Significance
MreB is a bacterial actin that supports cell wall synthesis and rod-like morphology in many bacteria. In the bacterium Myxococcus xanthus, MreB is also required for gliding motility. In this work, we studied the movement of MreB particles during M. xanthus gliding motility at high spatial and temporal resolution. We found that MreB provides a scaffold for the gliding motors while the gliding machinery drives the movement of MreB filaments. The interplay between MreB and the gliding machineries is analogous to the interdependent movements of myosin motors and actin in eukaryotic cells. Since their discovery, actin-like homologs have been implicated in many spatially organized cellular processes in bacteria. This study expands our knowledge of MreB function in bacterial gliding motility.
Keywords: bacterial cytoskeleton, flagella stator homolog, protein dynamics, super-resolution microscopy, peptidoglycan synthesis
Abstract
MreB is a bacterial actin that is important for cell shape and cell wall biosynthesis in many bacterial species. MreB also plays crucial roles in Myxococcus xanthus gliding motility, but the underlying mechanism remains unknown. Here we tracked the dynamics of single MreB particles in M. xanthus using single-particle tracking photoactivated localization microscopy. We found that a subpopulation of MreB particles moves rapidly along helical trajectories, similar to the movements of the MotAB-like gliding motors. The rapid MreB motion was stalled in the mutants that carried truncated gliding motors. Remarkably, M. xanthus MreB moves one to two orders of magnitude faster than its homologs that move along with the cell wall synthesis machinery in Bacillus subtilis and Escherichia coli, and this rapid movement was not affected by the inhibitors of cell wall biosynthesis. Our results show that in M. xanthus, MreB provides a scaffold for the gliding motors while the gliding machinery drives the movement of MreB filaments, analogous to the interdependent movements of myosin motors and actin in eukaryotic cells.
Actin, the most abundant protein in the human body, forms linear filaments to provide cells with mechanical support and participates in many cellular processes (1). In eukaryotic cells, myosin motors generate force along actin, enabling movements required for muscle contraction, motility, cell division, and intracellular transport (1). Actin-like proteins have also been discovered in bacteria. The bacterial actin MreB participates in a wide range of functions, including cell shape determination, cell wall biosynthesis, and motility (2–6). In rod-shaped bacteria, MreB guides the insertion of new cell wall material around the long axis of cells (7). In Bacillus subtilis and Escherichia coli, MreB filaments localize into punctate patterns and appear to form short filaments. Although the conformation of MreB filaments is still open to discussion, it is widely agreed that MreB is highly dynamic in most bacterial species (8–13). For example, MreB filaments rotate slowly (∼1 rpm, or 10–90 nm/s) around the long cell axis of B. subtilis and E. coli. Such movement is driven by the peptidoglycan (PG) synthesis machinery and blocked by the addition of antibiotics that inhibit PG synthesis (8–10).
MreB also plays a unique role in the gliding motility of Myxococcus xanthus, a rod-shaped, nonflagellated, Gram-negative soil bacterium. M. xanthus is able to glide along solid substrates without the aid of type IV pili. Its gliding motility is powered by the action of the Agl-Glt complex, which contains up to 17 proteins, including cytosolic, inner membrane, periplasmic, and outer membrane components (14, 15). Following its initial assembly at the leading cell pole, the Agl-Glt complex further assembles into a force-generating unit through interaction with AglRQS, a proton channel complex homologous to the E. coli flagella stator complex MotAB (5, 15–17) and GltG/I/J, which form a putative inner membrane platform. Assembled force-generating units move directionally toward the lagging cell pole following rotational trajectories and propel a rotational movement of the cell when they engage bacterial focal adhesions with the underlying surface (5, 17).
Visualized by regular fluorescent microscopy, proteins in the gliding machinery appeared either as blurry patches that moved in the cell envelopes or bright aggregates that remained relatively stationary at the focal adhesion sites (6, 15, 16, 18). Thus, single-particle tracking photoactivated localization microscopy (sptPALM) was used to clarify the dynamics of motility-related proteins at subdiffraction resolutions. sptPALM analyses of AglR, a MotA homolog, revealed that the motor subunits moved along helical trajectories (5). Consistent with this observation, GltD (AgmU) and AglR decorate a structure that appears helical in fixed cells, but the exact composition of this structure has remained elusive (5, 6). Recent studies suggest that the directionality of the motility complex depends on its interactions with three key cytoplasmic components: MreB, the Ras-like GTPase MglA, and a PilZ-like regulator, PlpA (19–21). Among these proteins, the direct interaction between MreB and MglA-GTP regulates the spatial assembly, disassembly, and directionality of the Agl-Glt complexes (19, 20).
M. xanthus gliding motility requires functional MreB filaments. The MreB inhibitor A22 [S-(3,4-dichlorobenzyl) isothiourea] blocks the gliding, but fails to do so in the cells that express an A22-resistent MreB variant (4–6). This result suggests that A22 inhibits gliding motility specifically through MreB. Nonetheless, the precise function(s) of MreB in gliding motility remain unclear. MreB could connect to the motility complex indirectly through MglA (19, 20); however, this connection has not been directly observed in motile cells. In this study, we constructed a functional photoactivatable MreB fusion and investigated its dynamics at single-particle resolution. We show that MreB provides a scaffold for the gliding motors while the gliding machinery drives the movement of MreB filaments. The interdependence between MreB and the gliding machineries reveal a direct function of MreB in myxobacterial gliding motility.
Results
Isolation of an M. xanthus Strain Expressing a Functional MreB-PAmCherry.
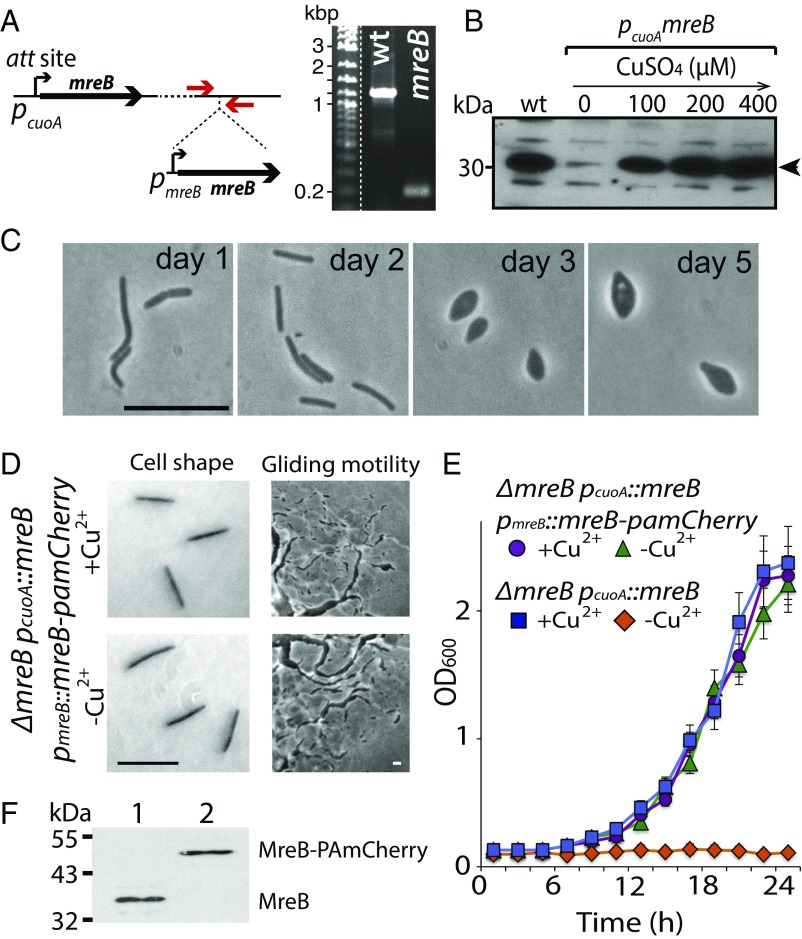
We constructed a M. xanthus strain that expresses MreB fused to photoactivatable mCherry (PAmCherry) to image the entire cellular MreB pool as well as single MreB particles. Because both the N and C termini of MreB participate in the polymerization of filaments (22, 23) and are sensitive to structural perturbation (SI Appendix, Fig. S1) (24), we inserted PAmCherry at various positions in the internal loops of MreB to minimize interference with function (SI Appendix, Table S1). To screen the MreB-PAmCherry constructs for functionality, we constructed a conditional mreB depletion strain, as mreB is essential for M. xanthus viability. In this depletion strain, mreB was expressed ectopically from a copper-inducible promoter (pcuoA::mreB), while the endogenous mreB was deleted (25) (Fig. 1A). When this deletion strain was subcultured in the absence of copper, mreB was not expressed (Fig. 1B); cells changed morphology and eventually lysed (Fig. 1C). We next integrated a single copy of each mreB-pamCherry construct, together with the endogenous mreB promoter, at the Mx8 prophage attachment site of the mreB depletion strain and compared the phenotypes of each strain in the presence or absence of 200 μM CuSO4. PAmCherry insertion between V229 and M230 showed wild-type phenotypes in terms of cell shape, growth rate, and gliding motility in the absence of copper, indicating that this MreB-PAmCherry fusion was functional (Fig. 1 D and E and SI Appendix, Table S1). This MreB-PAmCherry fusion appeared as a single band corresponding to its molecular weight (∼50 kDa), indicating that the fusion protein was stably expressed (Fig. 1F).
Fig. 1.
The MreB-PAmCherry construct rescues the mreB deletion phenotypes in M. xanthus. (A, Left) Genetic scheme of the MreB depletion strain. The copper-inducible copy of mreB is integrated into the M. xanthus chromosome at the Mx8-phage attachment (att) site, allowing deletion of the endogenous mreB. Red arrows show the position of the verification primers. (A, Right) PCR experiment demonstrating deletion of mreB in the depletion strain. (B) MreB can be completely depleted, and its expression depends on copper. Western blot of MreB in cell extracts from M. xanthus cells after 3 d of depletion or in the presence of various concentrations of CuSO4. The arrow points to the band of MreB. (C) Phase-contrast images of M. xanthus cells after 1, 2, 3, and 5 d of growth in the absence of copper, showing that 100% of the cells lost rod shape after 3 d, coinciding with the complete absence of MreB in cell extracts shown in B. (D and E) Expressed by the native mreB promoter (pmreB) and the sole source of MreB, the MreB-PAmCherry fusion fully rescues the mreB depletion phenotypes in cell shape maintenance (D), gliding motility (D), and growth (E). (F) Probed by the MreB antibodies, the MreB-PAmCherry fusion protein appears as a single band at the right molecular weight. Cell lysates from the wild-type DZ2 strain (lane 1) and the mreB depletion strain expressing MreB-PAmCherry (lane 2) were analyzed. (Scale bar: 10 μm.)
MreB Filaments Exhibit Rapid Dynamic Movements in M. xanthus Cells.
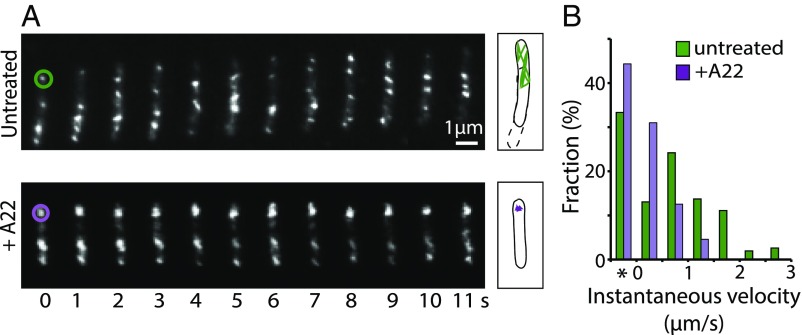
We used fluorescence microscopy to track MreB-PAmCherry in moving cells. When exposed to 405-nm excitation (0.2 kW/cm2) for 2 s, the majority of MreB-PAmCherry was photoactivated, appearing as small patches along the cell body, similar to the localization described for other bacteria (Fig. 2A and SI Appendix, Fig. S2). We recorded the dynamics of the fluorescent patches by time-lapse microscopy at 10 Hz using highly inclined and laminated optical sheet illumination (26). The MreB filaments in M. xanthus showed rapid irregular motion both across the cell width and along the long cell axis, in contrast to the slow circumferential movements observed in growing E. coli and B. subtilis cells (8–10) (Fig. 2A and Movie S1). Due to the high density and frequent merging and splitting of the fluorescent patches (Movie S1), few patches could be tracked continuously for more than 10 frames. Instantaneous velocities of such patches (Npatches = 35, Nvelocities = 342) showed a broad distribution from stationary (v = 0) to ∼3 μm/s (Fig. 2B).
Fig. 2.
MreB filaments move rapidly in M. xanthus. (A) Time-lapse images of MreB-PAmCherry filaments in representative untreated and A22-treated cells (Movies S1 and S2). Trajectories of the MreB patches that are marked by circles are displayed on the Right, where dotted and solid lines mark the outlines of the cell in the first and last frames, respectively. (B) Distribution of instantaneous velocities of MreB-PAmCherry filaments. *The stationary populations.
We then incubated the cells with 100 μg/mL A22 to inhibit the de novo polymerization of MreB filaments. Immediately after this treatment, the fluorescence signal of MreB-PAmCherry became diffuse, although filament-like patches were still visible. This suggests that while many MreB molecules were no longer polymerized after A22 treatment, a subpopulation of MreB remained polymerized (Fig. 2A). These filaments were significantly less dynamic. We tracked these patches at 2 Hz using time-lapse microscopy and calculated their displacement in between frames. We found that in A22-treated cells, 44% (Npatches = 100; Nvelocities = 1,162) of the MreB filaments remained stationary (v = 0). The remaining filaments moved with low velocities (v <1.5 μm/s), and movements faster than 1.5 μm/s were not observed (Fig. 2 and Movie S2). Since A22 blocks both the de novo polymerization of MreB (2, 27) and the gliding motility (4, 5), disruption of either of these processes can explain the absence of rapid MreB movement under these conditions.
A Subpopulation of MreB Particles Displayed Rapid Motion in a Directed Manner.
To track individual MreB particles, we reduced the time of the 405-nm excitation to 100 ms so that only a few MreB-PAmCherry particles were photoactivated at a given time in each cell. In images recorded at 10 Hz, photoactivated MreB-PAmCherry appeared as isolated spots. Since MreB has been shown to associate with the cell wall synthesis machinery (8–10) and attach to the membrane both in vivo and in vitro (28, 29), the major population of MreB particles should be restricted to the cylindrical cell envelope (30).
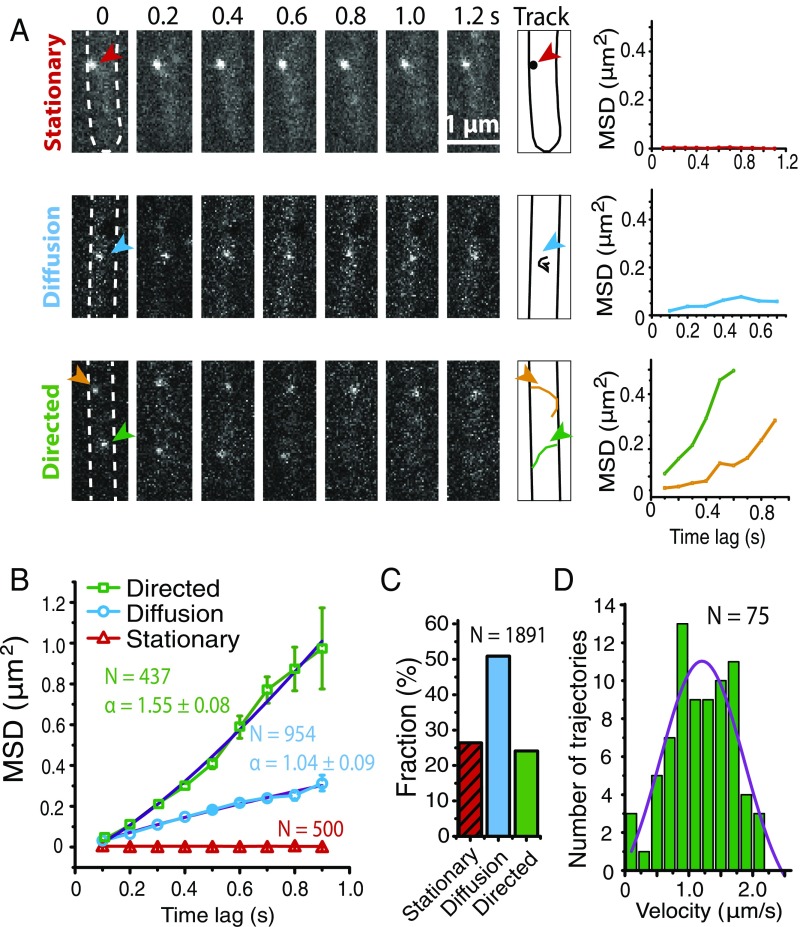
Under our illumination condition, many MreB particles entered and exited the focal plane. To analyze the dynamics of MreB, 1,891 fluorescent particles that remained in focus for 4–12 frames (0.4–1.2 s) were chosen for further analysis. Among these particles, 500 (26.4%) remained stationary within a single pixel (100 nm ×100 nm) in 1.2 s (Fig. 3 and Movie S3), and 1,391 particles showed detectable displacements. Three-dimensional displacements of these particles on the cylindrical cell envelopes were extracted from 2D data (SI Appendix, Methods and Fig. S3). Mean square displacements (MSDs) of these particles were calculated to determine their diffusion coefficient (D) and time exponent (α). We found that the trajectories with high α values (α > 1.5) also featured high D (0.15 < D < 1.4), while low α (α < 1.5) values often accompanied low D (D < 0.3). The distribution of α and D values of all 1,391 trajectories suggests that MreB particles might move in two distinct modes (SI Appendix, Fig. S4). To categorize the movements of these motile MreB particles, we performed computational simulation. We simulated the directed motion of hypothetical particles in an area of 0.6 × 5 μm, which mimicked the cell surfaces illuminated in our experiments (SI Appendix, Fig. S5A). Among the 1,081 particles stimulated, 1,038 (96%) showed α values between 1.5 and 2 (SI Appendix, Fig. S5B). Based on this simulation, the movements of motile MreB particles were grouped into two categories by their time exponent (α ≤ 1.5 and α > 1.5).
Fig. 3.
MreB displays three major types of movement characteristics in M. xanthus. (A) More than one-quarter (26.4%) of the MreB-PAmCherry particles remained stationary (Upper, also see Movie S3). The movements of the MreB-PAmCherry particles that showed detectable displacements were classified into two categories based on the values of α; α ≤ 1.5 indicates diffusion (Middle) (Movie S4), while α > 1.5 indicates directed motion (Lower) (Movie S5). (B) The MSDs of each population of MreB particles were plotted against time lag (Δt). α values of the particles showing diffusion and directed motion were calculated. (C) Percentage distribution of each MreB population. (D) The velocity distribution of 75 trajectories that crossed the midlines of cells.
One-half of the particles (50.4%; n = 954) showed small but detectable displacements and frequent directional changes, with calculated α ≤ 1.5 (Fig. 3 and Movie S4). The mean α value of the pooled data in this population is 1.04 ± 0.09 (Materials and Methods). This group may represent the diffusion of the MreB filaments. The D of MreB in this population was 0.10 ± 0.01 µm2/s, which might reflect the bulk of the MreB filaments.
Four hundred thirty-seven particles (23.1%) displayed significant displacements, with calculated α > 1.5 (Fig. 3 and Movie S5). The α value of the pooled data in this population is 1.55 ± 0.08, indicating directed motion. Similar to its homologs in other Gram-negative bacteria (29), M. xanthus MreB contains a putative amphipathic helix for direct membrane binding (SI Appendix, Fig. S1). Consistent with this prediction, the D of the directed motion of MreB is 0.22 ± 0.01 µm2/s, comparable to the D values of the proteins that diffuse freely in membranes (31). To verify the directed motion of these MreB particles, velocity autocorrelation [CV(τ)] of each trajectory was calculated. For a random motion, CV(τ) drops to 0 at the very first time step, while directed motion maintains a positive nonzero correlation due to the tendency to move in the same direction (30). The calculated CV(τ) of the particles moving in directed manner remained positive for at least 1 s (SI Appendix, Fig. S6). Taken together, these data indicate that a major population of MreB particles moves in a directed manner.
When 3D trajectories on cylindrical cell surfaces project onto 2D images, their velocities and trajectory angles in 3D are close to the projected velocities and angles in 2D near the midlines of cells. Among the 437 trajectories of directed motion, 75 trajectories that crossed the midlines of cells were identified for angle and velocity calculations (9). For each trajectory, its velocity was estimated by fitting the MSD data to MSD = y0 + 4DΔt + (vΔt)2, where v is velocity (9). The velocities of individual MreB particles showed a broad distribution. The average velocity of these MreB particles was 1.18 ± 0.50 μm/s, and the maximum velocity was around 2.5 μm/s (Fig. 3D). Thus, the directed motion of M. xanthus MreB particles is one to two orders of magnitude faster than MreB movements in B. subtilis and E. coli during cell wall synthesis (8–10). Although M. xanthus cells also rotate slowly around their long cell axes (17), the effect of this rotation (5–10 nm/s) is negligible compared with the rapid movement of MreB particles.
To verify that the observed trajectories were not from free PAmCherry generated by MreB degradation, we expressed PAmCherry alone in M. xanthus cells under the control of the copper-inducible promoter (25). Due to their rapid motion, single PAmCherry particles appeared as blurry objects that could not be followed at the 10-Hz acquisition used for MreB tracking. These particles became traceable at 67 Hz. Visualized at high time resolution, PAmCherry diffused quickly along the long cell axes at 3.12 ± 1.00 μm2/s (SI Appendix, Fig. S7 and Movie S6), comparable to that of free GFP in E. coli (32). With this diffusion rate, the behavior of free PAmCherry was easily distinguishable from that of the PAmCherry-tagged MreB particles.
The Directed Motion of MreB Particles Depends on the Gliding Motor.
The velocities of the MreB particles that moved in directed manner are similar to the velocities of individual motors in gliding motility (5). To test if MreB and gliding motors move in similar trajectories, we also calculated the angle between the trajectory and the cell axis for the MreB particles that crossed the midlines of cells. The average trajectory angle was 31.5 ± 21.5°, and the angular distribution peaked around 25° (Fig. 4A). We revisited the dynamics of gliding motors by imaging and analyzing single AglR-PAmCherry particles using the same method and found that the average trajectory angle of AglR is 35.8 ± 20.6°, and the angular distribution also peaked around 25° (Fig. 4A). Thus, besides their similar velocities, the directed movements of MreB and AglR also follow similar helical trajectories in M. xanthus, which are in sharp contrast to the circumferential trajectories of the slow MreB rotation in B. subtilis and E. coli, where angles peak around 90° (8–10).
Fig. 4.
The directed motion of MreB depends on the gliding motor. (A) The angular distribution histogram of MreB trajectories is similar to that of AglR, with the Pearson’s coefficient, r, of 0.81. (B) Time lapse (Upper) and kymograph (Lower) of the MreB aggregates in a ΔaglQS cell. The rapid motion of MreB is blocked by deletion of aglQ and aglS, which encode MotB homologs in the gliding motor (Movie S7). (C) Population analysis of the behaviors of MreB particles under different conditions. The number of MreB particles analyzed is shown on top of the bars. In the ΔaglQS strain, MreB aggregates, rather than single particles, were analyzed. (D) Velocities of the particles in untreated wild-type and ΔaglQS cells, and the cells treated by A22 and the antibiotics that inhibit cell wall synthesis.
To test whether the directed motion of MreB depends on the gliding motors directly, we tracked MreB-PAmCherry particles after the inhibition of gliding motility. In a ΔaglQS mutant, we deleted the genes for two MotB homologs, AglQ and AglS, which are critical parts of the gliding motor. In this case, multiple MreB molecules were activated in aggregates at polar and midcell locations, which remained fluorescent for >10 s (Fig. 4B). Almost all of these particles (99.7%; n = 328) remained stationary, and directed motion was not observed (Fig. 4C and Movie S6). Consistent with the observations on MreB filaments, in the presence of A22, >60% of MreB particles remained stationary, while the MreB population that moved in directed motion decreased significantly (Fig. 4C).
The Rapid Helical Motion of MreB Is Not Coupled to PG Biosynthesis.
To test if the rapid helical motion of MreB is also related to PG biosynthesis, we studied the dynamics of MreB-PAmCherry in the presence of fosfomycin, mecillinam, and vancomycin, antibiotics that inhibit PG biosynthesis (9). We investigated the rapid helical movements of MreB in the presence of these inhibitors at twice their minimum inhibitory concentrations (MICs; SI Appendix, Fig. S8). In these treated cells, approximately 20% of MreB particles moved in a directed manner, indistinguishable from the particles in untreated cells (Fig. 4C). Antibiotic treatment did not affect the velocities of the directed motion of MreB particles (n = 54, P = 0.78 for fosfomycin; n = 71, P = 0.42 for mecillinam; and n = 54, P = 0.93 for vancomycin). We concluded that the directed motion of MreB is not coupled to PG synthesis in M. xanthus (Fig. 4D).
The Slow Circumferential Motion of MreB.
In B. subtilis and E. coli, MreB filaments were reported to move circumferentially around the long cell axis with velocities of about 10–90 nm/s (8–11). To track such slow movement reliably in M. xanthus with a low photon budget of PAmCherry, we lowered the image acquisition rate to 0.2 Hz. Among 50 MreB-PAmCherry particles that appeared stationary at high temporal resolution (10 Hz), we detected only six particles (12%) to rotate using 0.2-Hz acquisition. None of these particles rotated >90° within 200 s (<0.08 rpm) (SI Appendix, Fig. S9 and Movie S8), which is significantly slower than that in B. subtilis and E. coli (8–10). We were not able to determine if the antibiotics that inhibit cell wall synthesis also inhibit the slow circumferential movement of MreB in M. xanthus due to its low occurrence and velocities.
Discussion
The mechanism of gliding motility in M. xanthus has remained elusive because it differs significantly from the well-characterized flagella and type IV pili-mediated motility. Although recent advances identified many of the proteins involved in gliding motility, the coupling of motor movements to surface translocation remains obscure (33–35). Previous studies showed that AglR exhibits rapid movements within the cell membrane following helical trajectories (5). In this study, we explored the relationship between MreB and the gliding motors. Surprisingly, we observed that MreB particles also show rapid helical movements, and that the movements of MreB and AglR are interdependent.
In M. xanthus, >20% of MreB particles show rapid directed motion. This motion is in sharp contrast with the MreB homologs in B. subtilis and E. coli. Not only do MreB particles move in helical trajectories, but also the velocity of their movement is one to two orders of magnitude higher than the circumferential movements of B. subtilis and E. coli MreB. In addition, this helical motion of M. xanthus MreB is not controlled by the PG biosynthesis machinery, but appears to function in gliding motility. Our results suggest that MreB filaments provide scaffolds for the gliding motors, which would be consistent with the following observations: (i) gliding motility is reversibly and rapidly blocked by A22, and this effect is not observed when an A22-resistant MreB allele is expressed (4); (ii) MreB interacts with MglA-GTP, and A22 blocks the recruitment of MglA to the motility machinery (20); (iii) MreB particles exhibited motor-dependent dynamics, proving that MreB is connected to the motors. One cluster of gliding motors may generate a force of ∼20 pN (36), which might be sufficient to drive the rotation of an MreB filament. The rapid helical motion of gliding motors may cause their MreB scaffolds to move in the same or opposite direction (6, 17). This is analogous to myosin and actin in eukaryotic cells, where myosin motors drive the movement of actins, while actin filaments also guide the motion of myosin molecules (1). The aggregated fluorescent signals of MreB particles in the ΔaglQS cells suggest rearrangement of the MreB filaments; however, both the shape and growth rate of ΔaglQS cells remained similar to those of the wild-type cells (SI Appendix, Fig. S10). Thus, both the architecture of MreB filaments and the mechanism by which ΔaglQS cells maintain cell shape remain to be investigated.
What is the function of the rapid motion of MreB? Both the motion of the gliding complexes and the growth of PG show helicity (5, 6, 17, 37). In a recent report, Faure et al. (17) hypothesized that the gliding machineries of M. xanthus might move along PG strands or grooves. Since MreB connects to both cell wall synthesis and motility in M. xanthus, it could guide the motility complex with respect to PG. The rapid motion of MreB might be important for M. xanthus to disassemble and assemble MreB scaffolds at very high rates to accommodate the gliding motors.
Besides the rapid helical motion, M. xanthus MreB also rotates slowly perpendicular to the long cell axis, with a rate comparable to the rotation of the cell body (SI Appendix, Fig. S9 and Movie S8) (17). However, the slow circumferential rotation of MreB is not likely a result of cell rotation, for two reasons. First, many MreB particles did not rotate within 200 s. Second, in the same cell, one MreB particle rotated while the other remained stationary (SI Appendix, Fig. S9 and Movie S8). Thus, the slow circumferential rotation might play a role in cell wall synthesis. The low occurrence and velocities could reflect the slow growth rate of M. xanthus cells (doubling time ∼4 h). Much of the mechanism by which MreB supports cell wall synthesis in M. xanthus remains unclear. Nevertheless, since all of the molecular machineries that associate with MreB have the potential to affect the dynamics of MreB filaments, the versatile nature of MreB might provide a mechanism for cells to coordinate multiple cellular functions.
Materials and Methods
Construction and Analysis of the mreB Depletion Strain.
To delete the endogenous mreB, we constructed the plasmid pMAT3-mreB, which when introduced in M. xanthus allowed the expression of mreB under the control of a copper-inducible promoter, pcuo (25). We then deleted the endogenous mreB in this genetic background in the presence of 0.1 mM CuSO4. The resulting strain was confirmed by PCR. The mreB depletion strain was cultured in CTT medium (38) to an OD600 of 0.8 in the presence of 0.1 mM CuSO4. The cells were harvested, washed, resuspended in CTT without CuSO4, and incubated for 5 d. Cell morphology was examined at various depletion times by microscopy, and the protein level of MreB was determined by Western blot analysis as described previously (4).
MreB-PAmCherry Construction.
PAmCherry was inserted between Val299 and Met230 of the MreB protein. The fusion protein was expressed at the Mx8 prophage attachment site of the mreB depletion strain under the control of the native mreB promoter. More details are provided in SI Appendix, Materials and Methods.
sptPALM and Data Analysis.
M. xanthus cells were grown in CYE to 4 × 108 cfu/mL. Imaging of single particles was performed on an inverted Nikon Eclipse-Ti microscope with a 100× 1.49 NA TIRF objective, and the images were collected using a Hamamatsu ImagEM ×2 EMCCD camera C9100-23B (effective pixel size, 160 nm). PAmCherry was activated using a 405-nm laser (0.3–3 W/cm2 for 1 s), and excited and imaged using a 561-nm laser (0.2 kW/cm2) under near-total internal reflection illumination (5). Images were acquired at 10 Hz for MreB-PAmCherry and at 67 Hz for free PAmCherry.
Single PAmCherry particles were localized using an algorithm written in MATLAB (MathWorks). Fluorescent spots were fit by a symmetric 2D Gaussian function, whose center was assumed to be the particle’s position (39, 40). Particles in consecutive frames were considered to belong to the same trajectory when they were within a user-defined distance of 320 nm. Data analysis was performed using standard methods (9, 30, 41–43). More detailed information is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank George Oster for helpful discussions. This work was supported by the Texas A&M Startup Fund (to B.N.), the National Institutes of Health (Grants GM020509, to D.R.Z. and GM094522, to A.Y.), European Research Council (Dissecting a Novel Mechanism of Cell Motility; Starting Grant 21105, to T.M.), and the National Science Foundation (Grant MCB-1617028, to A.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716441115/-/DCSupplemental.
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 4.Mauriello EM, et al. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nan B, et al. Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci USA. 2013;110:E1508–E1513. doi: 10.1073/pnas.1219982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nan B, et al. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 8.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 9.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olshausen PV, et al. Superresolution imaging of dynamic MreB filaments in B. subtilis: A multiple-motor-driven transport? Biophys J. 2013;105:1171–1181. doi: 10.1016/j.bpj.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defeu Soufo HJ, Graumann PL. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 2004;5:789–794. doi: 10.1038/sj.embor.7400209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luciano J, et al. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faure LM, et al. The mechanism of force transmission at bacterial focal adhesion complexes. Nature. 2016;539:530–535. doi: 10.1038/nature20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nan B, et al. The polarity of myxobacterial gliding is regulated by direct interactions between the gliding motors and the Ras homolog MglA. Proc Natl Acad Sci USA. 2015;112:E186–E193. doi: 10.1073/pnas.1421073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treuner-Lange A, et al. The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions. J Cell Biol. 2015;210:243–256. doi: 10.1083/jcb.201412047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogue CB, Zhou T, Nan B. PlpA, a PilZ-like protein, regulates directed motility of the bacterium Myxococcus xanthus. Mol Microbiol. 2018;107:214–228. doi: 10.1111/mmi.13878. [DOI] [PubMed] [Google Scholar]
- 22.van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 23.van den Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Santos N, et al. Comprehensive set of integrative plasmid vectors for copper-inducible gene expression in Myxococcus xanthus. Appl Environ Microbiol. 2012;78:2515–2521. doi: 10.1128/AEM.07502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 27.Bean GJ, et al. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda YT, et al. Assembly of MreB filaments on liposome membranes: A synthetic biology approach. ACS Synth Biol. 2012;1:53–59. doi: 10.1021/sb200003v. [DOI] [PubMed] [Google Scholar]
- 29.Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lommerse PH, Snaar-Jagalska BE, Spaink HP, Schmidt T. Single-molecule diffusion measurements of H-Ras at the plasma membrane of live cells reveal microdomain localization upon activation. J Cell Sci. 2005;118:1799–1809. doi: 10.1242/jcs.02300. [DOI] [PubMed] [Google Scholar]
- 32.Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S. Protein mobility in the cytoplasm of Escherichia coli. J Bacteriol. 1999;181:197–203. doi: 10.1128/jb.181.1.197-203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nan B, Zusman DR. Novel mechanisms power bacterial gliding motility. Mol Microbiol. 2016;101:186–193. doi: 10.1111/mmi.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. Bacteria that glide with helical tracks. Curr Biol. 2014;24:R169–R173. doi: 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balagam R, et al. Myxococcus xanthus gliding motors are elastically coupled to the substrate as predicted by the focal adhesion model of gliding motility. PLoS Comput Biol. 2014;10:e1003619. doi: 10.1371/journal.pcbi.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Furchtgott L, Huang KC, Shaevitz JW. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc Natl Acad Sci USA. 2012;109:E595–E604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu SS, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu G, et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS One. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 41.Lee TK, Meng K, Shi H, Huang KC. Single-molecule imaging reveals modulation of cell wall synthesis dynamics in live bacterial cells. Nat Commun. 2016;7:13170. doi: 10.1038/ncomms13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monnier N, et al. Bayesian approach to MSD-based analysis of particle motion in live cells. Biophys J. 2012;103:616–626. doi: 10.1016/j.bpj.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgenstein RM, et al. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci USA. 2015;112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.