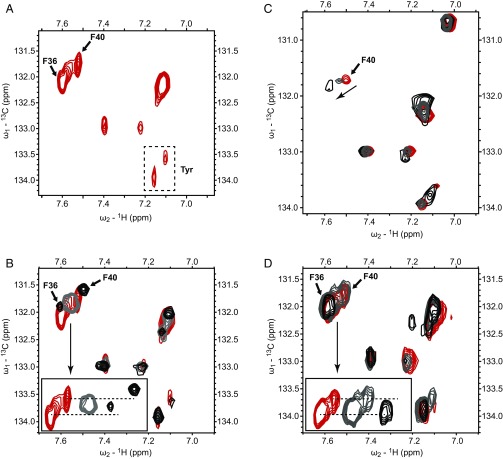
Fig. 4.
Phe/Tyr region of 1H/13C TROSY-HSQC spectra for 13C-labeled wild-type and F36A FlK with and without carba(dethia) substrate analogs bound. (A) Wild-type FlK with no ligand present. (B) Wild-type FlK in complex with 0 mM FAcCCoA (red), 0.56 mM FAcCCoA (gray, 50% bound), and 6.4 mM FAcCCoA (black, 95% bound). Inset shows shift of the three-peak cluster containing Phe-36 and Phe-40. (C) F36A FlK in complex with 0 mM FAcCCoA (red), 2.9 mM FAcCCoA (gray, 50% bound), and 50 mM FAcCCoA (black, 95% bound). The arrow indicates the shift of Phe-40. (D) Wild-type FlK in complex with 0 mM AcCCoA (red), 7.9 mM AcCCoA (gray, 50% bound), and 32.5 mM (black, 80% bound). Inset shows lack of shift of Phe-36 and disappearance of Phe-40 in the three-peak cluster containing Phe-36 and Phe-40.
The carba(dethia) analogs of fluoroacetyl–CoA (FAcCCoA) and acetyl–CoA (AcCCoA) were selected to examine binding because significant hydrolysis of the ester analogs would be expected over the timescale of the NMR experiment despite the low hydrolysis rate constants. These ligands were added to 13C-labeled wild-type FlK at two different concentrations, resulting in the protein being 50% and 95% bound with FAcCCoA and 50% and 80% bound with AcCCoA, respectively. These values were estimated from the KD values for the complexes determined by ITC. Upon addition of substrate analogs, only the three-peak cluster at the δ7.6-ppm region on the 1H axis, which encompasses Phe-36 and Phe-40, exhibited substantial chemical shift perturbation among Phe residue moieties. Remarkably, while Phe-36 displayed a significant change in chemical shift upon binding of FAcCCoA, the chemical shift of Phe-36 was not affected by binding to AcCCoA (Fig. 4D). This observation suggests that Phe-36 only interacts with the substrate when the fluorine substituent is present. In addition, we observe a decrease in linewidth for both the Phe-36 and Phe-40 peaks upon fluorinated ligand binding, indicating that dynamic motion on the NMR timescale has been reduced.
In comparison, Phe-40 is perturbed upon binding of both analogs, although it exhibits different degrees of change. The peak assigned to Phe-40 shifts upon binding to the fluorinated ligand, but simply broadens upon binding of AcCCoA. Phe-40 is not oriented toward the active site in the crystal structure of wild-type FlK (21). In this case, Phe-40 might be allosterically perturbed as it is within close proximity of Thr-42, a key residue believed to play a structural role in maintaining the active site (24). Interestingly, the decrease in linewidth observed with binding of the fluorinated ligand is not observed when the fluorine is absent.
The change in chemical shift observed for Phe-36 and Phe-40 upon binding of the fluorinated ligand indicates that the chemical environment of the side-chain is altered, arising through either conformational changes of these residues induced by ligand binding or direct interaction with the ligand itself. In addition, this binding event results in a change in the dynamics in these two residues where motion is reduced. Overall, the NMR studies demonstrate that Phe-36 is engaged in the ligand-binding process only in the presence of a fluorine substituent, in agreement with the thermodynamic and kinetic data showing that fluorine molecular recognition depends in large part on Phe-36. Furthermore, the different responses exhibited by Phe-40 upon binding to FAcCCoA versus AcCCoA suggests that the nature of the binding process, such as binding mechanism or binding rate, may be different when the fluorine moiety is not present.
Conclusions
Given the contribution of fluorine to the design and development of bioactive small molecules, understanding the interaction of fluorinated ligands with protein targets continues to be a key area of study. While synthetic interactions have provided much insight to the different modes by which fluorine interacts with macromolecules, the fluoroacetyl–CoA thioesterase FlK provides a unique platform to study a fluorinated ligand–protein pair that has been evolutionarily optimized for fluorine selectivity. In this context, we have designed a set of fluorinated and nonfluorinated substrate analogs to elucidate the molecular basis for fluorine molecular recognition by FlK. These analogs allow the impact of differential binding in fluorine discrimination to be isolated and studied. This is especially important for FlK, given that the differences in KM between fluorinated and nonfluorinated thioester substrates may not reflect the difference in binding affinity or KD since the first step in catalysis is irreversible enzyme acylation.
Previous crystallographic and modeling studies had suggested possible enthalpic and entropic contributions to selective fluorinated substrate binding (21, 29). Modeling of fluoroacetyl–CoA into the FlK active site suggested that a dipolar interaction between the C-F bond and Arg-120 could contribute enthalpically to fluorine-selective binding (29). However, the proposed interaction placed the C-F bond in a linear hydrogen bond geometry rather than with the C-F bond oriented perpendicular to the plane of the guanidinium side chain, as is most commonly observed in fluorine–arginine interactions in the Protein Data Bank (4). On the other hand, crystal structures of apo–FlK and the FlK–product complex suggested that release of water from the C-F unit and exclusion of water from the active site could contribute entropically to fluorinated substrate binding. Our ITC measurements with fluorinated and nonfluorinated oxa(dethia)-, aza(dethia)-, and carba(dethia)- substrate analogs show that entropic effects (∼1 kcal/mol) drive the preferential binding of fluorinated ligands to FlK. Interestingly, enthalpic interactions do not appear to play a major role in fluorine selectivity despite the proposed C-F–arginine interaction (29). While it remains possible that this particular interaction differs with the thioester substrates, our data suggest that it is either energetically insignificant compared with the observed entropic contributions, or that it functions in catalysis, perhaps in poising the substrate for reactivity given the seemingly weak oxyanion hole of FlK. The increase in affinity derived from the fluorine substituent is similar in magnitude to that observed for synthetic systems. However, the mode of fluorine recognition utilized in the evolved FlK–fluoroacetyl–CoA system is distinct from that observed in synthetic protein–ligand systems, in which enthalpic interactions (∼0.2–0.3 kcal/mol per interaction) typically serve as the main driver for fluorine selectivity (4).
Combined with structural information, the importance of entropic contributions in selective fluorinated substrate binding suggests that FlK’s hydrophobic lid, and Phe-36 in particular, are key determinants of fluorine molecular recognition. Consistent with this hypothesis, thermodynamic analysis of analog binding to the F36A FlK mutant shows that Phe-36 makes an important energetic contribution to fluorine-selective binding. This finding is consistent with previous mutagenesis studies which implicated an important role for Phe-36 in fluorine discrimination based on differences in KM for fluoroacetyl–CoA compared with acetyl–CoA (21). Presteady-state experiments show that the catalytic steps for F36A FlK hydrolysis are indistinguishable from wild type, supporting the hypothesis that differences in KM should directly be derived from substrate binding and recognition. A combination of enzyme inhibition and NMR studies suggests that the presence of Phe-36 slows the off-rate of the fluorinated substrate from FlK following initial binding. These studies also show that the increased residence time of the substrate requires both fluorine and Phe-36, and the effect is lost if either partner is not present.
To gain more molecular insight into this process, 2D-NMR experiments were carried out with FlK with both the fluorinated and unfluorinated carba(dethia)-substrate analog. These studies show that indeed a specific interaction exists between Phe-36 and the fluorine substituent, as the peak assigned to Phe-36 only shifts upon binding of the fluorinated inhibitor. Interestingly, not only does the Phe-36 chemical environment change, its dynamic motion is also reduced along with a second residue on the lid, Phe-40. As such, the slower off-rate of the fluorinated analog and reduced mobility of Phe-36 may be related to the reduction of the rate at which Phe-36 samples the open and closed conformations upon ligand binding. The reduced motion of lid residues upon binding suggests that ordered water may serve as the entropic contributor to fluorine discrimination. One possibility is that there is an increased release of ordered waters upon binding of the fluorinated ligand, as the C-F unit is predicted to be more hydrophobic than the corresponding C-H bond (21, 41, 42). Another possibility is that Phe-36 facilitates formation of a hydrophobic aggregate between the fluorinated ligand and the lid with an overall smaller surface area compared with individual noninteracting solutes (acetyl–CoA or F36A FlK). It has been found that the entropy of solvation of these aggregates is less unfavorable (42), which could account for the observed behavior of FlK. Both of these models are consistent with our analysis of Phe-36, which was found in this study to sample open and closed states, even in the apo form, that appear to alter the solvent accessibility of the active site.
Although there are numerous examples in which unnatural fluorinated substrates can initiate unusual enzymatic reaction pathways based on the high polarity of the C-F bond, there are few enzymes that have evolved to take advantage of fluorine’s unique properties to achieve substrate selectivity. FlK has evolved to exploit multiple distinct features of the C-F bond, including both its polarity and its hydrophobicity, to achieve its remarkable substrate selectivity. While our previous work has shown that FlK relies in part on the high inductive polarization of the C-F bond to discriminate fluoroacetyl–CoA from its nonfluorinated congener, the results reported here show that the increased hydrophobicity of the C-F unit is also an important component for entropically driven substrate selectivity. This mechanism of fluorine discrimination is distinct from that found in analyses of synthetic enzyme inhibitors, in which enhanced ligand affinity conferred by fluorine is typically mediated by multiple dipolar interactions (4, 41). Taken together, these studies provide insight into naturally selected molecular recognition of fluorine and identify mechanisms for the design of bioactive fluorinated small molecules and fluorine-selective biocatalysts.
Materials and Methods
Competitive Inhibition of FlK with Substrate Analogs.
Steady-state kinetic experiments were performed using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) to detect release of free CoA as described previously (21). Enzymatic reactions were initiated by addition of wild-type or F36A FlK (5 nM for fluoroacetyl–CoA or 10 µM for acetyl–CoA) to reaction mixtures containing either fluoroacetyl–CoA (5–100 µM) or acetyl–CoA (100–1,000 µM) as the substrate. When included, competitive inhibitors were added at a concentration at or above the KI. Each rate was measured in triplicate. Kinetic parameters (kcat and KM) were determined by fitting the data to the equation V0 = Vmax[S]/(KM + [S]), where V0 is the initial rate and [S] is the substrate concentration, using Origin 6.0 (OriginLab Corporation). The KI was calculated according to the equation KI = [I]/(α − 1), where [I] is the inhibitor concentration and α is KM,app/KM.
Isothermal Titration Calorimetry.
ITC experiments were performed on a Microcal AutoiTC200 (GE Healthcare). Wild-type and F36A FlK were dialyzed against 100 mM Tris⋅HCl, pH 7.6 at 4 °C for at least 16 h. Acyl–CoA analog ligands were dissolved in water, carefully neutralized with 1 M NaOH, and lyophilized. The lyophilized powder was dissolved in the same dialysis buffer used for protein buffer exchange immediately before the experiment. Titrations were performed under low c-value conditions (43) in which a large (10–100-fold) excess of ligand was titrated into the protein solution. Acyl–CoA analogs (7.5–25 mM) were titrated into the cell containing wild-type or F36A FlK (15–100 µM) using a preinjection (0.5 µL) followed by 13 additional injections (3.2 µL). The preinjection point was discarded, and data were baseline corrected and integrated. Nonlinear curve fitting using the one set of sites model in Origin 7.0 software (Microcal) was used to determine the binding constant (K), the heat change (ΔH), and the entropy change (ΔS). Due to the nature of the low c-value experimental design (43), stoichiometry of the interaction could not be fit and was held constant at 1:1 ligand:protein, a reasonable assumption based on the available structural information. Errors in the thermodynamic parameters are derived from nonlinear curve fitting.
1H/13C HSQC NMR Experiments.
The samples used for NMR experiments consisted of 160–280 µM of 13C-labeled FlK constructs in 50 mM sodium phosphate pH 7.6, 100 mM NaCl containing 10% deuterium oxide. Analog ligands (FAcCCoA and AcCCoA) were added from concentrated stock solutions in water. The 2D 1H/13C TROSY-HSQC experiments were performed on a Bruker Avance II 900 MHz spectrometer equipped with a TCI cryoprobe at 298 K, and the resulting spectra were processed using Topspin version 3.2.
Complete Materials and Methods.
Detailed procedures for the methods are described above, and additional experiments can be found in the SI Appendix, SI Materials and Methods. A full description of materials and methods, as well as supporting sequence alignments, NMR spectra, ITC, and biochemical data are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by the generous support of the National Institutes of Health (R01 GM123181). A.M.W. also acknowledges the support of a National Science Foundation Graduate Research Fellowship and an NIH NRSA (National Research Service Award) Training Grant (1 T32 GMO66698). The funds for the 900 MHz NMR spectrometer housed in the QB3 Institute in Stanley Hall at University of California, Berkeley were kindly provided by the NIH (GM68933).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717077115/-/DCSupplemental.
References
- 1.Jeschke P. The unique role of fluorine in the design of active ingredients for modern crop protection. ChemBioChem. 2004;5:571–589. doi: 10.1002/cbic.200300833. [DOI] [PubMed] [Google Scholar]
- 2.Purser S, Moore PR, Swallow S, Gouverneur V. Fluorine in medicinal chemistry. Chem Soc Rev. 2008;37:320–330. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- 3.Furuya T, Kamlet AS, Ritter T. Catalysis for fluorination and trifluoromethylation. Nature. 2011;473:470–477. doi: 10.1038/nature10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 5.O’Hagan D, Rzepa H. Some influences of fluorine in bioorganic chemistry. Chem Comm. 1997:645–652. [Google Scholar]
- 6.Smart BE. Fluorine substituent effects (on bioactivity) J Fluor Chem. 2001;109:3–11. [Google Scholar]
- 7.O’Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 8.Ippolito JA, Christianson DW. The contribution of halogen atoms to protein-ligand interactions. Int J Biol Macromol. 1992;14:193–197. doi: 10.1016/s0141-8130(05)80026-1. [DOI] [PubMed] [Google Scholar]
- 9.Finzel BC, et al. Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity. Protein Sci. 1998;7:2118–2126. doi: 10.1002/pro.5560071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairi M, Gerig J. Fluorine recognition at the active site of (N-(4-fluorophenyl)-N-phenylcarbamoyl)-alpha-chymotrypsin. J Am Chem Soc. 1984;106:3640–3643. [Google Scholar]
- 11.Gao J, Qiao S, Whitesides GM. Increasing binding constants of ligands to carbonic anhydrase by using “greasy tails”. J Med Chem. 1995;38:2292–2301. doi: 10.1021/jm00013a005. [DOI] [PubMed] [Google Scholar]
- 12.Wong DT, Bymaster FP, Engleman EA. Prozac (fluoxetine, Lilly 110140), the first selective serotonin uptake inhibitor and an antidepressant drug: Twenty years since its first publication. Life Sci. 1995;57:411–441. doi: 10.1016/0024-3205(95)00209-o. [DOI] [PubMed] [Google Scholar]
- 13.Roman DL, Walline CC, Rodriguez GJ, Barker EL. Interactions of antidepressants with the serotonin transporter: A contemporary molecular analysis. Eur J Pharmacol. 2003;479:53–63. doi: 10.1016/j.ejphar.2003.08.056. [DOI] [PubMed] [Google Scholar]
- 14.Olsen JA, et al. A fluorine scan of thrombin inhibitors to map the fluorophilicity/fluorophobicity of an enzyme active site: Evidence for C-F...C=O interactions. Angew Chem Int Ed Engl. 2003;42:2507–2511. doi: 10.1002/anie.200351268. [DOI] [PubMed] [Google Scholar]
- 15.Schweizer E, et al. A fluorine scan at the catalytic center of thrombin: C–F, C–OH, and C–OMe bioisosterism and fluorine effects on pKa and log D values. ChemMedChem. 2006;1:611–621. doi: 10.1002/cmdc.200600015. [DOI] [PubMed] [Google Scholar]
- 16.Drlica K, Malik M. Fluoroquinolones: Action and resistance. Curr Top Med Chem. 2003;3:249–282. doi: 10.2174/1568026033452537. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto J, et al. Pyridonecarboxylic acids as antibacterial agents. 2. Synthesis and structure-activity relationships of 1,6,7-trisubstituted 1,4-dihydro-4-oxo-1,8-naphthyridine-3-carboxylic acids, including enoxacin, a new antibacterial agent. J Med Chem. 1984;27:292–301. doi: 10.1021/jm00369a011. [DOI] [PubMed] [Google Scholar]
- 18.Clarke DD. Fluoroacetate and fluorocitrate: Mechanism of action. Neurochem Res. 1991;16:1055–1058. doi: 10.1007/BF00965850. [DOI] [PubMed] [Google Scholar]
- 19.Lauble H, Kennedy MC, Emptage MH, Beinert H, Stout CD. The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex. Proc Natl Acad Sci USA. 1996;93:13699–13703. doi: 10.1073/pnas.93.24.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang F, et al. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: A thioesterase confers resistance to fluoroacetyl-coenzyme A. Chem Biol. 2006;13:475–484. doi: 10.1016/j.chembiol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Weeks AM, Coyle SM, Jinek M, Doudna JA, Chang MC. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry. 2010;49:9269–9279. doi: 10.1021/bi101102u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker MC, Wen M, Weeks AM, Chang MC. Temporal and fluoride control of secondary metabolism regulates cellular organofluorine biosynthesis. ACS Chem Biol. 2012;7:1576–1585. doi: 10.1021/cb3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weeks AM, Chang MC. Catalytic control of enzymatic fluorine specificity. Proc Natl Acad Sci USA. 2012;109:19667–19672. doi: 10.1073/pnas.1212591109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weeks AM, Keddie NS, Wadoux RDP, O’Hagan D, Chang MCY. Molecular recognition of fluorine impacts substrate selectivity in the fluoroacetyl-CoA thioesterase FlK. Biochemistry. 2014;53:2053–2063. doi: 10.1021/bi4015049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song F, et al. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Wu R, Bryan TL, Dunaway-Mariano D. In vitro kinetic analysis of substrate specificity in enterobactin biosynthetic lower pathway enzymes provides insight into the biochemical function of the hot dog-fold thioesterase EntH. Biochemistry. 2009;48:511–513. doi: 10.1021/bi802207t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis MA, et al. Structure of YciA from Haemophilus influenzae (HI0827), a hexameric broad specificity acyl-coenzyme A thioesterase. Biochemistry. 2008;47:2797–2805. doi: 10.1021/bi702336d. [DOI] [PubMed] [Google Scholar]
- 28.Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry. 2009;48:1293–1304. doi: 10.1021/bi801879z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dias MVB, et al. Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK. J Biol Chem. 2010;285:22495–22504. doi: 10.1074/jbc.M110.107177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang PT, et al. Automated electron-density sampling reveals widespread conformational polymorphism in proteins. Protein Sci. 2010;19:1420–1431. doi: 10.1002/pro.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosin M, Spiteller D, Spencer JB. Malonyl carba(dethia)- and malonyl oxa(dethia)-coenzyme A as tools for trapping polyketide intermediates. ChemBioChem. 2009;10:1714–1723. doi: 10.1002/cbic.200900093. [DOI] [PubMed] [Google Scholar]
- 32.Nazi I, Koteva KP, Wright GD. One-pot chemoenzymatic preparation of coenzyme A analogues. Anal Biochem. 2004;324:100–105. doi: 10.1016/j.ab.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Clarke KM, Mercer AC, La Clair JJ, Burkart MD. In vivo reporter labeling of proteins via metabolic delivery of coenzyme A analogues. J Am Chem Soc. 2005;127:11234–11235. doi: 10.1021/ja052911k. [DOI] [PubMed] [Google Scholar]
- 34.Holmquist M. Alpha/Beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 35.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 36.Palmer AGI, Fairbrother WJ, Cavanagh J, Skelton N, Rance M. Protein NMR Spectroscopy, Second Edition: Principles and Practice. Academic; Cambridge, MA: 2006. [Google Scholar]
- 37.Teilum K, Brath U, Lundström P, Akke M. Biosynthetic 13C labeling of aromatic side chains in proteins for NMR relaxation measurements. J Am Chem Soc. 2006;128:2506–2507. doi: 10.1021/ja055660o. [DOI] [PubMed] [Google Scholar]
- 38.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pervushin K, Braun D, Fernández C, Wüthrich K. [15N,1H]/[13C,1H]-TROSY for simultaneous detection of backbone 15N-1H, aromatic 13C-1H and side-chain 15N-1H2 correlations in large proteins. J Biomol NMR. 2000;17:195–202. doi: 10.1023/a:1008399320576. [DOI] [PubMed] [Google Scholar]
- 40.Hattori Y, et al. Utilization of lysine 13C-methylation NMR for protein-protein interaction studies. J Biomol NMR. 2013;55:19–31. doi: 10.1007/s10858-012-9675-9. [DOI] [PubMed] [Google Scholar]
- 41.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Homans SW. Dynamics and thermodynamics of ligand–protein interactions. Top Curr Chem. 2007;272:51–82. [Google Scholar]
- 43.Turnbull WB, Daranas AH. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.