Significance
Bacterial infection is still a global threat to human health. Gene expression profiling of pathogenic bacteria during infection may aid in a better understanding of the pathophysiological events. Here, we present a reporter-based technology to identify in vivo repressed (ivr) genes of Vibrio cholerae. Our data demonstrate that constitutive expression of ivr genes reduces colonization fitness, indicating a pivotal role of gene silencing. A comprehensive characterization of an H+/Cl− transporter revealed a defined spatiotemporal expression pattern, being induced in the stomach to facilitate survival under low pH, but silenced under alkaline pH in the lower gastrointestinal tract to avoid detrimental effects. Thus, previously understudied ivr genes could reveal new insights into host–pathogen interaction and alternative therapeutic strategies.
Keywords: Vibrio cholerae, cholera, murine model, in vivo expression technology, acid tolerance response
Abstract
The facultative human pathogen Vibrio cholerae changes its transcriptional profile upon oral ingestion by the host to facilitate survival and colonization fitness. Here, we used a modified version of recombination-based in vivo expression technology to investigate gene silencing during the in vivo passage, which has been understudied. Using a murine model of cholera, we screened a V. cholerae transposon library composed of 10,000 randomly generated reporter fusions and identified 101 in vivo repressed (ivr) genes. Our data indicate that constitutive expression of ivr genes reduces colonization fitness, highlighting the necessity to down-regulate these genes in vivo. For example, the ivr gene clcA, encoding an H+/Cl− transporter, could be linked to the acid tolerance response against hydrochloric acid. In a chloride-dependent manner, ClcA facilitates survival under low pH (e.g., the stomach), but its presence becomes detrimental under alkaline conditions (e.g., lower gastrointestinal tract). This pH-dependent clcA expression is controlled by the LysR-type activator AphB, which acts in concert with AphA to initiate the virulence cascade in V. cholerae after oral ingestion. Thus, transcriptional networks dictating induction of virulence factors and the repression of ivr genes overlap to regulate in vivo colonization dynamics. Overall, the results presented herein highlight the impact of spatiotemporal gene silencing in vivo. The molecular characterization of the underlying mechanisms can provide important insights into in vivo physiology and virulence network regulation.
The Gram-negative bacterium Vibrio cholerae is the causative agent of the life-threatening secretory diarrheal disease cholera, which has an estimated global burden of 3 million cases per year (1). The rapid spread of cholera during an outbreak is highlighted by the current situation in Yemen, which has recorded more than 900,000 cases between October 2016 and November 2017 (2). As a facultative pathogen, V. cholerae transits between two very dissimilar environments, persisting in aquatic ecosystems between the outbreaks and colonizing the intestinal tract of the human host. Upon oral ingestion, V. cholerae passages through the stomach to reach the small intestine, its primary site of colonization. The induction of a complex regulatory cascade, summarized as the ToxR regulon, activates the expression of virulence factors. With the onset of acute secretory diarrhea, V. cholerae is released back into the aquatic environment. During interepidemic periods, V. cholerae is thought to persist outside of the host in estuarine and coastal aquatic reservoirs in a biofilm or viable but nonculturable state.
During its life cycle, V. cholerae has to cope with environmental and host-associated stressors (3). The fast adaptation of V. cholerae to different conditions is remarkable and represents a key feature for the transition fitness between the different stages of the V. cholerae life cycle allowing explosive spreads of cholera. Several studies have been conducted to analyze gene expression profiles of V. cholerae during the in vivo stage, in stool derived from cholera patients, in the transition from patient to aquatic environment, or during biofilm development (4–10). These studies have mainly focused on gene induction to reveal factors facilitating the fitness under the condition of interest. Less attention has been given to the genes silenced during a specific stage of the life cycle. One notable example is the work by Hsiao et al. (11), who demonstrated that repression of the mannose-sensitive hemagglutinin (MSHA) type IV pilus is essential for in vivo colonization. It is likely that MSHA represents just one example of a negatively regulated gene and that silencing the expression of many other genes occurs to allow proper colonization and achieve full virulence in vivo.
Diverse techniques are available for V. cholerae to investigate differential gene expression, including reporter-based screens such as in vivo expression technology (IVET) and recombination-based in vivo expression technology (RIVET), as well as microarrays or RNAseq. The latter have two main limitations, including the consequences of averaging heterogeneity in the bacterial population and the restriction to a snapshot analysis (12, 13). Any unique spatial or temporal gene expression patterns related to specific regions in the intestinal tract or a certain period during infection might be lost. However, recent studies indicate the presence of spatiotemporal expression profiles during V. cholerae in vivo colonization (5, 6, 14). In comparison with other technologies, the single cell-based reporter system RIVET is capable of detecting gene induction in subpopulations according to their spatial and temporal expression (14–16). RIVET resembles a temporally controlled reporter system allowing the detection of gene induction based on a resolvase (TnpR)-mediated excision of a reporter gene cassette (res cassette) flanked by TnpR-recognition sites (res). The induction of a gene transcriptionally fused with the promoterless tnpR results in the expression of TnpR and consequently leads to an irreversible excision of the res cassette containing two selectable marker genes. RIVET has been successfully applied in a variety of studies to identify genes induced during in vivo colonization or biofilm formation (14, 17–22) but currently does not allow the detection of genes that are transcriptionally silenced.
Here, we aimed to investigate genes repressed by V. cholerae during intestinal colonization using an advanced version of the recombination-based expression technology. Having identified 101 in vivo repressed genes, initial characterization of a subset revealed that, in the majority of cases, in vivo silencing is essential to achieve full colonization fitness. Furthermore, the detailed characterization of clcA revealed that expression is tightly controlled spatiotemporally. ClcA is involved in the acid tolerance response of V. cholerae, which facilitates survival upon oral ingestion and stomach passage but becomes detrimental for colonization in the alkaline environment of the intestine. Our findings also provide a physiologically relevant characterization of the proposed function of chloride channel (CLC) family members as an electrical shunt for an outwardly directed proton pump along pH gradients in the environment (23).
Results
Introducing the TetR-Controlled Recombination-Based in Vivo Expression Technology.
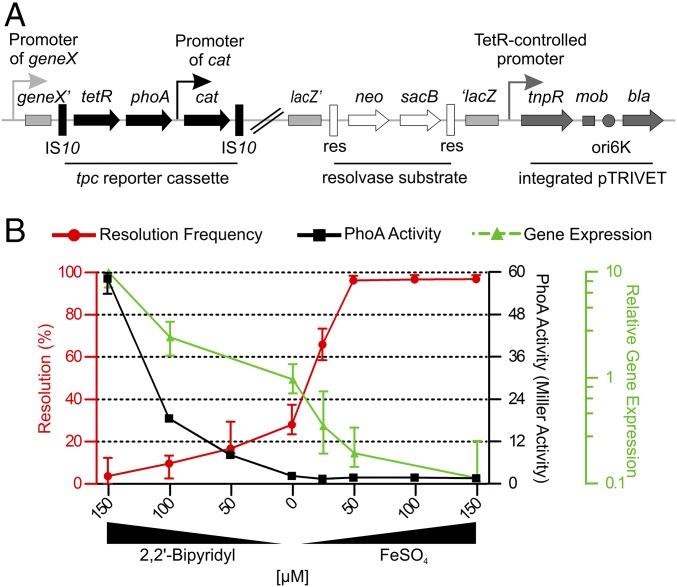
To comprehensively characterize gene repression during the in vivo stage of V. cholerae, we constructed a modified version of the resolvase-based technology (RIVET) using the transcriptional repressor TetR as an additional control element. TetR-controlled recombination-based in vivo expression technology (TRIVET) consists of three parts: a tetR-phoA-cat (tpc) reporter cassette, an integrated pTRIVET suicide plasmid carrying the TetR-controlled tnpR gene, and the TnpR target, the res cassette (Fig. 1A). The res cassette used for TRIVET, harboring neo and sacB genes, is identical to the latest RIVET version and integrates in the lacZ locus of the V. cholerae chromosome, conferring kanamycin resistance KmR and sucrose sensitivity SucS (18). The tpc cassette consists of promoterless tetR and phoA genes, encoding the TetR repressor and the alkaline phosphatase (PhoA), respectively. Furthermore, the tpc cassette contains a constitutively expressed cat cassette allowing direct selection via chloramphenicol resistance (CmR). When subcloned on a suicide vector, the tpc cassette can either be randomly inserted into the chromosome via transposon mutagenesis using pLOF (24) derivatives containing the Tn10 system, or integrated downstream of a specific promoter of interest via homologous recombination using pCVD442 (25) derivatives for gene allelic exchange in bacteria. In both cases, the expression of tetR and phoA is controlled by upstream chromosomal sequences carrying V. cholerae promoters. Finally, the suicide plasmid pTRIVET carries the TetR-controlled TnpR, as well as a homologous sequence for integration onto the V. cholerae chromosome downstream of the lacZ locus. The orientation of insertion thereof uses the transcriptional terminator of lacZ to prevent RNA polymerase read-through transcription of tnpR. Hence, the expression of tnpR solely relies on the TetR-controlled promoter. It should be noted that pLOF, pCVD442, and the pTRIVET share homologous sequences: e.g., the ori6K and RP4 mob region. Thus, the construction of the TRIVET library needs to follow an exact order to avoid interference of the individual suicide plasmids, as described in Methods (for details, see Construction of the V. cholerae TRIVET Libraries).
Fig. 1.
A resolvase-based screen to identify in vivo repressed genes. (A) Schematic illustration of the genetic components of the TetR-controlled in vivo expression technology (TRIVET). Chromosomal sequences are highlighted in light gray, tetR-phoA-cat (tpc) cassette in black, integrated pTRIVET in dark gray, and the res cassette parts in open shapes. The tpc cassette is integrated into a V. cholerae hypothetical geneX via Tn10 mutagenesis, resulting in a transcriptional fusion of tetR and phoA to the chromosomal promoter of geneX. Subsequently, pTRIVET is integrated downstream of lacZ. Thus, the expression level of TetR via the geneX promoter dictates the expression of TnpR, which catalyzes the excision of the res cassette. The IS10 elements and constitutive cat promoter of the tpc cassette, the TetR-controlled tnpR, mobilization (mob), origin of replication (oriR6K), and ApR (bla) regions of pTRIVET, as well as the gene for KmR (neo), SucS (sacB), and the target sites of resolvase (res) of the res cassette are indicated. (B) Shown are the effects of iron availability on resolution frequency (red) and alkaline phosphatase (PhoA) activity (black) of strain Vc_res1_TRIVET irgA::tpc, as well as the relative expression of irgA (green) in WT determined by qRT-PCR. LB was supplemented with varying amounts of 2,2′-bipyridyl or FeSO4 as indicated to mimic low and high iron concentrations, respectively. Resolution frequencies and PhoA activities were determined after 8 h while samples for gene expression analyses were taken during the exponential phase. Data are presented as the median with interquartile range (n ≥ 6).
Sufficient tetR expression via the integrated tpc cassette results in repression of tnpR and consequently a stable presence of the res cassette, which can be monitored via the resistance profile (KmR and SucS). In contrast, insufficient expression of tetR causes derepression of tnpR, resulting in excision and irreversible loss of the res cassette. Hence, the loss of the res cassette can be monitored by a phenotypic change to KmS and SucR, allowing the subsequent identification of resolved strains on sucrose plates similar to the latest RIVET screens (14, 17, 21, 22). The detectable gene repression spectrum was increased by modulation of the system sensitivity using variations of the res cassette and the tnpR-promoter element with the TetR-binding sites. In comparison with the WT res sites, the mutant res1 sequences contain a point mutation at their cross-over sites thought to result in a decreased recombination efficiency. Furthermore, the plasmids pTRIVET and pTRIVET1 differ in the tnpR allele promoter elements. The original promoter sequence of the tetracycline-resistance gene tetA was used for pTRIVET while a more tightly regulated version of the promoter is present in pTRIVET1, resulting in lower transcriptional efficacy of tnpR. Overall, the diverse combinations should allow a broad-spectrum identification of in vivo repressed genes altering expression levels from high to low, high to moderate, and moderate to low.
To confirm the functionality of TRIVET, the tpc reporter cassette was transcriptionally fused to the iron-regulated irgA promoter, which is highly expressed at low iron levels and repressed at high concentrations of iron (26–29). The combinations of res cassettes and pTRIVET versions resulted in the following four strains: Vc_res_TRIVET irgA::tpc, Vc_res_TRIVET1 irgA::tpc, Vc_res1_TRIVET irgA::tpc, and Vc_res1_TRIVET1 irgA::tpc. The extent of resolution and the PhoA activity was determined after 8 h growth in Luria–Bertani (LB) broth supplemented with varying amounts of the iron chelator 2,2′-bipyridyl or FeSO4 to mimic differential iron availability in vitro. Consistent with the reported transcriptional regulation of irgA, the level of PhoA activity decreased as the concentration of ferrous iron increased (Fig. 1B, black line). In contrast to variants of the res cassette and the tnpR promoter, all four strains harbored the same tpc reporter cassette and consequently an identical phoA allele. Thus, the measured PhoA activity for each of the four strains was similar at a given iron concentration. That is why the curve presented in Fig. 1B (black line) reflects the PhoA activities for all four strains at a given iron concentration. The PhoA activity reached very low levels, preventing detection of irgA gene silencing, especially after addition of FeSO4. Thus, the relative expression levels of irgA were additionally determined by quantitative real-time RT-PCR (qRT-PCR) along the tested iron concentrations. Consistent with the PhoA activity, the relative expression level of irgA decreased with increasing iron concentrations, but the decline was more steady and allowed monitoring of irgA repression along the range of iron concentration used in this study (Fig. 1B, green line). Conversely, the level of resolution for each of the strains increased with increasing the iron concentration, although with a different dose–response for each strain (Fig. 1B, red line and Fig. S1). The strains Vc_res_TRIVET irgA::tpc and Vc_res1_TRIVET irgA::tpc harboring the relatively strong TetR-controlled promoter upstream of tnpR resolved more readily than the corresponding strains Vc_res_TRIVET1 irgA::tpc and Vc_res1_TRIVET1 irgA::tpc, which contained pTRIVET1 with a more tightly regulated promoter element (Fig. S1). Strains with the more sensitive res cassette showed a marginally higher (only in a few cases significantly higher) resolution throughout the tested iron concentrations compared with isogenic strains, which contained the res1 mutant cassette (Fig. S1). In summary, all four strains followed dose-dependent resolution frequencies, although with different sensitivities.
To determine the dynamics of resolution upon shifting bacteria from low to high iron concentrations, we examined the kinetics of resolution frequencies for strain Vc_res_TRIVET irgA::tpc, which showed the greatest increase in resolution frequency between low and high iron concentrations. In parallel, repression of irgA was indirectly determined by assaying PhoA activity. Vc_res_TRIVET irgA::tpc was grown under iron-limiting conditions (LB supplemented with 150 µM 2,2′-bipyridyl) to logarithmic phase and then transferred into high iron media (LB supplemented with 25 µM FeSO4) and further cultivated for up to 8 h. After a lag time of ∼40 min, we observed a steep rise in resolution that reached a plateau at ∼240 min, with maximum resolution of ∼70% for the tested iron concentration. PhoA activities showed similar induction kinetics, but a more linear rise in activity, reaching the limit of detection also at ∼240 min (Fig. S2). As discussed previously (30), a direct comparison of these two reporter assays is limited by the enzymatic differences: i.e., resolvase-mediated DNA recombination of few substrate molecules in the cytoplasm vs. PhoA cleavage of an excess of substrate molecules in the periplasm. However, our results demonstrated that repression of irgA after transfer of the bacteria to high iron growth conditions resulted in a detectable increase of resolution from 60 to 240 min, which represents a suitable temporal window to identify repressed genes during in vivo colonization.
In Vivo Repressed Genes (ivr) Identified by in Vivo Library Screening.
The final TRIVET library with sufficient TetR expression to stabilize the res/res1 cassette in vitro comprised 20 independent pools with ∼500 independent transposon insertions per pool for each of the four different combinations Vc_res_TRIVET::pLOFtpc, Vc_res1_TRIVET::pLOFtpc, Vc_res_TRIVET1::pLOFtpc, and Vc_res1_TRIVET1::pLOFtpc.
The pools were used to intragastrically inoculate infant mice, followed by harvest of small intestines ∼22 h postinfection and plating of serial dilutions of the intestinal homogenates on LB-Sm/Suc plates. Resolved strains were subsequently screened for high PhoA activity (>10 Miller units) during in vitro growth to reduce the false positive rate. The exact insertion site of the tpc cassette was identified by sequence analyses of 186 resolved strains (Methods). In total, we identified 101 unique gene fusions that were repressed during in vivo colonization. A comprehensive list of the identified genes can be found in Table S3. Since our reporter system is based on detection of infection-repressed promoters, the table also indicates the predicted operons containing repressed genes. Genes involved in cellular and metabolic processes (40 genes) or associated with transport systems (11 genes) constitute the major groups. Previous reports focusing on in vivo induced genes describe similar results in terms of the gene distribution in these categories, indicating distinct changes in the cellular physiology during the in vitro to in vivo transition. This supports the hypothesis that V. cholerae faces differences in the nutrient content and availability inside the host compared with in vitro cultivation. Importantly, three ivr genes belong to the biosynthesis cluster of the MSHA pilus, which was recently reported to interfere in mucosal penetration and epithelial cell attachment if not silenced during in vivo colonization (11). The identification of the MSHA biosynthesis gene cluster thus validates the functionality of the TRIVET screen. Five ivr genes involved in regulatory systems have been identified, including the key regulator for the flagellar biogenesis FlrA. An additional four ivr genes are related to bacterial motility and chemotaxis. Based on the current model, flagellar motility is activated upon entrance in the human host to allow proper attachment and penetration through the mucosal layer. After the initial stage of infection and in agreement with our results, down-regulation of flagellar synthesis is essential to relieve the repression on virulence factors and enhance colonization fitness (31, 32). In contrast, bacteria initiate an RpoS-dependent mucosal escape response at later stages, where the bacteria up-regulate chemotaxis and motility genes, detach from the epithelial surface into the fluid-filled lumen, and eventually exit the host (5, 33). Notably, four ivr genes encode GGDEF/EAL proteins, which modulate levels of c-di-GMP, a secondary messenger that represses motility and virulence in V. cholerae (34). Hence, lowering c-di-GMP levels during infection is essential for colonization fitness (35).
Overexpression of ivr Genes Decreases in Vivo Fitness.
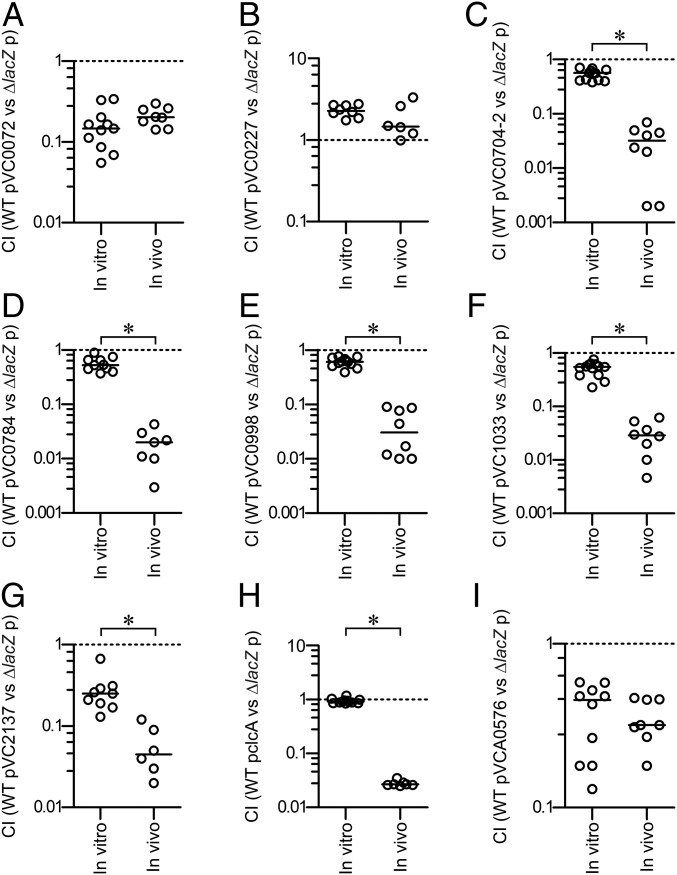
It is tempting to speculate that constitutive expression of ivr genes may have adverse effects on colonization fitness in the host. To test this hypothesis, we constructed strains constitutively expressing ivr genes by replacing the tetA gene of pBR322 with the ivr gene of interest. The chosen ivr genes represented diverse functional groups, including transporters, motility, c-di-GMP modulation, and LPS-biosynthesis. Subsequently, the colonization fitness of the resulting strains was analyzed in direct competition to the parental WT carrying the empty vector. Our results revealed that six of the nine strains constitutively expressing ivr genes showed a significant attenuation in vivo compared with the in vitro control (Fig. 2).
Fig. 2.
Constitutive expression of ivr genes reduces in vivo colonization fitness. Results are presented as the competitive index (CI) representing the output ratio of strains constitutively expressing the gene(s) of interest from a pBR322c derivative versus a fully virulent LacZ− derivative of the WT harboring the empty vector in LB broth (in vitro) and in vivo using the infant mouse model. The following competitions are shown: (A) WT pVC0072 vs. ∆lacZ p; (B) WT pVC0227 vs. ∆lacZ p; (C) WT pVC0704-2 vs. ∆lacZ p; (D) WT pVC0784 vs. ∆lacZ p; (E) WT pVC0998 vs. ∆lacZ p; (F) WT pVC1033 vs. ∆lacZ p; (G) WT pVC2137 vs. ∆lacZ p; (H) WT pclcA vs. ∆lacZ p; (I) WT pVCA0576 vs. ∆lacZ p. Each circle represents the CI from a single assay. Horizontal bars indicate the median of each dataset. The asterisks indicate significantly different medians of the in vivo compared with the respective in vitro dataset (n ≥ 6, P < 0.05, using a Mann–Whitney U test).
It should be taken into account that constitutive expression of ivr genes resulted, in some cases, in decent attenuation in vitro: e.g., in case of VC0072, VC2137, or VCA0576. In general, the pBR322 and its derivatives for constitutive expression of ivr genes were stably maintained for at least 16 h, even in the absence of antibiotic selection, while plasmids constitutively expressing VC0072 or VCA0576 represent two notable exceptions (Fig. S3). It should be mentioned that placing an ivr gene under control of the tetA promoter may rather cause an overexpression than a constitutive expression. Thus, we cannot exclude that interference in the natural expression level could induce stress, affect plasmid maintenance, and cause severe attenuation in vitro. Thus, for future studies, alternative strategies should be considered: e.g., expression of ivr genes from the chromosome using an in vivo active promoter.
The ivr genes with adverse effects on colonization fitness include the VC0702-4 operon, encoding the c-di-GMP phosphodiesterase MbaA and the polyamine sensor NspS, both involved in biofilm regulation, and a predicted NTPase with unknown function (36, 37). VC0998 (HubP) and VC2137 (FlrA) are involved in motility, with FlrA acting as the master regulator of V. cholerae flagellar biosynthesis. The transmembrane protein HubP is necessary for the proper polar localization of ParA-like proteins, such as the flagella gene regulator FlhG, thereby controlling the chemotactic machinery and the flagellum (38). Constitutive expression of transporter associated ivr genes [i.e., VC0784 (sodium/alanine symporter), VC1033 (zinc/cadmium/mercury/lead-transporting ATPase), and VCA0526 (chloride channel protein, ClcA)] also resulted in reduced colonization fitness in vivo, respectively. We found only three cases where constitutive expression did not result in a significant attenuation in vivo compared with in vitro condition: i.e., VC0072, VC0227, and VCA0576 (Fig. 2).
In summary, our results support the hypothesis that silencing of several ivr genes is crucial for V. cholerae to achieve proper colonization fitness.
ClcA Enhances Survival Fitness in the Stomach but Becomes Detrimental in the Lower Gastrointestinal Tract.
In particular, constitutive expression of ClcA resulted in a robust 50-fold attenuation in vivo compared with in vitro. ClcA belongs to the H+/Cl− transporters of the CLC family, which consists of integral membrane proteins responsible for translocating chloride through cell membranes. It was recently proposed that CLC family members in Escherichia coli may contribute to the acid resistance mechanism and function as an electrical shunt for an outwardly directed proton pump that is linked to arginine/agmatine (Arg+/Agm2+) or glutamate/GABA amino acid decarboxylation systems (23). In general, these systems move a positive charge outward (e.g., Arg+ exchanging for Agm2+). Consequently, an electrical shunt is needed to prevent the excessive inner membrane hyperpolarization that would otherwise paralyze the system. A similar situation would apply for V. cholerae, which uses the lysine decarboxylase CadA along with the lysine/cadaverin antiporter CadB to detoxify the protons leaking into the cell at low pH. Indeed, Ding and Waldor (39) reported in 2003 that a V. cholerae clcA mutant exhibits reduced acid resistance and enhanced in vivo fitness without further characterization of these observations.
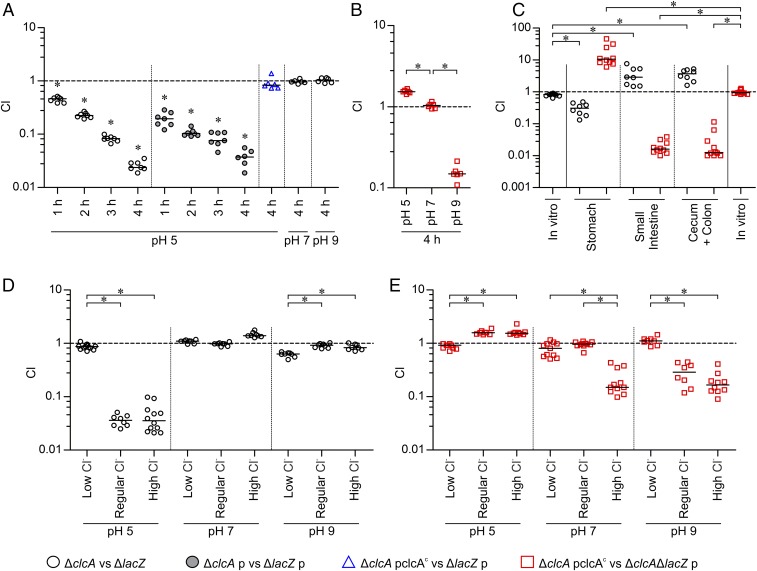
Intrigued by these facts, we compared the fitness of strains with altered ClcA expression in LB broth adjusted to different pH using HCl or NaOH, as well as during colonization of the mouse model using competition assays. Compared with the fully virulent ΔlacZ, the clcA mutant showed a significant defect in acidic conditions (pH 5, reflecting stomach pH of infant mice) (Fig. S4) after only 1 h of incubation, which steadily increased and resulted in severe attenuation within 4 h (Fig. 3A, black circles). In contrast, no disadvantage was observed under neutral (pH 7) and alkaline (pH 9) conditions. Presence of an empty vector (pBR322) in both strains (ΔclcA p vs. ΔlacZ p) did not massively impact the observed attenuation under acidic conditions (Fig. 3A, gray filled circles), excluding the possibility that the presence of a plasmid abolishes the phenotype due to altered bacterial growth. Constitutive expression of ClcA from a plasmid in trans completely restored the defect of ΔclcA (Fig. 3A, blue triangles).
Fig. 3.
ClcA affects survival fitness in acidic and alkaline environments in a chloride-dependent manner. Results are shown as the competitive index (CI) for colonization experiments comparing the following strains: ∆clcA to a fully virulent ∆lacZ (black circles), ∆clcA with empty vector (p) to ∆lacZ with empty vector (p, gray filled black circles), ∆clcA constitutively expressing clcA from a plasmid (∆clcA pclcAc) to ∆lacZ carrying the empty vector (p, blue triangles), or ∆clcA pclcAc to ∆clcA∆lacZ with empty vector (p, red squares). (A and B) In vitro competitions were performed for 1 to 4 h as indicated in LB adjusted to pH 5, 7, or 9 using HCl or NaOH, respectively. (C) Competitions were performed in vitro (LB, pH 7) or in vivo using the infant mouse model. (D and E) In vitro competitions were performed for 4 h in LB with various chloride concentrations adjusted to pH 5, 7, or 9 using H3PO4 or NaOH, respectively. Different chloride concentrations were achieved by addition of no NaCl (low chloride), 171 mM NaCl (regular), or 750 mM NaCl (high chloride), respectively. Each circle represents the CI from a single assay (n ≥ 6), the horizontal bars indicate the median of each dataset, and the asterisks indicate significant differences (P < 0.05) using either a Wilcoxon signed-rank test to indicate CIs significantly different to hypothetical value of 1 (A) or a Kruskal–Wallis test followed by post hoc Dunn’s multiple comparisons (B–E).
Next, we competed a strain constitutively expressing ClcA against the clcA mutant under acidic, neutral, and alkaline conditions. In comparison with a complete lack of ClcA, the constitutive expression of ClcA resulted in a slight, but significant, advantage at acidic pH, equal fitness under neutral pH, and a severe defect at alkaline pH (Fig. 3B). Consistent with the in vitro results, deletion of clcA resulted in a defect compared with the WT in the stomach, reflecting acidic environments (Fig. S4), but higher colonization fitness along the small intestine and cecum with colon, representing alkaline environments (Fig. 3C, black circles). In contrast, the strain constitutively expressing ClcA had a significant advantage over the clcA mutant in the stomach but exhibited severe attenuation in the small intestine and cecum with colon (Fig. 3C, red squares). These results highlight the essential role of ClcA for survival at low pH, its dispensability at neutral pH, and a detrimental effect when present at alkaline pH.
Notably, the low pH condition was achieved by addition of HCl. Thus, the chloride levels also changed within the in vitro cultivation and may affect the fitness of strains with altered ClcA expression. To decipher the impact of chloride levels and pH on the observed fitness defects, we performed competitions between the strains with altered ClcA expression (ΔclcA vs. ΔlacZ or ΔclcA pclcAc vs. ΔclcAΔlacZ p) in LB broth adjusted to different pH (using H3PO4 or NaOH) and varying Cl− concentrations, respectively (Fig. 3 D and E). Consistent with previous results obtained by adjusting low pH using HCl, the clcA mutant showed a pronounced defect in broth supplemented with regular or high chloride levels (Fig. 3 A and D). In contrast, low chloride levels negated the growth defect of ΔclcA to WT levels under acidic conditions. At neutral or alkaline conditions, the clcA mutant and WT competed equally for almost all chloride concentrations tested. Only under alkaline condition and low chloride was a slight growth defect of ΔclcA observed (Fig. 3D).
In full concordance with previous results, the constitutive expression of ClcA resulted in a marked attenuation at alkaline pH with regular or high chloride levels (Fig. 3E). However, the defect was absent in alkaline broth with low chloride levels. Furthermore, the results indicate that constitutive expression of clcA can be disadvantageous even at neutral pH, if chloride levels are elevated. High chloride levels are only tolerated under acidic conditions by a strain constitutively expressing clcA, and, under such conditions, the clcA allele can confer a slight advantage over the WT. Thus, the initially observed effects of ClcA on growth fitness under acidic and alkaline environments can be refined. Sufficient chloride concentrations are necessary to cause a ClcA-dependent fitness advantage under acidic conditions or cause a defect under alkaline conditions. In low chloride concentrations, ClcA-dependent disadvantages are negligible, probably due to the limiting amounts of chloride acting as counter ion for the H+/Cl− transporter.
Expression of clcA Is pH-Dependent and Regulated by AphB.
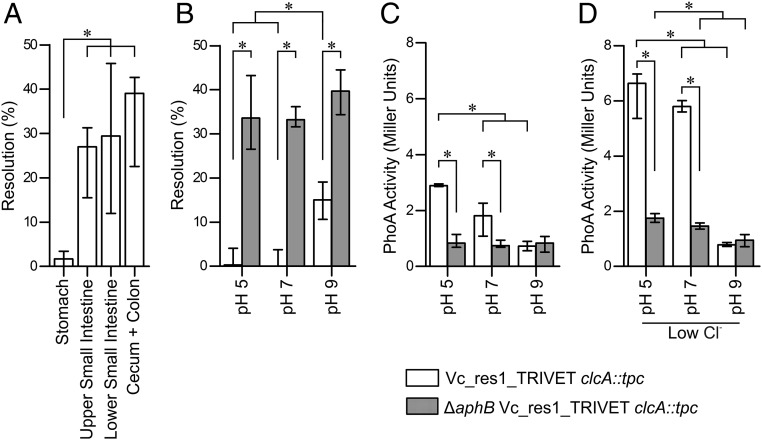
Upon oral ingestion by the host, V. cholerae has to passage through the low pH of the stomach, which implies the induction of factors mediating acid resistance: e.g., ClcA. Thus, the silencing of clcA to allow its identification as an ivr gene should occur in the lower gastrointestinal tract. To elucidate the spatial silencing expression of clcA in vivo and further investigate transcriptional regulation in vitro, the unresolved strain Vc_res1_TRIVET clcA::tpc was reconstructed. This strain harbors the tpc cassette transcriptionally fused to the promoter of clcA together with a TetR-controlled resolvase and the res1 cassette. Thus, the expression level of ClcA directly correlates with the PhoA activity while the resolution frequency follows an inverse relationship. To identify the spatial repression of clcA in vivo, resolution frequencies of Vc_res1_TRIVET clcA::tpc were assayed in different parts of the murine gastrointestinal tract (Fig. 4A). While almost no resolution could be detected in the stomach, resolution frequencies significantly increased in the upper and lower small intestine, to reach the highest median levels of ∼40% in the colon. These data suggest that clcA is expressed in the stomach, followed by a silencing of the gene expression in the small intestine and cecum with colon. Concordant with these in vivo results, the in vitro cultivation of the same strain in LB adjusted to pH 5, pH 7, and pH 9 confirmed elevated resolution frequencies at alkaline conditions, compared with neutral or acidic conditions (Fig. 4B, open bars). Conversely, PhoA activity was highest under low pH and significantly dropped with increasing pH to reach the lowest levels during growth in alkaline conditions (Fig. 4C, open bars). Based on the measured PhoA activities, clcA expression is even enhanced under low chloride conditions at acidic and neutral pH (Fig. 4 C and D). Thus, the absence of the growth defect of ΔclcA cannot simply be explained by a repression of clcA in the WT at low chloride (Fig. 3D).
Fig. 4.
Transcriptional regulation of clcA is pH-dependent mediated via AphB. (A) Shown are the in vivo resolution frequencies of strain Vc_res1_TRIVET clcA::tpc in the stomach, upper/lower small intestine, and colon with cecum. (B–D) In vitro resolution frequencies and PhoA activities of the strains Vc_res1_TRIVET clcA::tpc (open bars) and ∆aphB Vc_res1_TRIVET clcA::tpc (gray bars) are shown after overnight cultivation in LB adjusted to various pH as indicated. Differential chloride concentrations were achieved by addition of no NaCl (low chloride) or 171 mM NaCl (regular). (A–D) Results are shown as median with interquartile range and asterisks indicate significant differences between the datasets (n ≥ 6, P < 0.05, using a Kruskal–Wallis test followed by post hoc Dunn’s multiple comparisons).
Expression patterns of clcA in the WT were confirmed by qRT-PCR, with relatively high expression of clcA during growth in acidic conditions and low during growth in alkaline conditions in comparison with neutral conditions, respectively (Fig. S5, black circles). Unfortunately, we failed to detect any signal for clcA from in vivo derived samples, indicating that the in vivo expression levels of clcA are below the limit of detection, which corresponds to an at least 10-fold lower expression level compared with neutral in vitro conditions (Fig. S5).
Notably, a recent study by Kovacikova et al. identified the LysR-type activator protein AphB as a key regulator for genes involved in the V. cholerae response to anaerobic conditions and acidic pH. AphB was shown to activate several genes by binding to their promoter regions, including clcA and cadC, encoding the activator for the cadBA operon, which encodes a lysine/cadaverine antiporter and lysine decarboxylase relevant for survival in acidic environments (40). To verify whether AphB is responsible for the pH-dependent expression pattern of ClcA, we assessed the resolution frequency, the PhoA activity, and the clcA expression level in a ∆aphB background (Fig. 4 B and C, gray bars, as well as Fig. S5, red circles). The observed high-resolution frequencies, low PhoA activities, and low clcA expression levels even in cultivation of acidic conditions indicate a loss of induction upon deletion of AphB. This confirms that AphB is essential for the clcA induction upon low pH. Furthermore, neither the resolution frequency, the PhoA activity, nor the clcA expression level significantly changed upon variation of the pH in a ∆aphB background. Thus, AphB is essential for the pH-dependent expression pattern of ClcA.
Furthermore, AphB still acts as an activating element under low chloride levels. Notably, at acidic and neutral pH, the overall PhoA activities under low chloride were generally higher, compared with regular chloride in both strains tested: i.e., Vc_res1_TRIVET clcA::tpc and ∆aphB Vc_res1_TRIVET clcA::tpc, respectively (Fig. 4 C and D). This result suggests that clcA expression is activated under low chloride levels in addition to AphB by a currently unknown factor.
In summary, the results imply an AphB-dependent activation of clcA expression in acidic environments, as well as a chloride-dependent regulation resulting in high clcA expression at low chloride levels. Concordantly, ClcA is crucial for survival fitness of V. cholerae in the presence of chloride. Interestingly, under alkaline conditions, ClcA not only becomes dispensable, but its expression is even detrimental for survival fitness in the presence of chloride.
Discussion
A hallmark of the V. cholerae life cycle is the transition between the aquatic reservoir and the gastrointestinal tract of the human host. Both are complex environments with diverse niches and varying conditions. V. cholerae adapts to the diverse conditions presented during different aspects of its life cycle by changing the transcriptional profile. Therefore, V. cholerae encodes a large number of transcriptional regulators (157), two component systems [59 according to ref. 41], and complex signaling pathways (e.g., cAMP, c-di-GMP, or quorum sensing).
While several studies have focused on genes induced by V. cholerae during a defined stage in the life cycle to identify factors facilitating survival fitness, silencing of genes has been somewhat neglected. It could be hypothesized that repression of certain genes during the in vivo colonization is as important as the induction of virulence factors. In this study, we successfully established TRIVET, representing a modified variant of the recombination-based technology, and identified 101 ivr genes of V. cholerae. As seen with other techniques, a recombination-based reporter technology has certain limitations, but also unique advantages, such as single cell expression profiling and detection of transient gene silencing during the entire infection period. The implementation of phoA as a second reporter was used to eliminate false positives by enabling confirmation of substantial expression levels during growth in vitro for each ivr candidates. Moreover, phoA is a valuable reporter to analyze ivr gene expression under diverse in vitro conditions, as exemplified for clcA in this study. In six out of nine cases, constitutive expression of an ivr gene resulted in significant attenuation in vivo compared with the WT. Thus, silencing of ivr genes seems to be crucial to achieve full colonization fitness. For most ivr genes we identified, additional studies are needed to elucidate whether their constitutive expression results in a fitness disadvantage in vivo, including the underlying molecular mechanisms relevant for these phenotypes. One notable exception is the identified ivr genes of the MSHA biosynthesis gene cluster, as it has been recently reported that MSHA binds secreted immunoglobulins in a non–antigen-specific manner, which blocks mucosal penetration and epithelial cell attachment of the pathogen (11). As mucosal penetration is required for proper virulence gene induction (42), evasion of the host innate immune response via repression of MSHA during early stages of infection is critical for V. cholerae to achieve full colonization fitness.
Within this study, we focused on the ivr gene clcA, encoding an H+/Cl− exchange transporter, proposed to exchange two chloride ions for one proton (43). Recently, Iyer et al. showed that E. coli uses the CLC family of chloride channels to survive low pH conditions in vitro. The proposed mechanism links the chloride channels to the bacterial acid resistance response mediated via an amino acid decarboxylation system and functions as an electrical shunt for an outwardly directed proton pump (23). The authors already suggest that such a chloride channel should be beneficial in the context of the HCl-rich stomach but did not present direct evidence. Our study links the ClcA function to its physiological role in the stomach, using V. cholerae as a model organism, which needs to pass through the acidic stomach to reach the primary colonization site in the small intestine (Fig. 5). The CadAB lysine decarboxylation system of V. cholerae, which consumes lysine and a proton and produces carbon dioxide and cadaverine, plays a major role in the acid tolerance response and is critical for full colonization fitness (44). Our results indicate that ClcA enhances survival fitness in acidic environments like the stomach but becomes detrimental at neutral conditions with high chloride levels or alkaline environments like the intestinal tract. Fitness defects of clcA mutants in acidic conditions and strains constitutively expressing clcA in alkaline conditions are negated at low chloride levels, highlighting the activity of ClcA as an H+/Cl− exchange transporter. Notably, Iyer et al. (23) previously hypothesized that such chloride channels must be tightly regulated to prevent H+/Cl− exchange at neutral or alkaline conditions, which would be disruptive to energy metabolism. As shown in this study, clcA is transcriptionally regulated by AphB in a pH-dependent manner, being highly expressed in acidic conditions and expressed at low levels in alkaline conditions. Interestingly, the transcriptional regulatory protein AphB initiates the expression of the virulence cascade upon transition into the host by activating the expression of the membrane-bound transcriptional regulatory complex TcpPH (40). We note with great interest that AphB was shown to be pH- and oxygen-responsive, with an active conformation at low pH or under low oxygen tension, but it remains inactive at alkaline pH (45, 46). In addition, the AphAB complex is repressed by the quorum-sensing regulator HapR, reported to be activated during the course of infection (5, 47). These structural and regulatory observations of AphB activity are consistent with pH-dependent regulation and the resulting repression of clcA in lower parts of the gastrointestinal tract.
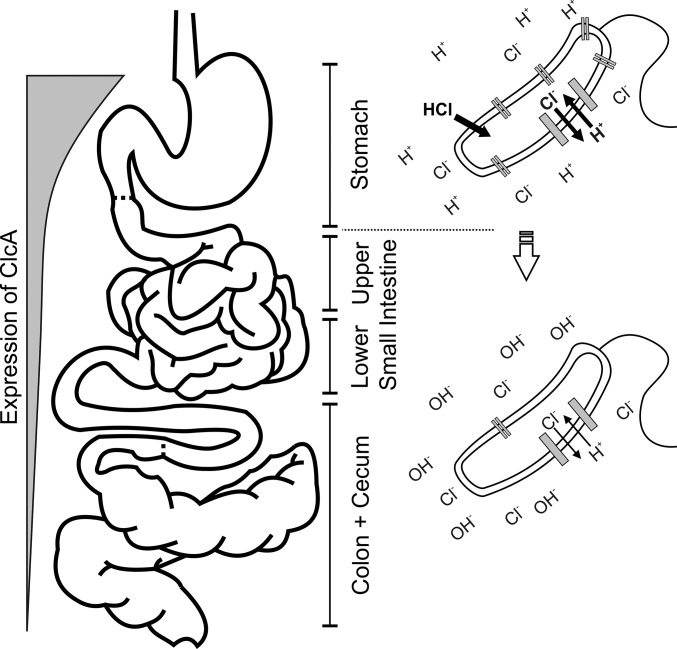
Fig. 5.
Model of the H+/Cl− transporter (ClcA) requirements along the gastrointestinal tract. Upon oral ingestion, V. cholerae passages through the stomach (acidic environment), and clcA is activated by AphB. ClcA is essential to detoxify the bacteria from chloride and acts as an electrical shunt to prevent the excessive membrane hyperpolarization. The expression of clcA decreases once V. cholerae passes to the lower gastrointestinal tract (alkaline environment), which is crucial to achieve full colonization fitness. Under alkaline conditions, activity of ClcA would exploit the proton motive force and disrupt energy metabolism.
The investigation of in vivo repressed genes identified by TRIVET adds a fundamental perspective to understanding bacterial transcriptional response upon host contact. The results of this study emphasize the impact of regulated silencing of genes by a bacterial pathogen during in vivo colonization. The importance of such conditional expression is exemplified by the spatial on–off state of clcA along the gastrointestinal tract to accommodate the changing needs of the pathogen within different niches, resulting in an overall in vivo fitness advantage. Interestingly, clcA expression is controlled by AphB, which is also involved in the initiation of the virulence cascade. This indicates that positive and negative regulation is necessary to define such complex networks like virulence gene regulation during host colonization. Although previously poorly investigated, our studies contribute to an understanding of gene silencing that will likely reveal new insights in host–pathogen interactions and physiology for a variety of pathogens during in vivo colonization.
Methods
Bacterial Strains and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in Table S1. If not otherwise noted, the strains were cultured in Luria–Bertani (LB) broth at 15 × g with aeration at 37 °C. Supplements were used in the following final concentrations: streptomycin (Sm, 100 µg/mL), ampicillin (Ap, 100 µg/mL or 50 µg/mL in combination with other antibiotics), kanamycin (Km, 50 µg/mL), chloramphenicol (Cm, 2 µg/mL for V. cholerae; 10 µg/mL for E. coli), sucrose (10%), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal, 40 µg/mL).
Construction of the TRIVET System.
The TRIVET system required the construction of several elements, partially based on components of the recombination-based in vivo expression technology. To construct the tetR-phoA-cat (tpc) cassette harboring promoterless tetR and phoA, but constitutively expressed cat (CmR), respective fragments were amplified as follows: tetR using chromosomal DNA from XL-1 and oligonucleotide pairs tetR-5′-BamHI and tetR-3′-Kpnl, phoA using chromosomal DNA from SM10λpir oligonucleotide pairs phoA-5′-KpnI and phoA-3′-XbaI, and cat from pAC1000 and oligonucleotide pairs cat-5′-XbaI and cat-3′-BamHI. PCR fragments were digested with the appropriate enzymes given by the name of the oligonucleotide and ligated into a BamHI-digested pTrc99A-Km to obtain pTrc-tpc. From there, the tpc cassette was amplified using oligonucleotide pairs tetRphoAcat-5′-NotI and tetRphoAcat-3′-NotI, and the obtained PCR fragment was digested with NotI and ligated into a similarly digested pLOFKm to obtain pLOF::tpc, which now harbored the tpc cassette within the transposable element. The vectors pTRIVET and pTRIVET1 are derivatives of pGP704, which contain an oriR6K origin that requires the host-encoded protein π (pir gene product) for replication, the broad-host-range mobilizable region mobRP4, bla for ampicillin resistance, an ∼800-bp fragment of the 3′ end of lacZ derived from V. cholerae allowing insertion into the chromosome via homologous recombination, and a TetR-controlled allele of tnpR. For the construction of these vectors, the lacZ fragment was amplified from chromosomal DNA from V. cholerae WT using oligonucleotide pairs lacZ-5′-SacI and lacZ-3′-NheI, and, as well, tnpR was amplified from pGOA1193 using oligonucleotide pairs tnpR-5′-BgIII and tnpR-3′-XbaI and digested with the appropriated enzymes given by the names of the oligonucleotides. Furthermore, oligonucleotide pairs PtetA-5′-3′ and PtetA-3′-5′, as well as Ptet1-5′-3′ and Ptet1-3′-5′, were designed to comprise the promoter sequence of tetA or a variant, resulting in a more stringent transcriptional regulation, respectively (48, 49). As these oligonucleotide pairs form a DNA fragment with compatible NheI and BglII sites upon hybridization, they were directly used for ligation together with the digested lacZ and tnpR fragment into a SacI/XbaI-opened pGP704, resulting in pTRIVET (containing promoter PtetA) and pTRIVET1 (containing promoter Ptet1). In general, ligation products were transformed into DH5αλpir, and colonies with the appropriate antibiotic resistance profile were characterized by PCR and/or restriction analyses for the correct construct.
Construction of the V. cholerae TRIVET Libraries.
Suicide plasmids pRES and pRES1 were mobilized into the V. cholerae WT by conjugation, and allelic exchange was used to place each cassette into the lacZ locus as previously described (18) to obtain Vc_res and Vc_res1. For transposon mutagenesis of Vc_res and Vc_res1, pLOF::tpc was transferred into the strain via filter mating with an E. coli SM10λpir mating strain harboring the respective plasmid (24). From each mating, ∼500 colonies of transposon mutants were selected for SmR, KmR, CmR, and ApS and pooled together. To obtain several independent transposon pools for Vc_res and Vc_res1, the mating and selection process was repeated 20 times for each recipient strain. Finally, pTRIVET or pTRIVET1 were mobilized into each pool via conjugation and integrated downstream of the lacZ locus via homologous recombination. Unresolved colonies of these TRIVET libraries were selected for SmR, KmR, CmR, and ApR, which resulted in four different combinations of library pools: Vc_res_TRIVET::pLOFtpc, Vc_res1_TRIVET::pLOFtpc, Vc_res_TRIVET1::pLOFtpc, and Vc_res1_TRIVET1::pLOFtpc. A prescreening step to eliminate fusion strains with low levels of resolution in vitro and thereby reducing the number of false positives was performed before the in vivo screening. This was achieved by growing each pool in LB broth to late log-phase in the absence of Km selection and then plating serial dilutions on LB-Sm/Km/Ap plates. After overnight incubation, colonies from plates with defined single colonies (at least 1,000 per pool) were recombined. In total, the TRIVET library consisted of 20 independent pools with ∼500 independent transposon insertions for each of the four combinations. Heterogeneity of transposon insertions before and after conjugation with pTRIVET or pTRIVET1, as well as after the prescreening, was verified by Southern blot analyses of 50 randomly picked colonies from each pool.
It should be emphasized that pRES variants, pLOF, the pTRIVET and pCVD442 derivatives for deletion mutagenesis share homologous sequences: e.g., the ori6K and RP4 mob region. Thus, the construction of the TRIVET library or strains harboring a TRIVET reporter system need to follow the order described above to avoid interference of the individual suicide plasmids.
Construction of Suicide Plasmids, Deletion Mutants, and Expression Plasmids.
DNA manipulations, including purification of chromosomal, plasmid, or PCR product DNA, PCRs, as well as construction of in-frame deletion mutants and expression plasmids, were carried out as described previously using derivatives of pCVD442 or pBR322, respectively (14, 50). Oligonucleotides used in this study are listed in Table S2. Briefly, constructions of in-frame deletion mutants and tpc-cassette insertions were carried out as described by Donnenberg and Kaper (25). In general, ∼800-bp PCR fragments located upstream and downstream of the gene of interest were amplified using the oligonucleotide pairs A_B_1 and A_B_2 or A_B_3 and A_B_4, in which A stands for the gene and B for the restriction site used (Table S2). After digestion of the PCR fragments with the appropriate restriction enzyme (New England Biolabs) indicated by the name of the oligonucleotide, they were ligated into pCVD442, which was digested with the appropriate restriction enzymes. Suicide plasmids for construction of transcriptional fusions of the tpc cassette to the irgA or clcA promoter on the V. cholerae chromosome were obtained as follows: The tpc cassette was amplified from pTrc-tpc using the oligonucleotides tetRphoAcat-5′-SacI and tetRphoAcat-5′-SacI. Eight hundred bp PCR fragments located upstream and downstream of irgA or clcA were amplified using the oligonucleotides listed in Table S2 (indicated by †). After digestion of the PCR fragments with the appropriate restriction enzyme (New England Biolabs) indicated by the name of the oligonucleotide, they were ligated into an SphI/XbaI-digested pCVD442. After transformation of the ligation products in DH5αλpir, ApR colonies (and CmR if the tpc cassette was present) were characterized by PCR and/or restriction analysis. In the case of the suicide plasmid derivatives, the correct constructs were transformed into SM10λpir and mobilized into V. cholerae by conjugation, which was achieved by cross-streaking donor and recipient on LB agar plates, followed by incubation for 6 h at 37 °C. V. cholerae conjugants were purified via selection for SmR/ApR (and CmR if the tpc cassette was present) colonies. In the case of pCVD442 derivatives, sucrose selection was used to obtain Aps colonies. Chromosomal deletions of tpc-cassette insertions were confirmed by PCR, respectively. All plasmids allowing constitutive expression of the genes were constructed in a similar manner by insertion of the respective gene into the tet gene of pBR322. PCR fragments of the interested gene spanning from the Shine–Dalgarno sequence to the stop codon were amplified using the oligonucleotide pairs A_5′_B and A_3′_B, in which A stands for the gene and B for the restriction site used (Table S2). PCR fragments were digested with the appropriate restriction enzyme (New England Biolabs) indicated by the name of the oligonucleotide and ligated into a similar digested pBR322. After transformation of the ligation products in DH5αλpir, ApR/TetS colonies were characterized by PCR and/or restriction analysis.
Evaluation of Plasmid Maintenance in the Absence of Antibiotic Selection.
The stability of pBR322 or derivates used for overexpression of ivr genes was monitored by growing appropriate strains (WT harboring p, pVC0072, pVC0227, pVC0704-2, pVC0784, pVC0998, pVC1033, pVC2137, pclcA, or pVCA0576) in LB-Ap to late exponential phase to be used as inoculum for a test culture (LB-Sm; starting OD600 = 0.05) incubated overnight. Maintenance of plasmids was determined by plating appropriate dilutions of the inoculum and the test culture on LB-Sm and LB-Ap. Stability for each plasmid derivative is given as the ratio of ApR/SmR cfu normalized for the ApR/SmR ratio of the inoculum.
Screening for in Vivo Repressed (ivr) Genes of V. cholerae.
An aliquot of each pool of the library was spread in triplicate on LB-Sm/Km/Ap plates. After overnight incubation, ∼5,000 colonies were collected from each plate and diluted in LB to ∼106 cfu/mL, and 50 µL were used to intragastrically inoculate 5-d-old CD-1 mice (anesthetized by isoflurane) as previously described (14, 30). Approximately 22 h postinfection, mice were euthanized, and their small bowels were removed and homogenized in 1 mL of LB plus 20% Gly. Serial dilutions of the homogenate were plated on LB agar lacking NaCl and supplemented with 10% Suc and Sm to select for resolved strains lacking the res or res1 cassette. After incubation overnight at 30 °C, eight SucR KmS colonies were picked from each mouse output, grown in LB, and stored at −80 °C in 96-well plates (Greiner) in LB plus 20% Gly.
Resolvase-based screens have a false positive rate of approximately 15% based on previous reports (14, 17, 22). In RIVETscreens, such false positives are eliminated by reconstruction of the original unresolved strains and assaying the resolution frequencies in vitro and in vivo. A ≥twofold increase in resolution during cultivation of the test condition compared with control condition has been generally applied as a very stringent criterion for bona fide induced genes (14, 22), which so far allowed confirmation by quantitative real time RT-PCR of all bona fide categorized genes tested. As the TRIVET system has more components, a reconstruction for all strains is relatively complicated and time-consuming. By reanalysis of the combined data acquired by previous resolvase-based screens (14, 17, 22), it appears that strains with in vitro resolution frequencies below 30% have only a 5% chance to be false positive. As the tpc cassette remains stably integrated in the chromosome, PhoA activity of in vivo resolved strains can be measured under in vitro conditions and correlate the obtained activity to a resolution frequency. According to the results obtained for the irgA-fusion test strain, an in vitro resolution frequency of 30% or lower correlates with PhoA activities of 10 Miller units or higher (Fig. 1). Thus, a simple PhoA activity assay of resolved TRIVET strains after in vivo passage is an efficient tool to reduce the number of false positives. Importantly, all of the nine identified in vivo resolved strains (Tables S1 and S3) were subjected to this validation step and exhibited an in vitro PhoA activity higher than 10 Miller units, which reinforces the overall feasibility of the TRIVET system.
To identify the exact insertion of the tpc cassette, chromosomal DNA was isolated from overnight cultures of the respective resolved strains, digested with EcoRI, and ligated into an EcoRI-opened pBR322. After transformation of the ligation products in DH5αλpir, selection for CmR allowed the identification of colonies harboring a plasmid with the functional cat gene of the tpc cassette, as well as adjacent chromosomal DNA of the V. cholerae genome. Isolated plasmids were used directly as templates in sequencing reactions with the oligonucleotide seqPrimer-cat that reads out from the 3′ end of cat. Sequences were compared with the V. cholerae N16961 genome database with blastN (blast.jcvi.org/cmr-blast/) to identify the exact position of the tpc-cassette insertion. We considered transcriptional fusions of tetR to any annotated ORF within which it had inserted in the same orientation, as well as any annotated ORF in the same orientation lying ≤100 bp from the ribosomal binding site (RBS) of tnpR as long as no factor-independent transcriptional terminators were present.
Stomach pH Measurement of Infant Mouse.
Five-day-old CD-1 mice were separated from the dam for 1 h and injected with 50 µL of saline by gavage. After 20 min, the mice were killed, and stomach content was taken into 1 mL of saline. The mixture pH was measured by the Metrohm 827 pH laboratory. Results are presented as the logarithmic scale of pH.
Resolution Assay.
To quantify resolution, strains were first grown on LB-Sm/Km/Ap plates overnight and adjusted in LB-Sm/Km/Ap to OD600 = 1, which was used as inoculum. To determine in vitro resolution frequencies, the inoculum was diluted 1:100 in 5 mL of LB-Sm/Ap, LB-Sm/Ap with various pH or LB-Sm/Ap supplemented with different concentrations of 2,2′-bipyridyl or FeSO4 and incubated for up to 8 h. To determine the in vivo resolution, the inoculum was diluted 1:500 in LB, and anesthetized 5-d-old CD-1 mice were intragastrically inoculated with 50 μL of this dilution (∼106 cfu per mouse). Mice were euthanized at 22 h infection time, and V. cholerae were recovered as described above. At the given time point, the amount of resolution in vitro and in vivo was determined by plating appropriate dilutions on LB-Sm/Km and LB-Sm/Ap plates. Results were expressed as the percent resolution, calculated as SmR/KmS cfu (SmR/ApR cfu minus SmR/KmR cfu) divided by SmR/ApR cfu.
Alkaline Phosphatase Activity Assay.
To find out the activity of gene transcription, V. cholerae cultures were incubated in LB with varying supplements or pH (e.g., 2,2′-bipyridyl, FeSO4, pH 5, pH 7, or pH 9), and PhoA activity was determined as previously described (51). The activities were expressed in Miller units: 1,000 × [A405/(A600 × mL × min)].
Quantitative Real-Time RT-PCR.
Expression of irgA and clcA was determined by quantitative real-time RT-PCR (qRT-PCR). For in vitro studies, WT or ΔaphB were grown to an OD600 of 0.5 to 0.8 in LB either supplemented with varying amounts of 2,2′-bipyridyl and FeSO4 or adjusted to a different pH value. Bacterial RNA extraction, DNase digestion, cDNA synthesis, and qRT-PCR were performed as described previously (52). For in vivo studies, anesthetized 5-d-old CD-1 mice were intragastrically inoculated with 50 μL of WT adjusted to OD600 of 0.002 in LB, corresponding to an infection dose of ∼106 cfu per mouse. After 22 h, mice were euthanized, and small intestine and cecum with colon were removed by dissection and mechanically homogenized in 1.5 mL of TRIzol Reagent (ThermoFisher Scientific). RNA was isolated using the Monarch Total RNA Miniprep Kit (New England Biolabs) according to the manufacturer’s protocol, and chromosomal DNA was digested by using RQ1 RNase-Free DNase (Promega). Synthesis of cDNA and qRT-PCR were performed as described previously using the iScript Select cDNA Synthesis Kit (Bio-Rad) and the SYBR GreenER qPCR SuperMix for ABI PRISM (ThermoFisher Scientific) (53). Each cDNA sample was tested in triplicate. The sequences of the primers used for qRT-PCR starting with “qPCR-” are listed in Table S2. Results were analyzed using StepOne Software v2.1, and relative gene expression comparisons were calculated by the mean cycle threshold of samples, which were normalized to the housekeeping gene 16S rRNA (VCr001) and to one randomly selected reference sample. For expression analysis of irgA, the reference sample was selected from the WT grown in LB without addition of 2,2′-bipyridyl or FeSO4 while a sample derived from WT grown in LB pH 7 served as reference for the clcA expression analysis.
Competition Assays.
Competition assays for both in vivo and in vitro were performed by using in-frame deletion mutants (lacZ+) competed against the isogenic, fully virulent Vc_res strain (lacZ−) to allow differentiation between the strains on X-Gal plates as previously described (14, 18, 51). In case of strains with constitutive expression of ivr genes, WT (lacZ+) with the respective pBR322 expression derivative were competed against Vc_res (lacZ−) containing the empty pBR322, or deletion mutants (lacZ+) with the respective pBR322 expression derivative were competed against deletion mutants (lacZ−) containing the empty pBR322, respectively. Briefly, strains for competition were grown overnight on LB plates supplemented with the appropriate antibiotics, resuspended in LB, and mixed in a 1:1 ratio. For in vivo competitions, 5-d-old CD-1 mice were separated from their dams 1 h before infection. Subsequently, they were anesthetized by inhalation of isoflurane gas and then inoculated by oral gavage with 50 µL of an appropriate dilution of the 1:1 mixture (e.g., mutant and isogenic WT strain), resulting in an infection dose of ∼106 cfu per mouse. To determine the exact inputs, appropriate dilutions of the inocula were plated on LB-Sm/X-Gal plates. After 22 h, the mice were killed, and the stomach, small intestine, and colon with cecum from each mouse were collected by dissection. The individual parts of the gastrointestinal tract were mechanically homogenized in LB broth with 15% glycerol, and appropriate dilutions were plated on LB-Sm/X-Gal. In vitro competitions, simultaneously performed using the same inoculum of in vivo, were done by inoculation of 2 mL of LB with 1:1,000 of the inoculum (∼105 cfu). The cultures were incubated overnight at 37 °C with aeration and subsequently diluted in LB, and appropriate dilutions were plated on LB-Sm/X-Gal. For the assay of acid tolerance response, competition mixtures were adjusted to OD600 = 0.5 in LB with indicated pH. At indicated time points, appropriate dilutions were plated on LB plates supplemented with the appropriate antibiotics. The colonies of mutant and WT were determined by counting the cfu and back-calculation to the original volume of the homogenized tissue or LB culture. In general, results are presented as the competition index (CI), which is the ratio of mutant strain cfu to Vc_res cfu normalized for the input ratio.
Ethics Statement.
CD-1 mice (Charles River Laboratories) were used in all experiments in accordance with the rules of the ethics committee at the University of Graz and the corresponding animal protocol, which has been approved by Austrian Federal Ministry of Science and Research Ref. II/10b. Mice were housed with food and water ad libitum and monitored under the care of full-time staff.
Statistics.
Generally, the Mann–Whitney U test or Kruskal–Wallis followed by Dunn’s posttest was used for single or multiple comparisons, respectively. In case of competition assays in acidic environments, a Wilcoxon signed-rank test was used to indicate significant differences to a hypothetical value of 1. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Andrew Camilli (Tufts University, Boston) for providing the original RIVET components and helpful discussions to establish the TRIVET system; and Michelle Dziejman for critically reading the manuscript. This work was supported by Austrian Science Fund (FWF) Grants W901 (DK Molecular Enzymology) (to F.C., F.G.Z., M.M., J.R., and S.S.) and P29405 (to J.R.), as well as Grants P27654 and P25691 (to S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716973115/-/DCSupplemental.
References
- 1.Conner JG, Teschler JK, Jones CJ, Yildiz FH. Staying alive: Vibrio cholerae’s cycle of environmental survival, transmission, and dissemination. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0015-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization 2017 Cholera Situation in Yemen, November 2017. Available at applications.emro.who.int/docs/EMROPub_2017_EN_20214.pdf?ua=1. Accessed January 5, 2018.
- 3.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moorthy S, Watnick PI. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol. 2005;57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen AT, et al. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen AT, et al. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog. 2010;6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bina J, et al. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci USA. 2003;100:2801–2806. doi: 10.1073/pnas.2628026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandlik A, et al. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson EJ, et al. Transmission of Vibrio cholerae is antagonized by lytic phage and entry into the aquatic environment. PLoS Pathog. 2008;4:e1000187. doi: 10.1371/journal.ppat.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao A, Liu Z, Joelsson A, Zhu J. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci USA. 2006;103:14542–14547. doi: 10.1073/pnas.0604650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 13.Beloin C, Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Schild S, et al. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelichio MJ, Merrell DS, Camilli A. Spatiotemporal analysis of acid adaptation-mediated Vibrio cholerae hyperinfectivity. Infect Immun. 2004;72:2405–2407. doi: 10.1128/IAI.72.4.2405-2407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelichio MJ, Spector J, Waldor MK, Camilli A. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun. 1999;67:3733–3739. doi: 10.1128/iai.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osorio CG, et al. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect Immun. 2005;73:972–980. doi: 10.1128/IAI.73.2.972-980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilli A, Mekalanos JJ. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SH, Angelichio MJ, Mekalanos JJ, Camilli A. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J Bacteriol. 1998;180:2298–2305. doi: 10.1128/jb.180.9.2298-2305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Hava DL, Waldor MK, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 21.Lombardo MJ, et al. An in vivo expression technology screen for Vibrio cholerae genes expressed in human volunteers. Proc Natl Acad Sci USA. 2007;104:18229–18234. doi: 10.1073/pnas.0705636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seper A, et al. Identification of genes induced in Vibrio cholerae in a dynamic biofilm system. Int J Med Microbiol. 2014;304:749–763. doi: 10.1016/j.ijmm.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer R, Iverson TM, Accardi A, Miller C. A biological role for prokaryotic ClC chloride channels. Nature. 2002;419:715–718. doi: 10.1038/nature01000. [DOI] [PubMed] [Google Scholar]
- 24.Herrero M, de Lorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg MB, DiRita VJ, Calderwood SB. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg MB, Boyko SA, Calderwood SB. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyckoff EE, Mey AR, Payne SM. Iron acquisition in Vibrio cholerae. Biometals. 2007;20:405–416. doi: 10.1007/s10534-006-9073-4. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg MB, Boyko SA, Calderwood SB. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J Bacteriol. 1990;172:6863–6870. doi: 10.1128/jb.172.12.6863-6870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camilli A, Beattie DT, Mekalanos JJ. Use of genetic recombination as a reporter of gene expression. Proc Natl Acad Sci USA. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pressler K, et al. AAA+ proteases and their role in distinct stages along the Vibrio cholerae lifecycle. Int J Med Microbiol. 2016;306:452–462. doi: 10.1016/j.ijmm.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Syed KA, et al. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol. 2009;191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wurm P, et al. Stringent factor and proteolysis control of sigma factor RpoS expression in Vibrio cholerae. Int J Med Microbiol. 2017;307:154–165. doi: 10.1016/j.ijmm.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Tischler AD, Camilli A. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect Immun. 2005;73:5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karatan E, Duncan TR, Watnick PI. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol. 2005;187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bomchil N, Watnick P, Kolter R. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J Bacteriol. 2003;185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaichi Y, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Y, Waldor MK. Deletion of a Vibrio cholerae ClC channel results in acid sensitivity and enhanced intestinal colonization. Infect Immun. 2003;71:4197–4200. doi: 10.1128/IAI.71.7.4197-4200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovacikova G, Lin W, Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol. 2010;192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc Natl Acad Sci USA. 2008;105:9769–9774. doi: 10.1073/pnas.0802241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 44.Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002;43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, et al. Differential thiol-based switches jump-start Vibrio cholerae pathogenesis. Cell Rep. 2016;14:347–354. doi: 10.1016/j.celrep.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor JL, et al. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol Microbiol. 2012;83:457–470. doi: 10.1111/j.1365-2958.2011.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 48.Bertram R, Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol. 2008;1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seper A, et al. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation. Mol Microbiol. 2011;82:1015–1037. doi: 10.1111/j.1365-2958.2011.07867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moisi M, et al. A novel regulatory protein involved in motility of Vibrio cholerae. J Bacteriol. 2009;191:7027–7038. doi: 10.1128/JB.00948-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lichtenegger S, et al. Characterization of lactate utilization and its implication on the physiology of Haemophilus influenzae. Int J Med Microbiol. 2014;304:490–498. doi: 10.1016/j.ijmm.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seper A, et al. Vibrio cholerae evades neutrophil extracellular traps by the activity of two extracellular nucleases. PLoS Pathog. 2013;9:e1003614. doi: 10.1371/journal.ppat.1003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 55.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vance RE, Zhu J, Mekalanos JJ. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect Immun. 2003;71:2571–2576. doi: 10.1128/IAI.71.5.2571-2576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolivar F, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 59.Hava DL, Hemsley CJ, Camilli A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J Bacteriol. 2003;185:413–421. doi: 10.1128/JB.185.2.413-421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 61.Fengler VH, et al. Disulfide bond formation and ToxR activity in Vibrio cholerae. PLoS One. 2012;7:e47756. doi: 10.1371/journal.pone.0047756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.