Significance
Inducible protein dimerization methods are powerful tools for the investigation of protein function. Unlike chemical dimerization systems, light-dependent systems can trigger dimerization at cellular and subcellular resolution. Here, we present a systematic comparison of the ability of three blue-light-dependent dimerization tools, Cry2/CIB1, iLID, and Magnets, to confine dimerization to small subcellular volumes. Our study highlights the parameters that contribute to defining the subcellular volume where dimer formation occurs, including switch-off kinetics of the photoreceptor(s). The highest confinement is achieved by Magnets and by iLID variants with very fast switch-off kinetics, although spatial resolution with these systems comes at the expense of the total level of dimers formed.
Keywords: optogenetics, protein–protein interaction, optical dimerizer, Cry2/CIB1, LOV domain
Abstract
Methods to acutely manipulate protein interactions at the subcellular level are powerful tools in cell biology. Several blue-light-dependent optical dimerization tools have been developed. In these systems one protein component of the dimer (the bait) is directed to a specific subcellular location, while the other component (the prey) is fused to the protein of interest. Upon illumination, binding of the prey to the bait results in its subcellular redistribution. Here, we compared and quantified the extent of light-dependent dimer occurrence in small, subcellular volumes controlled by three such tools: Cry2/CIB1, iLID, and Magnets. We show that both the location of the photoreceptor protein(s) in the dimer pair and its (their) switch-off kinetics determine the subcellular volume where dimer formation occurs and the amount of protein recruited in the illuminated volume. Efficient spatial confinement of dimer to the area of illumination is achieved when the photosensitive component of the dimerization pair is tethered to the membrane of intracellular compartments and when on and off kinetics are extremely fast, as achieved with iLID or Magnets. Magnets and the iLID variants with the fastest switch-off kinetics induce and maintain protein dimerization in the smallest volume, although this comes at the expense of the total amount of dimer. These findings highlight the distinct features of different optical dimerization systems and will be useful guides in the choice of tools for specific applications.
The physiological function of proteins within cells is critically dependent on their subcellular distribution and local regulation. Therefore, methods that allow manipulation of protein localization at the subcellular level, with high spatial and temporal precision, are powerful tools for the exploration of cell functions. These methods are typically based on genetic manipulations (1). While extremely powerful, genetic perturbations have limitations due to their slow timescales (hours to days) relative to protein dynamics of intracellular signaling networks (seconds) and many posttranslational physiological processes (minutes).
Chemically inducible dimerization tools, usually based on the FK506-binding protein (FKBP12) and its binding partner FRB in the presence of rapamycin or analogs (2), provide a versatile approach to acutely control protein recruitment to intracellular compartments (3–5) and modulate cellular activities (6–11). However, chemically inducible dimerization does not allow for spatial control of dimer formation at the subcellular level, and effects are difficult to titrate (12). The identification of light-switchable proteins was therefore a landmark discovery in cell biology. Light-induced dimerization based on these photoreceptors allows for protein perturbation with high spatial precision and on a timescale consistent with the speed and reversibility of intracellular reactions.
Several genetically encoded light-dependent protein dimerization systems have been generated and optimized (13–27). Upon light excitation, the photosensitive protein undergoes a conformational change and interacts with its binding partner. Since the photosensitive protein naturally reverts to its ground state over time, the duration of the interaction between the two components depends on the duration of the photostimulation and on the switch-on and switch-off kinetics of the photoreceptor. The switch-off kinetic describes how long the photoreceptor remains in its activated state once the excitation light is withdrawn from the system. This time can vary from seconds to hours (13–27). In a typical experimental setup, one component of the dimer (the bait) is targeted to a cellular subcompartment, while the other component (the prey) is expressed as a cytosolic protein. Light induces dimer formation, thus triggering the recruitment of the prey to the organelle. The optimal system for the regulation of biological processes at the subcellular level would allow redistribution of the desired amount of protein with the highest spatial precision and fastest on and off kinetics.
The light-dependent dimerization toolbox currently available has been extensively reviewed (28, 29) and some studies have provided a comparison of optical dimerizers (30, 31). However, a systematic evaluation of the spatial control of protein dimerization achieved by different tools under the same experimental conditions, and on a timescale consistent with fast intracellular processes, is still missing.
The greatest spatial control of protein dimerization described to date is that achieved with the Arabidopsis thaliana phytochrome B and PIF system. In this system, spatial localization is controlled by the simultaneous irradiation of different cell regions with an activating red wavelength and an inactivating far-red wavelength (15, 32, 33). The recruitment of soluble prey to specific regions of the plasma membrane (15, 33–35) or to selected subcellular compartments (32) was achieved using a microscope that simultaneously activates the photoreceptor in the region of interest (ROI) and deactivates it outside of this area (15). This dimerization system requires an exogenous cofactor, a tetrapyrrole chromophore (15), which has absorption peaks in both the red and the blue portion of the visible spectrum (21, 26), thus limiting the number of fluorescent proteins that can be used to follow intracellular dynamics. Additionally, the cofactor has to be administered (36) or expressed after cell-line engineering (37).
The use of an optical dimerization tool where photoactivation is achieved by a single wavelength of light allows for easier control of protein dimerization since it only requires spatial control by a single laser beam. Here we report a systematic investigation of the light-dependent spatial and temporal control of protein dimerization that can be achieved at the subcellular level with three of the most widely used blue-light-dependent dimerization systems with rapid switch-on and switch-off kinetics: Cry2-CIB1, iLID, and Magnets.
The Cry2-CIB1 system was one of the first to be developed (17), and is probably the most widely used in cell biology (38–42). The most commonly used variant of this system (25) is based on the photolyase homology region of the A. thaliana photoreceptor cryptochrome 2 (amino acids 1–498) and the 170 N-terminal amino acids of its interacting partner, the transcription factor CIB1. The iLID system (22) is based on the interaction between the Escherichia coli peptide SsrA fused to the C-terminal portion of the photosensitive LOV2 domain of Avena sativa phototrophin 1 and its interacting partner, SspB. Among the optical dimerizers based on the LOV2 domain (19, 20, 22), iLID has proven to be the most efficient at recruiting proteins to the membranes of intracellular compartments (31). The Magnets dimerization system was recently developed by Sato and coworkers (24) through the multidirectional engineering of the Neurospora crassa photoreceptor Vivid. A defining feature of Magnets is that both components of the dimer are photoreceptors, and both must be activated by light to achieve dimerization.
Our results highlight the parameters that control spatial confinement of dimer formation in small cytoplasmic volumes and emphasize the importance of the switch-off kinetic of the photoreceptor(s).
Results and Discussion
Subcellular Confinement of Dimer Formation with Blue-Light-Dependent Dimerization Systems.
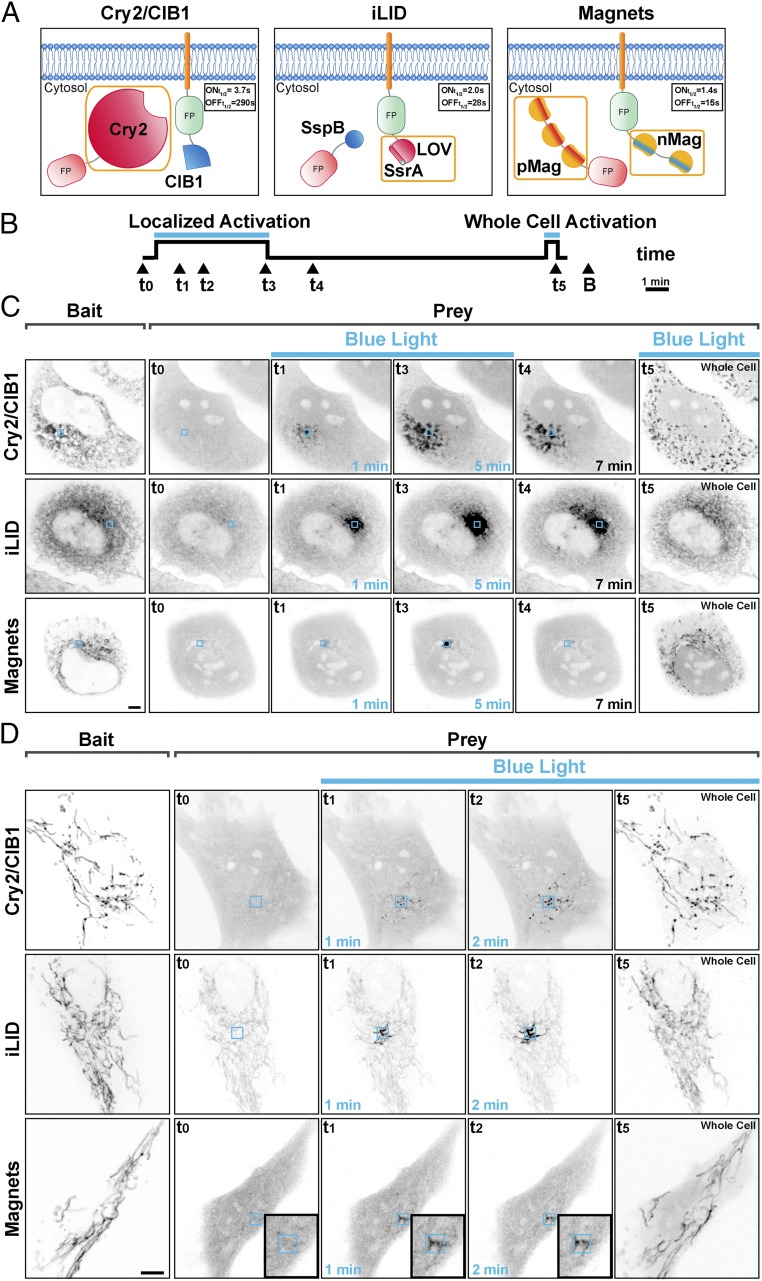
To determine the spatial localization of dimer formation with the three blue-light-dependent dimerization systems we expressed constructs encoding a cytosolic prey and a bait targeted to either the endoplasmic reticulum (ER) or mitochondria in fibroblastic cells (Fig. 1A). We then irradiated a 3-μm × 3-μm ROI for 5 min with 200-ms-long blue-light pulses every 2 s (488 nm; 7.07 W/cm2) using a confocal microscope. We observed the behavior of the prey for each dimerization system during and after local light irradiation and, as a control, after irradiation of the entire cell (Fig. 1B).
Fig. 1.
Spatial confinement of light-dependent protein dimerization achieved with blue-light-dependent dimerization systems. (A) Optical dimerization systems used. Protein photoreceptors are outlined by an orange box. The on and off kinetics of dimer formation, measured as the rate of recruitment and release of the prey protein to a mitochondria-anchored bait, in living cells at 37 °C is shown. (B) Experimental paradigm: Cells were illuminated within a 3-µm × 3-µm ROI with 200-ms blue-light pulses at 0.5 Hz for 5 min, allowed to recover in the absence of blue light for 12 min, and then illuminated over the entire surface with 200-ms blue-light pulses at 0.5 Hz for 30 s. The localization of the bait (B) was assessed at the end of the illumination period. (C and D) Comparison of the three systems in their property to confine bait–prey dimers to the photoexcitation area. Human fibroblastic cells expressing a soluble prey and an ER-targeted (C) or a mitochondria-targeted (D) bait, respectively. The left columns of each row show the expression of the bait. The other images in each row show the localization of the prey at the indicated time. With the Cry2/CIB1 and with the iLID/SspB systems dimers can be observed outside the illumination ROI in an area that increases with time. With Magnets the bait–prey interaction is mainly limited to the area of photoexcitation, remains confined to this region, and is rapidly reversible. (Magnification: D, Insets, 1.7×.) (Scale bars: 5 μm.)
We used the most widely employed configuration of the Cry2-CIB1 system (17, 25, 31), where the photoreceptor (the photolyase homology region of Cry2: CRY2PHR, amino acids 1–498; refs. 17 and 25) is the cytosolic prey and CIBN (the 170 N-terminal amino acids of the transcription factor CIB1; refs. 17 and 25) is the membrane-bound bait (Fig. 1A). We expressed a cytosolic mCherry-Cry2 (mCh-Cry2) fusion protein as the prey and CIBN as bait. The latter was fused to both EGFP and either the N-terminal ER-targeting sequence of cytochrome b5 (ER-CIBN) or the C-terminal mitochondrial targeting sequence of the mitochondrial protein OMP25 (CIBN-Mito) (see Tables S1 and S2 for a detailed description of all constructs). We observed that mCh-Cry2 rapidly populated large portions of the ER or mitochondria, despite the restricted area of blue-light illumination (Fig. 1 C and D and Movies S1 and S2). This can be explained by the slow (several minutes) switch-off kinetics of the Cry2-CIBN dimer ( = 3.7 ± 0.9 s, = 290 ± 30 s in vivo at 37 °C, Fig. S1 and Table S3). The activated prey (mCh-Cry2) can therefore diffuse over a large area of the ER or the mitochondrial network, away from the illuminated area, both as part of a locally generated dimer or as a soluble protein that can bind to the membrane-bound bait outside of the irradiated region. Thus, the switch-off kinetics not only limit temporal resolution but also spatial resolution. A more spatially restricted formation of the Cry2-CIBN dimer can in principle be achieved by using Cry2 as part of the bait fusion protein rather than as a component of the cytosolic prey. However, even in this case, diffusion of the dimer outside of the illuminated volume would limit spatial resolution due to the slow switch-off kinetics. Additionally, we and others (31, 43) have found that Cry2 is less effective in CIBN recruitment when bound to membranes. Fig. 1 C and D, Fig. S2, and Movies S3 and S4 highlight an additional problem of the Cry2-CIB1 system: Activated Cry2 has the propensity to oligomerize, both in solution (44, 45) and on target membranes (43), leading to the formation of clusters. These aggregates, which are particularly well visible after whole-cell activation, result in a delayed (or irreversible) dissociation of the dimer (Fig. S2).
The same experiments were performed using the iLID (22) system, where the dimerization is based on the interaction between the E. coli peptide SsrA, fused to the C-terminal portion of the LOV2 domain, and its interacting partner, SspB. In this case, the ER-tethered or mitochondria-tethered bait was the photoreceptor (ER-iLID or iLID-Mito, respectively) and the prey was the cytosolic protein tgRFPt-SspB (Fig. 1A). With this system, dimer accumulation remained spatially localized but not completely restricted to the illuminated volume due to the diffusion of dimers or photoactivated ER-iLID or iLID-Mito monomers to nonilluminated areas of the ER or outer mitochondrial membrane (Fig. 1 C and D and Movies S5 and S6). The tighter spatial confinement observed with iLID compared with Cry2-CIB1 can be explained both by the faster dissociation rate of the iLIDs [based on our measurement in cells at 37 °C = 2.0 ± 0.3 s and = 28.0 ± 1.0 s measured with the Nano variant (SspB wt); = 1.5 ± 0.2 s and = 34 ± 5 s with the Micro variant (SspB R73Q) (Fig. S1)] and by the diffusion of the iLID photoreceptor within a membrane rather than within the cytosol. As a control, the entire cell was photoactivated at the end of the experiment, showing recruitment of the prey throughout the entire ER or mitochondrial network, with a less intense fluorescent signal per unit surface area of these organelles than with the localized recruitment, as the prey was distributed to binding sites over a much larger area. It is also worth noting that because of the high basal affinity of SSpB for the interacting partner (130 nM–4.7 μM for the Nano and 800 nM–47 μM for the Micro variant; ref. 22) the interaction between the two components of the dimer is nonnegligible before the photoactivation, consistent with previous studies (31). This can explain the significant basal level of prey signal localized at the ER or mitochondria before photoactivation (second column in Fig. 1 C and D). However, this drawback could potentially be reduced by using recently engineered variants with higher molar affinity (3 μM–125 μM, Milli variant; ref. 27) and with further protein engineering.
We next tested Magnets. An important feature of Magnets is that both components of the dimer are photoreceptors (24) and that both of them need to be simultaneously activated by light to achieve dimerization (Fig. 1A). In this study, the dimerization pair was represented by a photoreceptor with a negatively charged dimerization interface (nMagHigh1) and a slow off kinetic ( of 2 h) (24), which was used as the membrane-bound bait, and a photoreceptor with a positively charged dimerization interface (pMagFast2) and a very fast off kinetic ( of 6.5 s) (24). As reported by Kawano et al. (24), and confirmed in our experiments, the dissociation rate of the dimer is dominated by the component with the fastest off kinetic (Fig. S1 and Table S3). As in the case of iLID, Magnets are well-suited to achieve protein dimerization with high spatial resolution, because the dimer will dissociate rapidly as it diffuses outside of the illumination volume. Moreover, given the small size of the photoreceptors (147 aa), it is possible to generate concatamers of Magnets components, thereby increasing the efficiency of protein recruitment in small cytoplasmic volumes by increasing the avidity of binding without considerably affecting the switch-on kinetic of the system (Figs. S3 and S4 and Table S3). For most of our experiments (see figure legends) we chose to use the dimerization pair represented by nMagHigh1(2x) and pMagFast2(3x) (Fig. 1A), whose kinetics parameters in living cells at 37 °C were experimentally determined: of 1.4 ± 0.1 s and of 15 ± 3 s (Fig. S1 and Table S3). The property of Magnets to confine protein dimerization is exemplified by the two experiments shown in Fig. 1 (Fig. 1 C and D), which demonstrate the recruitment of a prey protein to small regions of the ER (Fig. 1C) or of the mitochondrial network (Fig. 1D). In the first experiment, the “negative” Magnet (nMagHigh1) (24) fused to EGFP and to the N-terminal ER-targeting sequence of cytochrome b5 (ER-nMag) was used as the bait, while the “positive” Magnet (pMagFast2) (24) fused to tgRFPt [pMagFast2(3x)-tgRFPt] was used as the prey. Upon illumination, the prey was selectively recruited to the illuminated regions of the ER. This restricted localization persisted throughout the entire photoactivation period and the prey rapidly dissociated from the ER when the activating light was turned off (Fig. 1 C and D and Movie S7). As with iLID, photoactivation of the entire cell at the end of the experiment resulted in the recruitment of the prey, at a reduced concentration compared with the spatially localized case, across the whole ER. An even more localized recruitment could be obtained upon irradiation of a 1-μm2 ROI (Fig. S5). Similar results were obtained in cells expressing the negatively charged Magnet fused to the C-terminal mitochondrial targeting sequence of OMP25 [Mito-nMagHigh1(2x)] as bait and mCherry [pMagFast2(3x)-mCherry] as the prey (Fig. 1D and Movie S8). In order for Magnets to dimerize properly cells need to be incubated at 28 °C (24) for a few hours before the illumination experiments. We found that 6 h of incubation at this temperature allows for optimal functionality and we performed all of the imaging experiments after reincubating the cells to 37 °C for at least 1 h. This approach also works for cells that are highly sensitive to culture conditions, such as neurons (Fig. S6 and Movie S9).
Quantification of Dimer Formation and Spread Outside the Illumination Volume.
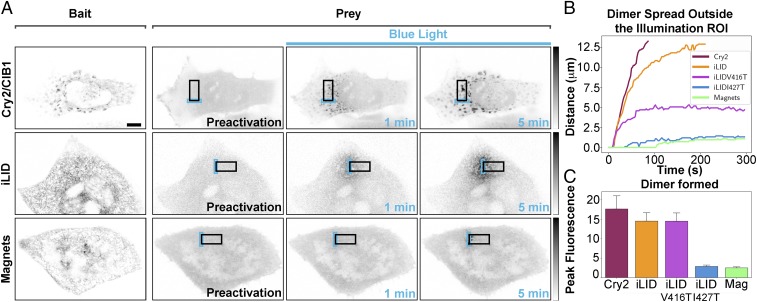
We quantified the degree of spatial localization provided by each optical dimerizer in HeLa cells expressing a soluble prey and an ER-tethered bait (Fig. 2A). To this aim we irradiated the cells within a 3-μm × 10-μm ROI (Illumination ROI, blue rectangles in Fig. 2 and Fig. S7A) for 5 min with 200-ms-long blue-light pulses every 2 s (488 nm; 7.07 W/cm2) with a confocal microscope and observed the distribution of prey for each dimerization system. By choosing a relatively large ROI (Fig. 2A and Fig. S7A) we minimized variability resulting from uneven distribution of the ER in cells. With the Cry2/CIB1 system the activated Cry2 (soluble prey) rapidly diffused outside the region of blue-light radiation and within a few minutes interacted with CIB1 molecules localized throughout the entire ER (Fig. 2A and Movie S10). With the iLID system dimerization was more spatially restricted than for Cry2/CIB1 (Fig. 2A and Movie S11) and the use of LOV2 domain variants (46) with faster off rates would be expected to allow for further confinement of protein dimerization using this system. Therefore, we tested two recently identified variants of the LOV2 domain (46) with faster switch-off kinetics: one harboring substitution V416T ( of 2.7 ± 0.3 s and = 5 ± 5 s, based on our measurement in cells at 37 °C) and another one harboring substitution I427T ( = 1.2 ± 0.3 s and = 4 ± 1 s) (Fig. S1 and Movies S12 and S13). As expected, these two variants enabled higher confinement of dimer formation, with the confinement of the I427T variants being similar to the one achieved by Magnets (Fig. 2A and Movie S14).
Fig. 2.
Quantification of the spatial confinement of dimer achieved by the three dimerization systems used. (A) HeLa cells expressing a soluble prey and an ER-tethered bait were photoexcited within a 3-µm ×10-µm ROI with 200-ms blue-light pulses at 0.5 Hz for 5 min. The area of prey–bait dimer localization for each dimerization system at 1 and 5 min after stimulation is shown. Fluorescence intensity in each time lapse is minimum–maximum-normalized (grays inverted lookup table), background-subtracted, and filtered with Gaussian blur. (Scale bar: 10 μm.) (B) Plots indicating the spatial spread of dimers away from the illuminated ROI for each system. The lines denote the maximum distance at which the normalized intensity has doubled over that of the intracellular basal intensity (n = 12 cells for Cry2/CIB1, 13 for iLID, 10 for iLID V416T, 6 for iLID I427T, and 17 for Magnets; n = 3). (C) Histograms showing the peak normalized fluorescence intensity reached by each system in the illuminated ROI after 5 min of light excitation [17.6 ± 3.4 for Cry2/CIB1, n = 12; 14.5 ± 2.2 for iLID, n = 13; 14.5 ± 2.1 for iLID V416T, n = 10; 2.9 ± 0.3 for iLID I427T, n = 6; 2.5± 0.3, n = 17 cells for Magnets (Mag); n = 3].
To quantify dimer presence outside of the blue-light illumination area we analyzed the amount of prey recruited to the ER membrane as a function of the distance from the border of the illumination area (Fig. 2 A and B and Fig. S7A). From these measurements we extracted the distance at which the fluorescence intensity had at least doubled over the basal level (more details in Fig. S7 A and B and SI Materials and Methods). The time and distance at which this threshold is reached is indicated by the plot shown in Fig. 2B (additional analysis is shown in Fig. S7C). The striking difference in dimer formation and spread with the different systems is clear from inspection of these figures. With the Cry2/CIB1 and the iLID systems a fluorescence intensity increase of 100% over the basal value was reached within seconds from the beginning of the delivery of the excitation light pulses in the ROI, peaking at >10 times the basal level (Fig. 2C), yielding substantial dimer formation over a wide surface of the ER. After 5 min of persistent pulsed illumination the dimer-positive distance for Cry2/CIB1 and iLID had spread over 13.5 μm. With Magnets dimerization outside the ROI was minimal and occurred only after about 100 s of illumination (Fig. 2B). After 5 min the dimer-positive distance for Magnets had spread to only about 1.1 μm outside the ROI (Fig. 2B). Interestingly, despite the facts that iLID V416T and iLID I427T have a three and four times faster than Magnets, respectively, they showed a spreading of dimer outside the illumination area of 4.7 μm for iLID V416T and of 1.3 μm for iLID I427T after 5 min of radiation (Fig. 2B). The ratios between the dimer-positive area for each dimerization system and the area of light radiation are shown in Fig. S7D.
These results suggest that the higher spatial confinement of the dimer generated by Magnets cannot simply be explained by the faster switch-off kinetic of this system. Furthermore, with Magnets the same spatial confinement of protein dimerization was observed both upon illumination of the ROI after prolonged dark exposure, or after a brief whole-cell illumination followed by a few minutes of darkness, that is, under conditions when the entire pool of the bait [nMagHigh1(2x)] is still active given its switch-off kinetic of hours (24) (Fig. S8). Thus, an important contributor to the high spatial confinement achieved by our Magnets configuration is the limited amount of activated prey (i.e., the amount of cytosolic prey present in the area of light radiation).
A higher degree of spatial confinement of the induced dimer comes at the expense of the total amount of locally recruited prey, as both with iLID I427T and with Magnets only a two- to threefold intensity increase in the amount of prey in the illumination area was observed (Fig. 2C and Fig. S7C). Importantly, we ruled out that inefficient optical activation by Magnets and of the iLID with fast off kinetics contributed to the lower amount of dimers formed under local illumination conditions. Local irradiation, in fact, was perfomed delivering blue-light power per surface (7.07 W/cm2) 3,000 times higher compared with that sufficient to achieve a complete activation of the systems in whole-cell activation experiments (3 × 10−3 W/cm2).
Comparison of Optical Dimerization Systems in Whole-Cell Activation Experiments.
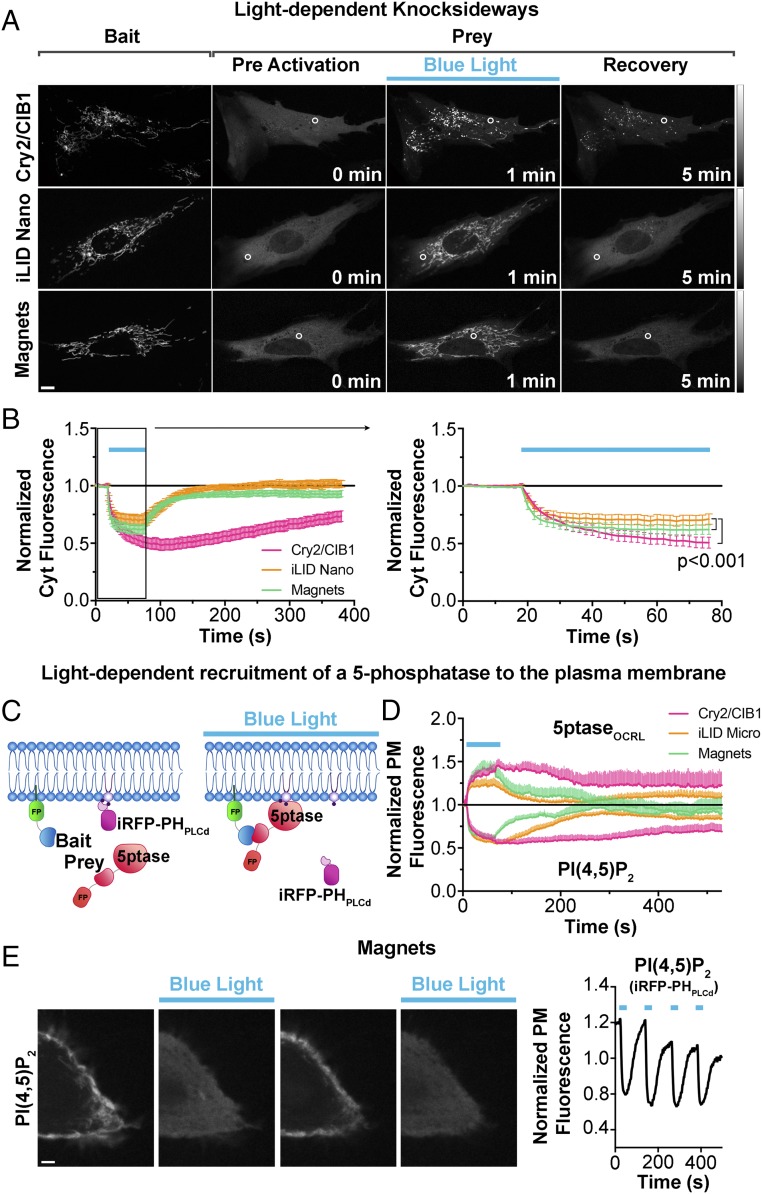
To compare the efficiency of these three optical dimerization tools in whole-cell activation conditions we perfomed optically dependent knocksideways (47–49) experiments: acute depletion of the prey from the cytosol by global illumination of cells expressing the bait at the surface of mitochondria (Movie S15). Human primary fibroblasts were transfected with the same constructs described above, irradiated for 1 min with 200-ms-long blue-light pulses every 2 s (488 nm; 3 × 10−3 W/cm2) using a confocal microscope, and loss of prey from a cytosolic ROI excluding mitochondria was measured (Fig. 3 A and B). Magnets were almost as effective as the Cry2/CIB1 system and more efficient than iLID in inducing the redistribuition of prey to mitochondria. Note that with iLID and Magnets the interruption of illumination is associated with a faster release and redistribution of the prey into the cytosol due to their faster switch-off kinetics compared with the Cry2/CIB1 system.
Fig. 3.
Comparison of the property of the three systems to recruit a prey to a membrane-bound bait upon global cell illumination. (A and B) Comparison of the efficiency of the three light-dependent dimerization systems in the blue-light-dependent (200-ms pulses at 0.5 Hz for 1 min) removal of a protein from the soluble cytosolic phase by sequestering it to the outer surface of mitochondria (so-called knocksideways; ref. 47) in human primary fibroblasts. Confocal microsopy images are shown in A (Scale bar: 10 μm; grays lookup table) and the quantification of the data is shown in B. Curves in B show a soluble prey’s removal from, and then reappearance into, the cytosol (the cytosolic ROI is indicated with a white circle). The region enclosed in the black box (Left) is shown at an expanded timescale (Right). n = 23 for Cry2/CIB1, 27 for iLID Nano, and 20 for Magnets; three independent experiments. Data were examined with a two-way ANOVA followed by Tukey’s multiple comparison test, both performed with GraphPad Prism. (C and D) Comparison of the efficiency and speed of the three light-dependent dimerization systems in the recruitment to the plasma membrane of an inositol 5-phosphstase (5ptaseOCRL) to induce loss of PI(4,5)P2, as detected by the PI(4,5)P2 probe iRFP-PHPLCδ in this membrane in COS7 cells. The cartoon in C shows the experimental setup. (D) The three systems have similar efficiency in depleting PI(4,5)P2, but recovery is much faster with iLID and Magnets (n = 10 for Cry2/CIBN, 12 cells for iLID, and 12 cells for Magnets; three independent experiments). An example of the speed and reversibility of Magnets is further illustrated in E, which shows HeLa cell expressing PM-nMag(3x), mCh-pMagFast2(3x)-5ptaseOCRL, and the PI(4,5)P2 reporter iRFP-PHPLCδ. Blue-light illumination cycles were applied as 200-ms pulses at 0.5 Hz for 60 s every 1.5 min. iRFP fluorescence detected by confocal microscopy (Left) and its quantification at the cell periphery [thus reflecting PI(4,5)P2 dynamics] (Right) are shown. (Scale bar: 5 μm.)
The three systems were also similarly efficient in the recruitment of an enzyme to the plasma membrane to catalyze a metabolic reaction. This was shown by cotransfection of COS7 cells with the phosphatidylinositol-4,5-biphosphate [PI(4,5)P2] reporter iRFP-PHPLCδ, a plasma membrane bait and an inositol 5-phosphatase prey (see Fig. 3C and SI Materials and Methods for construct details for each dimerization system). As shown in Fig. 3D, Cry2/CIB1, iLID, and Magnets are equally efficient and share a similar on rate in the recruitment of the phosphatase and in the dephosphorylation of plasma membrane PI(4,5)P2. However, the recovery of PI(4,5)P2 levels at the plasma membrane is much faster with iLID and even more with Magnets (Fig. 3D) because of the faster switch-off kinetics. An example of the fast on and off cycling of PI(4,5)P2 dephosphorylation (followed by resynthesis) achieved by Magnets is shown in Fig. 3E (Movie S16).
Light-Dependent Regulation of Protein Function at Single-Organelle Resolution.
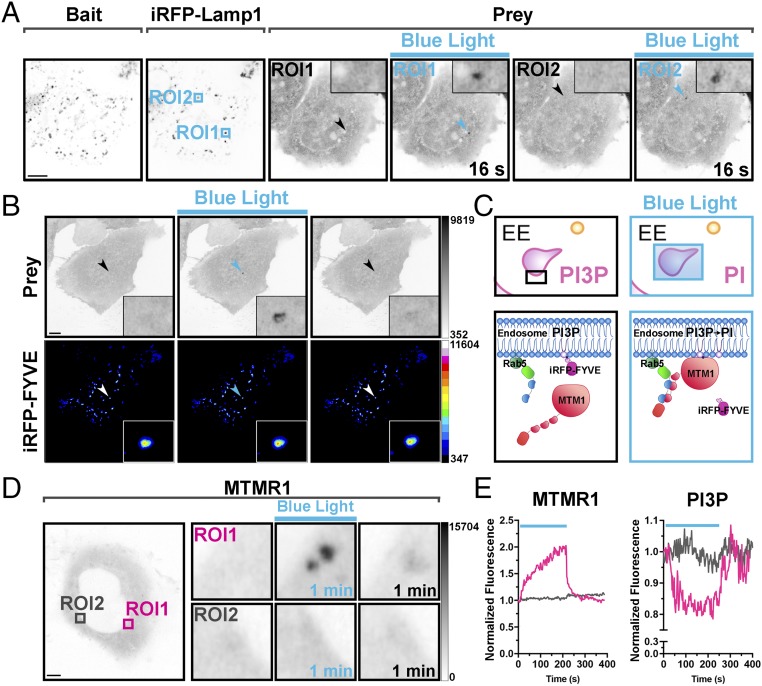
Since iLID and Magnets are functional when the photoreceptor is targeted to intracellular membranes they are suitable tools to control protein dimerization at the single-organelle level. This is not possible with the Cry2/CIB1 system in its most widely used configuration (photoreceptor as the prey) because of the rapid diffusion of the prey as shown in Fig. 1 C and D. As an example, we show that Magnets not only enables control of protein recruitment on microdomains of large organelles (Fig. 1) but can also be used for experimental manipulations at the level of a single small organelle, such as a single lysosome or endosome, as is shown in Fig. 4 A and B and Movies S17 and S18. When using an endosome-targeted Magnet as the bait (Rab5-nMag) and a Magnet fusion protein of the inositol 3-phosphatase myotubularin 1 [mCherry-pMagFast2(3x)-MTMR1] as the prey (Fig. 4C), levels of PI3P (an identity tag of early endosomes) could be reduced selectively and reversibly (within less than 1 min) on a single endosome, with the same efficiency as when the enzyme is recruited to endosomes via whole-cell irradiation (Fig. 4 D and E and Fig. S9 A and B).
Fig. 4.
Light-dependent recruitment of a prey protein at single-organelle resolution. (A) Localized protein recruitment to a single lysosome. HeLa cell expressing Lys-nMag (bait), pMagFast2(3x)-tgRFPt (prey), and iRFP-Lamp1 (lysosomal marker). iRFP fluorescence was used to select the area of blue-light illumination (arrows). Blue boxes indicate two areas subjected to sequential 200-ms blue-light pulses at 0.5 Hz for 2 and 1 min, for ROI1 and ROI2, respectively. The prey is recruited selectively to the illuminated lysosome as shown (Inset). (Magnification: Insets, 5×.) (Scale bar: 10 μm.) (B) Localized protein recruitment to a single endosome. HeLa cell expressing nMag-Rab5 (not shown), pMagFast2(3x)-mCherry (prey), and iRFP-FYVE [PI3P binding endosome marker]. iRFP fluorescence was used to select the endosome to be illuminated (arrows). (Insets) Prey recruitment at that endosome. (Magnification: Insets, 6×.) (Scale bar: 10 μm.) (C–E) Selective reduction of PI3P levels on a single endosome (magenta box in the left micrograph of D), without affecting PI3P on the other endosomes (e.g., gray box), by the specific recruitment of the PI3P phosphatase MTMR1 to that endosome. Schematic cartoon depicting the experiment is shown in C, and a quantification of the recruitment to endosomes of MTMR1 (mCherry fluorescence) and of the levels of PI3P (iRFP fluorescence) on the same endosomes is shown in E. (Magnification: D, Right Insets, 7×.) (Scale bar: 5 μm.)
All experimental procedures involving the use of mice were performed in agreement with the Yale University Institutional Animal Care and Use Committee.
Conclusions
The discovery and characterization of new photoreceptors in plants, fungi, and bacteria (50–52), combined with the directional engineering of these photoreceptors (20, 22, 25, 27), provides us with a growing palette of optical dimerization tools. These tools have demonstrated their utility in manipulating intracellular signaling pathways (40, 53, 54) and in investigating protein function at the subcellular (15, 32, 41, 55–57) and cellular level (13, 14, 16–19, 23, 24, 38–40, 53) in cell culture and in living organisms (26, 34, 58, 59). Since the spatial and temporal scale of biological processes is highly variable, it is important to choose for each application an optical dimerization pair that achieves the optimal recruitment pattern, spatial confinement, and speed.
In this study we have compared the properties of three dimerization systems, with emphasis on their ability to control protein interaction with high temporal resolution and spatial subcellular precision. While our study is focused on only three of several dimerization systems developed in recent years (refs. 13–27, 60; see ref. 28 for a complete review), the factors affecting dimer formation and spread revealed by our experiments are expected to apply also to other systems. Our results reveal that when protein dimerization is elicited in the entire cell all of the blue-light-dependent dimerization tools investigated, Cry2/CIB1, iLID, and Magnets, are similarly effective, and the temporal regulation of the biological response depends on the switch-off kinetics of each system. For applications that require protein relocation not only on a fast timescale (seconds) but also in a small subcellular region the iLID system, and to an even greater extent the Magnets system, are well-suited tools, once again because of their fast switch-off kinetics relative to the Cry2/CIB1. Results may vary depending on which of the two elements of the dimerization pairs are used as bait and prey. With the iLID configuration used in our study (photoreceptor as the bait on the membrane) the dimer rapidly dissociates as it diffuses outside the area of light radiation due to the very fast switch-off kinetic of the photoreceptor. The same applies to the Magnets system used (the photoreceptor with slow switch-off kinetics is the membrane-bound bait, while the cognate photoreceptor with fast switch-off kinetics is the prey). While with the iLID system the entire cytosolic pool of prey could participate in dimer formation even after local irradiation, with the Magnet system only the pool of cytosolic photoreceptor present in the irradiated volume could interact with the prey, so that a lower amount of dimer was formed, restricting the presence of dimer outside the illuminated volume. As demonstrated by the experiments described in this paper (Figs. 3 and 4), at least for some applications, the amount of dimerization achieved is sufficient to elicit the desired effect.
Consideration of how different parameters of each optical dimerization system are tightly interrelated is critical to achieve optimal results for any given application.
Supplementary Material
Acknowledgments
We thank F. Wilson, S. Wilson, and M. Lessard for superb technical assistance and E. B. Kromann for insightful discussions. This work was supported by Cell Biology Core of the Yale Diabetes Research Center Grant P30DK045735 and the G. Harold & Leila Y. Mathers Foundation. P.D.C. was supported in part by NIH Grants NS36251 and DK 82700 and by the Kavli Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713845115/-/DCSupplemental.
References
- 1.Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. RNAi screening comes of age: Improved techniques and complementary approaches. Nat Rev Mol Cell Biol. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 3.Fegan A, White B, Carlson JCT, Wagner CR. Chemically controlled protein assembly: Techniques and applications. Chem Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]
- 4.Putyrski M, Schultz C. Protein translocation as a tool: The current rapamycin story. FEBS Lett. 2012;586:2097–2105. doi: 10.1016/j.febslet.2012.04.061. [DOI] [PubMed] [Google Scholar]
- 5.DeRose R, Miyamoto T, Inoue T. Manipulating signaling at will: Chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013;465:409–417. doi: 10.1007/s00424-012-1208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Várnai P, Thyagarajan B, Rohacs T, Balla T. Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J Cell Biol. 2006;175:377–382. doi: 10.1083/jcb.200607116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Várnai P, Tóth B, Tóth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- 8.Zoncu R, et al. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng S, et al. A rapidly reversible chemical dimerizer system to study lipid signaling in living cells. Angew Chem Int Ed Engl. 2014;53:6720–6723. doi: 10.1002/anie.201402294. [DOI] [PubMed] [Google Scholar]
- 10.Schifferer M, Feng S, Stein F, Tischer C, Schultz C. Reversible chemical dimerizer-induced recovery of PIP2 levels moves clathrin to the plasma membrane. Bioorg Med Chem. 2015;23:2862–2867. doi: 10.1016/j.bmc.2015.03.048. [DOI] [PubMed] [Google Scholar]
- 11.Schifferer M, Feng S, Stein F, Schultz C. Reversible chemical dimerization by rCD1. Methods Enzymol. 2017;583:173–195. doi: 10.1016/bs.mie.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 12.Komatsu T, et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 14.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 15.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 19.Strickland D, et al. TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lungu OI, et al. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller K, et al. Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res. 2013;41:e124. doi: 10.1093/nar/gkt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guntas G, et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci USA. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 24.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 25.Taslimi A, et al. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol. 2016;12:425–430. doi: 10.1038/nchembio.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaberniuk AA, Shemetov AA, Verkhusha VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods. 2016;13:591–597. doi: 10.1038/nmeth.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerman SP, et al. Tuning the binding affinities and reversion kinetics of a light inducible dimer allows control of transmembrane protein localization. Biochemistry. 2016;55:5264–5271. doi: 10.1021/acs.biochem.6b00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rost BR, Schneider-Warme F, Schmitz D, Hegemann P. Optogenetic tools for subcellular applications in neuroscience. Neuron. 2017;96:572–603. doi: 10.1016/j.neuron.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K, Cui B. Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 2015;33:92–100. doi: 10.1016/j.tibtech.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak GP, Strickland D, Vrana JD, Tucker CL. Benchmarking of optical dimerizer systems. ACS Synth Biol. 2014;3:832–838. doi: 10.1021/sb500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. Correlating in vitro and in vivo activities of light-inducible dimers: A cellular optogenetics guide. ACS Synth Biol. 2016;5:53–64. doi: 10.1021/acssynbio.5b00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang X, Jost AP-T, Weiner OD, Tang C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell. 2013;24:2419–2430. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graziano BR, et al. A module for Rac temporal signal integration revealed with optogenetics. J Cell Biol. 2017;216:2515–2531. doi: 10.1083/jcb.201604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley CE, et al. Reversible optogenetic control of subcellular protein localization in a live vertebrate embryo. Dev Cell. 2016;36:117–126. doi: 10.1016/j.devcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toettcher J, Gong D, Lim WA, Weiner O. A light-based feedback control system for generating user-defined intracelullar signaling dynamics. Biophys J. 2012;102:41a. [Google Scholar]
- 36.Toettcher JE, Gong D, Lim WA, Weiner OD. Light control of plasma membrane recruitment using the Phy-PIF system. Methods Enzymol. 2011;497:409–423. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller K, et al. Synthesis of phycocyanobilin in mammalian cells. Chem Commun (Camb) 2013;49:8970–8972. doi: 10.1039/c3cc45065a. [DOI] [PubMed] [Google Scholar]
- 38.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Nan D, Fan J, Bogan JS, Toomre D. Optogenetic activation reveals distinct roles of PIP3 and Akt in adipocyte insulin action. J Cell Sci. 2016;129:2085–2095. doi: 10.1242/jcs.174805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong D, et al. Frequency and amplitude control of cortical oscillations by phosphoinositide waves. Nat Chem Biol. 2016;12:159–166. doi: 10.1038/nchembio.2000. [DOI] [PubMed] [Google Scholar]
- 41.Sinnen BL, et al. Optogenetic control of synaptic composition and function. Neuron. 2017;93:646–660.e5. doi: 10.1016/j.neuron.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan L. Optogenetic control of molecular motors and organelle distributions in cells. Biophys J. 2016;110:317a. doi: 10.1016/j.chembiol.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Che DL, Duan L, Zhang K, Cui B. The dual characteristics of light-induced cryptochrome 2, homo-oligomerization and heterodimerization, for optogenetic manipulation in mammalian cells. ACS Synth Biol. 2015;4:1124–1135. doi: 10.1021/acssynbio.5b00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 45.Taslimi A, et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, et al. LOVTRAP: An optogenetic system for photoinduced protein dissociation. Nat Methods. 2016;13:755–758. doi: 10.1038/nmeth.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MS, Hirst J. Rapid Inactivation of Proteins by Knocksideways. Wiley; Hoboken, NJ: 2013. [DOI] [PubMed] [Google Scholar]
- 48.Hirst J, et al. Contributions of epsinR and gadkin to clathrin-mediated intracellular trafficking. Mol Biol Cell. 2015;26:3085–3103. doi: 10.1091/mbc.E15-04-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garay C, et al. Epidermal growth factor-stimulated Akt phosphorylation requires clathrin or ErbB2 but not receptor endocytosis. Mol Biol Cell. 2015;26:3504–3519. doi: 10.1091/mbc.E14-09-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoltowski BD, et al. Conformational switching in the fungal light sensor vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science. 2016;354:343–347. doi: 10.1126/science.aaf9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bugaj LJ, O’Donoghue GP, Lim WA. Interrogating cellular perception and decision making with optogenetic tools. J Cell Biol. 2017;216:25–28. doi: 10.1083/jcb.201612094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC. Optogenetic control of organelle transport and positioning. Nature. 2015;518:111–114. doi: 10.1038/nature14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi-Takagi A, et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–338. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guglielmi G, Barry JD, Huber W, De Renzis S. An optogenetic method to modulate cell contractility during tissue morphogenesis. Dev Cell. 2015;35:646–660. doi: 10.1016/j.devcel.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harterink M, et al. Light-controlled intracellular transport in Caenorhabditis elegans. Curr Biol. 2016;26:R153–R154. doi: 10.1016/j.cub.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 60.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.