Significance
Ion channels are proteins that mediate the flow of ions across cell membranes. Human genetic mutations of one type of ion channel, called hTRPM4, underlie a form of progressive familial heart block. Its distribution among many tissues, however, suggests that its functions are broad. We have solved the atomic structure of hTRPM4 to an overall resolution of 3.7 Å. The channel is composed of four identical subunits surrounding a central pore. We show the path of Na+ ions through the channel and point out aspects of the channel’s internal machinery that may affect its function. The structure will enable more directed experiments to understand the physiological function of this channel.
Keywords: ion channel, transient receptor potential channel, cardiac arrhythmia, cryomicroscopy
Abstract
Transient receptor potential melastatin subfamily member 4 (TRPM4) is a widely distributed, calcium-activated, monovalent-selective cation channel. Mutations in human TRPM4 (hTRPM4) result in progressive familial heart block. Here, we report the electron cryomicroscopy structure of hTRPM4 in a closed, Na+-bound, apo state at pH 7.5 to an overall resolution of 3.7 Å. Five partially hydrated sodium ions are proposed to occupy the center of the conduction pore and the entrance to the coiled-coil domain. We identify an upper gate in the selectivity filter and a lower gate at the entrance to the cytoplasmic coiled-coil domain. Intramolecular interactions exist between the TRP domain and the S4–S5 linker, N-terminal domain, and N and C termini. Finally, we identify aromatic interactions via π–π bonds and cation–π bonds, glycosylation at an N-linked extracellular site, a pore-loop disulfide bond, and 24 lipid binding sites. We compare and contrast this structure with other TRP channels and discuss potential mechanisms of regulation and gating of human full-length TRPM4.
Transient receptor potential (TRP) channels are permeable to cations, with most conducting both monovalent and divalent ions (1). TRP melastatin subfamily member 4 (TRPM4) and TRPM5 have the distinction among TRPM channels of being activated by, but impermeable to, Ca2+ (2, 3), with preferred conduction for Na+ > K+ > Cs+ > Li+ >> Ca2+, Cl−. Under physiological conditions its single-channel conductance is 25 pS (4, 5). Activation is modulated by PI(4,5)P2, PKC phosphorylation, calcium, and calmodulin (6, 7). TRPM4 channel blockers include intracellular nucleotides such as ATP, ADP, AMP, and AMP-PNP (adenylyl-imidodiphosphate), with IC50s of 1.3 to 1.9 μM (8).
TRPM4 is widely expressed in many tissues, and appears to be an important regulator of dendritic cell migration, mast cells, lymphocytes, pancreatic β-cells, neurons, and smooth muscle cells in the vasculature and bladder (9–13). TRPM4 has also been shown to be important for proper activation in heart conduction pathways (14, 15); it is active in the late phase of repolarization of the cardiac ventricular action potential and enhances beta adrenergic-mediated inotropy. Mutations are associated with conduction defects (16–18), resembling those associated with mutations in the cardiac voltage-gated Na+ channel NaV1.5 (SCN5A) (19–22).
During preparation of this manuscript, three cryo-EM structures of the TRPM4 channel were reported (23–25): hTRPM4 in lipid nanodiscs with and without calcium bound at ∼3 Å (25), and another bound to decavanadate at ∼3.5 Å (23). Here, we present the cryo-EM structure of full-length human TRPM4 at an overall resolution of 3.7 Å, with putative Na+ ions in the conduction pathway. We provide additional information to the recently published structures, with details on subunit interactions, domain arrangement, sodium selectivity, and binding sites in the structure of TRPM4.
Structure of Human TRPM4
Full-length human TRPM4 (1,214 residues) was expressed using the BacMam expression system (Materials and Methods). The TRPM4 construct used for expression retained the key functional properties of the wild-type channel, such as permeability to Na+ and K+ and activation by intracellular calcium (Fig. S1A). The protein, obtained in the absence of exogenous calcium ions, eluted as a monodisperse peak in 0.1% (wt/vol) digitonin during gel filtration; the peak fraction was subjected to structure determination by cryo-EM (Fig. S1B). We calculated the structure of the TRPM4 tetramer from 61,653 particles selected from an initial dataset of 244,216 particle images recorded on a direct electron detector. From these particles, we obtained a map of TRPM4 with an overall resolution of 3.7 Å, as estimated by a Fourier shell correlation of 0.143 (Fig. S2 and Table S1). The local resolution of the transmembrane domain is 3.5 Å. At this resolution, we were able to build an atomic model for the S1 to S6 region and the N- and C-terminal domains de novo, without reference to other TRP channel structures (Fig. S3).
The tetrameric TRPM4 ion channel overall dimensions were 150 × 120 × 120 Å, consisting of N-terminal cytosolic domains, a transmembrane domain with six membrane-spanning helices per monomer, and C-terminal domains (Fig. 1 A–D). The first four transmembrane domain helices (S1 to S4) are followed by a pore domain with two long helices (S5 and S6) linked by a pore helix. The transmembrane domain has four prominent features: (i) The TRP domain contains two helices, one long following S6 and one short connecting with the C-terminal domain (Fig. 1 E and F); (ii) the voltage-sensor domain (VSD) lacks the four arginine residues on S4 that are responsible for the strong voltage sensing in classic voltage-gated sodium and potassium channels (26, 27). The two conserved arginine residues (Arg892 and Arg905) found at either end of S4 contribute little to voltage sensitivity (Figs. S4 and S5); (iii) a long pore loop between S5 and S6 (66 residues, amino acids 951 to 1,016); and (iv) a short helix between S2 and S3 (S2–S3 linker), lying parallel to the inner layer of the plasma membrane (Fig. 1 E and F). The density of the intracellular S2–S3 loop in the overall TRPM4 structure is poorly resolved, indicating potential dynamic motion. Beyond these features, the most striking difference between TRPM4 and other TRP channel subfamily members is an ∼700-amino acid-long N-terminal domain that interacts with the TRP domain, pre-S1 helix, and C-terminal domain. The C-terminal domain is also unique in that a connecting helix bridges the TRP domain and the conserved coiled-coil domain (Fig. S5).
Fig. 1.
Overall structure of human full-length TRPM4 in the apo state. (A and B) Side (A) and top (B) views of the cryo-EM reconstruction density map of human TRPM4 at 3.7-Å overall resolution; each subunit is color-coded. (C and D) Ribbon diagrams representing the same orientation and colors with the channel’s dimensions indicated. (E) Structural details of a single human TRPM4 subunit. (F) Linear diagram depicting the major structural domains, color-coded to match the ribbon diagram. The N-linked N992 glycosylation site (N-G) and the Cys993–Cys1011 disulfide bond are indicated.
Six cholesteryl hemisuccinates (CHSs) per monomer are evident in the tetrameric TRPM4, embedded in the hydrophobic transmembrane regions. As expected, CHS hydrophilic regions face extracellularly or intracellularly, while their hydrophobic tails face the transmembrane region (Fig. S6). We also observe an extracellular glycosylation site at N992 on each monomer (Fig. S7) and two cysteines, C993 and C1011, forming a disulfide bond in the pore loop (Fig. S7). The disulfide bond is adjacent to the glycosylation site and near the initiation of S6, perhaps indicating a site for small-molecule recognition or redox sensitivity.
Domain Interactions
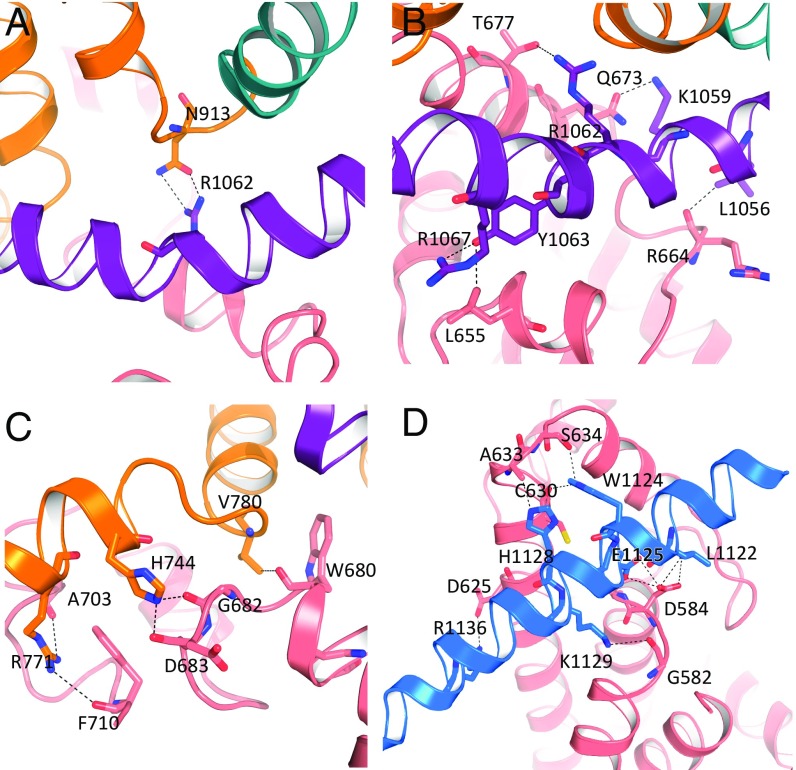
The TRP domain commonly plays a role in channel activation and allosteric modulation (28, 29). In TRPM4, the TRP domain makes contact with the S4–S5 linker via hydrogen bonding, similar to that in TRPV1 and TRPA1 (30–32). N913 on the S4–S5 linker forms hydrogen bonds with R1062 on the TRP domain (Fig. 2A). The polar interactions between the TRP domain and N-terminal domain are extensive, involving three positively charged residues (K1059, R1062, and R1067) and two hydrophobic residues (L1056 and Y1063) on the TRP domain, and L655, Q673, T677, and R664 of the N-terminal domain (Fig. 2 B and C). The N-terminal domain also interacts with the connecting helix in the C-terminal domain. This helix bridges the N- and C-terminal domains (Fig. 2D), indicating its potential importance for TRPM4 subunit assembly.
Fig. 2.
Detailed domain interactions of human TRPM4. Interactions between (A) the TRP domain and S4–5 linker, (B) TRP domain and N terminus, (C) N terminus and pre-S1 helix, and (D) N and C termini. Residues at domain interfaces are labeled with hydrogen bonds and electrostatic interactions as dashed lines.
TRPM4: Aromatic Interactions
Aromatic interactions, such as π–π interactions, are important in protein folding and thermal stability (33). In particular, two ordered aromatic π–π stacks are present in TRPM4’s transmembrane domain. The aromatic ring planes of F793 on S1 and F902 on S4 form an edge-to-face π–π interaction (Fig. 3A). A face-to-face π–π stack, 4.2 Å from the ring centroid, is observed in Y944 near the end of S5 and F1027 on S6 (Fig. 3B). Similar π–π interactions are not observed in TRPV1, but in TRPA1 the S5 helix’s F884 forms π–π interactions with F944 on the S6 helix in a similar position as in TRPM4, suggesting relevant interhelical interactions (30).
Fig. 3.
Aromatic interactions of human TRPM4. Ribbon diagrams showing aromatic interactions of human TRPM4: π–π bonds (A and B) and cation–π bonds (C–E). Residues and helices at these interactions are labeled.
Cation–π interactions are as important as hydrogen bonding, hydrophobic effects, and ion pairing in determining protein structures (34). Several cation–π interactions are present in TRPM4 structures. In the S1–S2 loop, F805 interacts with R882 in the S3–S4 loop (Fig. 3C), and R955 interacts with W1000 within the pore loop (Fig. 3D). Another two pairs of cation–π interactions are located on the TRP domain (Y1063/R1067, F1069/R1072), with Y1063/R1067 making polar interactions with the N-terminal domain (Fig. 3E). These interactions with the TRP domain should facilitate allosteric coupling.
Cytosolic Domains
The N- and C-terminal domains of ion channels are usually cytosolic. In TRPM4, the four C termini form a homotetramer via parallel coiled coils, sitting directly below the pore. Interestingly, preceding each coiled-coil helix is an ∼25-amino acid connecting helix that bends ∼120° to form an inverted “L” with the coiled-coil helix (Fig. 4A, Left). The four connecting helix subunits are perpendicular to each other when viewed from the top, and link with the coiled coils to form a central hole (Fig. 4A, Right). At the top of this hole, the side chain of S1143 forms a hydrogen bond with the backbone of D1144 on the adjacent helix, with the four hydrogen bonds stabilizing the coiled coil. Two leucines and one glutamate interaction “boxes” in the coiled-coil hole stabilize the structure. These consist of leucines (L1148) forming hydrophobic interactions at 3.6 Å, L1162 forming hydrophobic interactions at 3.5 Å, and E1169 forming a hydrogen bond at 2.9 Å (Fig. 4B).
Fig. 4.
Cytosolic domains of human TRPM4. (A) Side (Left) and top (Right) views of the C-terminal domain. (B) Top views along the enlarged central hole formed by coiled-coil helices are shown with the key residues labeled. Hydrogen bonds and electrostatic interactions are shown as dashed lines and distances are indicated. (C) Side and top views of the N-terminal domain. (D) Side views of a single subunit of the N-terminal domain. ALR, ankyrin-like repeat; ISD, isolated subdomain. The arrows in C and D are pointing to a helix of each N-terminal domain. These hydrophobic regions are anchored to the inner leaflet of the plasma membrane.
The N-terminal region of TRPM4 shields the coil-coiled domain (Fig. 4C). The proximal N terminus contains five β-sheets and six α-helices forming a relatively independent subdomain (Fig. 4D), followed by ankyrin-like repeats (Fig. 4D). Two long helices and one short helix form a stable connecting subdomain that interacts with the C terminus and supports the transmembrane domain. Three short helices and several loops form a linker subdomain extending to the transmembrane domain (Fig. 4D). Notably, a short helix in the linker subdomain (amino acids P689 to A696; PIWALVLA) is hydrophobic and anchors to the inner leaflet of the plasma membrane (Fig. 4 C and D, arrows).
The Ion Conduction Pathway
Two gates control TRPM4’s conduction: 975FGQ977 in the selectivity filter and I1040 at the intracellular gate (Fig. 5 A and B). In the apo state, both gates appear to be closed (Fig. 5 A and B). The three-residue motif (975FGQ977) of the TRPM4 selectivity filter is highly conserved in TRPM2, TRPM5, and TRPM8, while FGE/YGE are the corresponding residues in TRPM1, TRPM3, TRPM6, and TRPM7 (Fig. S5). In TRPM4, the side chains of Q977 and the backbone carbonyls of G976 and F975 pointing into the ion pathway form the external entrance to the central cavity (Fig. 5A). The external entrance of the ion conduction pathway is electronegative, presumably concentrating cations (Fig. 5 C and D). The pore selectivity filter dimensions, defined by the diagonal side-chain carbonyl oxygens of Q977 and the main-chain carbonyl oxygens of G976, are 7.4 and 6.0 Å, respectively (Fig. S8). In our structure, the most restricted site in the ion permeation pathway is the lower gate (Fig. 5 A and B), formed by the diagonal side chains of S6’s I1040 at 5.1 Å and S1044 at 5.2 Å (Fig. S8).
Fig. 5.
TRPM4’s ion conduction pathway. (A) The ion conduction pathway is shown as dots and mapped using HOLE. (B) Pore radius along the central axis. The side chains of Q977 to F975 form a narrow constriction at the selectivity filter, while I1040 is the most restricted site of the lower gate. Side (C) and bottom (D) views of the electrostatic map. The surface is colored according to the calculated electrostatic potential, revealing the tetrameric distribution of charge. Blue indicates positive potential, red indicates negative, with transparent white being neutral.
At the top of the permeation pathway, in the linker between the pore helix and S6, are an N-linked N992 glycosylation site and a conserved disulfide bond (C993–C1011; Fig. 1F and Fig. S7). The extracellular entrance to the ion conduction pathway is electronegative, as noted above (Fig. 5 C and D). Four spherical nonprotein densities in the map are present in the ion conduction pathway and at the periplasmic cytosolic exit of the channel. Surprisingly, the fifth and strongest spherical density is found in the entrance of the coiled-coil domain in the C terminus. Since the only cation in the purification buffer is sodium, we suspect that the densities in the 40-Å-long pore are hydrated Na+ (Na1 to 5; Fig. 6), but we cannot exclude the possibility that potassium ions could normally occupy the site at the cytosolic surface. Na1 and Na2 are surrounded by the side chains of Q977 and the main-chain pore-loop carbonyls of G976 and F975 (Fig. 6B, Upper). The distance between Na1 and Na2 and the carbonyls of G976 is 3.4 and 4.9 Å, respectively, consistent with a partially hydrated Na+ (hydrated radius, 2.4 Å). The four backbone carbonyl oxygens of G976 coordinate the putative Na+ in the Na2 site just below the selectivity filter. Na3 and Na4 are located within the lower gate (Fig. 6B, Middle). Na3 is recognized by the backbone carbonyl groups of I1040 and the side chains of S1044. Just below Na3, Na4 interacts with the main-chain carbonyl groups and the side chains of S6’s S1044. The isoleucine in the lower gate of the ion conduction pathway is highly conserved within the TRPM subfamily, whereas the serine, S1044, is only found in the monovalent-preferring channels, TRPM4 and TRPM5 (Fig. S5). Thus, S1044 might contribute to specific TRPM4/5 gating. Finally, Na5 is located in the C-terminal coiled-coil entrance and is recognized by the backbone carbonyl groups of S1143 and the side chains of the negatively charged D1144 (Fig. 6B, Lower).
Fig. 6.
Putative sodium binding sites of human TRPM4. Overall (A) and enlarged (B) views of the putative Na+ binding sites; side views of TRPM4. Five nonprotein densities, consistent with partially hydrated Na+ ions, along the pore and at the entrance of the coiled-coil domain, are indicated as purple spheres and labeled as Na1 to Na5 (Top to Bottom). (B) Enlarged densities of partially hydrated Na+ (Na1 to Na5) are shown at the binding sites. The key amino acid residues are labeled, and interactions between the ions and binding sites are shown as dashed lines.
Discussion
We describe human full-length TRPM4’s apo structure with densities in its conduction pathway that are consistent with Na+ ions. Confirmed human disease-related mutations associated with progressive familial heart block type IB [PFHB1B; OMIM (Online Mendelian Inheritance in Man) 604559] are mapped onto the structure in Fig. S9. There are several phenotypes in TRPM4-deficient mice, including abnormal cardiac conduction (17, 35), hypertension (36), more severe IgE-mediated acute immune responses (13), and inflammation-induced neurodegeneration and spinal cord injury in the central nervous system (12, 37).
Overall, the structures in the four independent publications (23–25) are similar, with variability, as expected, in the more flexible and thus lower-resolution N terminus (Fig. S10). We attribute the five central path densities to sodium ions, although structure–function studies and/or crystallography should be carried out to confirm this hypothesis (Fig. S10A). Six cholesteryl hemisuccinates, a lipid anionic detergent used in purification, are seen per monomer in the region between the VSD and pore domain (Fig. S10A), indicating potential lipid binding sites. We also point out interactions between domains (Fig. 2) and aromatic (π–π and cation–π) interactions (Fig. 3) that were not described in detail in the other publications. Our primary aim was to shed light on the mechanism of the TRPM4 channel’s relative selectivity for monovalent cations compared with other TRP channels. TRPM4 and TRPM5 are ∼40% identical and the most closely related of the TRPM family. Both are permeable to monovalent ions such as Na+ and K+, but poorly conduct divalent ions.
TRPM4’s ion selectivity filter and lower gate are identified as G976 and I1040, respectively. In mammalian voltage-gated Na+-selective channels, positively charged lysine residues in the pore’s “DEKA” motif mediate sodium selectivity by a combination of size restriction and charge. Interestingly, there are two positively charged residues, Arg965 and Arg969, located in the pore helix of TRPM4/5, which are not found in the other, nonselective, TRPMs. Since the serine at 1044 is only found in the Na+-selective TRPM4 and TRPM5 channels (e.g., asparagine in TRPM1 to TRPM3, TRPM6, and TRPM7), this residue should be tested for effects on gating. Given its exposed location, it might also be phosphorylated to alter conduction.
Human TRPM4’s structure also offers interesting insight into its assembly and regulation, including details of domain interactions and aromatic interactions in TRPM4’s apo state, TRP domain interactions with the S4–S5 linker and the N-terminal domain, N–C termini interactions, and the Cys993–Cys1011 disulfide bond in the pore loop. These all provide interesting starting points to examine potential regulators of gating.
Materials and Methods
Protein Expression and Purification.
The full-length human TRPM4 construct (amino acids 1 to 1,214) was cloned into the pEG BacMam vector (38), and a maltose-binding protein tag was added to its N terminus. P3 baculovirus was produced in the Bac-to-Bac Baculovirus Expression System (Invitrogen). HEK293S GnTI− cells were infected with 10% (vol/vol) P3 baculovirus at a density of 2.0 to 3.0 × 106 cells per mL for protein expression. After 12 to 24 h, 10 mM sodium butyrate was added and the temperature was reduced to 30 °C. Cells were harvested at 72 h after transduction, and resuspended in a buffer containing 30 mM Hepes, 150 mM NaCl, and 1 mM DTT (pH 7.5) with EDTA-free protease inhibitor mixture (Roche) for 30 min followed by solubilization for 2 to 3 h in a buffer containing 1.0% (wt/vol) N-dodecyl-beta-d-maltopyranoside (Affymetrix), 0.1% (wt/vol) cholesteryl hemisuccinate (Sigma), 30 mM Hepes, 150 mM NaCl, and 1 mM DTT (pH 7.5) with EDTA-free protease inhibitor mixture (Roche). The supernatant was isolated by 100,000 × g centrifugation for 60 min, followed by incubation in amylose resin (New England BioLabs) at 4 °C overnight. The resin was washed with 20 column volumes of washing buffer containing 25 mM Hepes, 150 mM NaCl, 0.1% (wt/vol) digitonin, 0.01% (wt/vol) CHS, and 1 mM DTT (pH 7.5) with EDTA-free protease inhibitor mixture (Roche). The protein was eluted with four column volumes of washing buffer with 40 mM maltose. The protein was then concentrated to 0.5 mL with a 100-kDa molecular mass cutoff concentrator (Millipore) before further purification on a Superose 6 column in a buffer composed of 25 mM Hepes, 150 mM NaCl, 0.1% (wt/vol) digitonin, and 1 mM DTT (pH 7.5). The peak, corresponding to tetrameric TRPM4, was collected and concentrated to 7.8 mg/mL for electron cryomicroscopy.
Electron Microscopy Data Collection.
Purified human TRPM4 protein (3.5 µL) in digitonin buffer at 7.8 mg/mL was applied onto a glow-discharged, 400-mesh copper Quantifoil R1.2/1.3 holey carbon grid. Grids were blotted for 7 s at 100% humidity and flash-frozen by liquid nitrogen-cooled liquid ethane using an FEI Vitrobot Mark I. The grid was then loaded onto an FEI TF30 Polara electron microscope operating at 300 kV accelerating voltage. Image stacks were recorded on a Gatan K2 Summit direct detector set in superresolution counting mode using SerialEM (39), with a defocus range between 1.5 and 3.0 μm. The electron dose was set to 8 e−⋅physical pixel−1⋅s−1 and the subframe time to 200 ms. A total exposure time of 10 s resulted in 50 subframes per image stack. The total electron dose was 52.8 e− per Å2 (∼1.1 e− per Å2 per subframe).
Image Processing and 3D Reconstruction.
Image stacks were gain-normalized and binned by 2× to a pixel size of 1.23 Å before drift and local movement correction using MotionCor2 (40). The images from the sum of all frames with dose weighting were subjected to visual inspection and poor images were removed before particle picking. Particle picking and subsequent bad particle elimination through 2D classification were performed using Python scripts/programs (41) with minor modifications in the 8× binned images. The selected 2D class averages were used to build an initial model using the common lines approach implemented in SPIDER (42) through Maofu Liao’s Python scripts (41), which was applied to later 3D classification using RELION (43). The contrast transfer function (CTF) parameters were estimated using CTFFIND4 (44) using the sum of all frames without dose weighting. Quality particle images were then boxed out from the dose-weighted sum of all 50 frames and subjected to RELION 3D classification. RELION 3D refinements were then performed on selected classes for the final map. The resolution of this map was further improved by using the sum of subframes 1 to 14.
Model Building, Refinement, and Validation.
For the full-length protein, a polyalanine model was first built in Coot (45). Taking advantage of the defined geometry of helices and clear bumps for Cα atoms in the transmembrane domain, amino acid assignment was subsequently achieved based primarily on the clearly defined side-chain densities of bulky residues. Resolution of the first part of the N-terminal domain was insufficient for backbone tracing, and hence the polyalanine model was used for that region. The refined atomic model was further visualized in Coot. A few residues with side chains moving out of the density during the refinement were fixed manually, followed by further refinement. The TRPM4 model was then subjected to global refinement and minimization in real space using the PHENIX (46) module phenix.real_space_refine (47), and geometries of the model were assessed using MolProbity (48) in the comprehensive model validation section of PHENIX. The final model exhibited good geometry, as indicated by the Ramachandran plot (preferred region, 90.42%; allowed region, 9.33%; outliers, 0.25%). The pore radius was calculated using HOLE (49).
Electrophysiology.
Whole-cell currents were recorded from the same cells used for protein expression as described above. Recordings were conducted at room temperature with an Axopatch 200B patch-clamp amplifier controlled via a Digidata 1440A (Molecular Devices). Patch pipettes of 2 to 5 MΩ contained 156 mM CsCl, 1 mM MgCl2, 10 mM CaCl2, 10 mM EGTA, and 10 mM Hepes, yielding 10 µM free calcium (calculated with https://web.stanford.edu/∼cpatton/webmaxcS.htm) (pH 7.4). The saline bath solution contained 140 mM NaCl, 4.8 mM KCl, 1.2 mM MgCl2, 10 mM glucose, and 10 mM Hepes (pH 7.4). TRPM4 current was inhibited using 9-phenanthrol at 10 µM. Cells were held at 0 mV, and 200-ms ramps from −100 to 100 mV were applied every 2 s. Currents were digitized at 10 kHz and low-pass-filtered at 2 kHz.
Supplementary Material
Acknowledgments
We thank Dr. Steve Harrison and the Cryo-EM Facility (Harvard Medical School) for use of their microscopes. We thank Dr. Maofu Liao for providing the Python scripts and help in image processing. We thank members of the D.E.C. laboratory for productive discussions. J.Z. was supported by Thousand Young Talents Program of China and National Natural Science Foundation of China Grant 31770795. This work was supported by funds from the Howard Hughes Medical Institute (to D.E.C.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6BWI) and Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb (EMDB code EMD-7299).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722038115/-/DCSupplemental.
References
- 1.Clapham DE, Runnels LW, Strübing C. The TRP ion channel family. Nat Rev Neurosci. 2001;2:387–396. doi: 10.1038/35077544. [DOI] [PubMed] [Google Scholar]
- 2.Ehara T, Noma A, Ono K. Calcium-activated non-selective cation channel in ventricular cells isolated from adult guinea-pig hearts. J Physiol. 1988;403:117–133. doi: 10.1113/jphysiol.1988.sp017242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Launay P, et al. TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell. 2002;109:397–407. doi: 10.1016/s0092-8674(02)00719-5. [DOI] [PubMed] [Google Scholar]
- 4.Nilius B, et al. The selectivity filter of the cation channel TRPM4. J Biol Chem. 2005;280:22899–22906. doi: 10.1074/jbc.M501686200. [DOI] [PubMed] [Google Scholar]
- 5.Nilius B, et al. Voltage dependence of the Ca2+-activated cation channel TRPM4. J Biol Chem. 2003;278:30813–30820. doi: 10.1074/jbc.M305127200. [DOI] [PubMed] [Google Scholar]
- 6.Nilius B, et al. Regulation of the Ca2+ sensitivity of the nonselective cation channel TRPM4. J Biol Chem. 2005;280:6423–6433. doi: 10.1074/jbc.M411089200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Okawa H, Wang Y, Liman ER. Phosphatidylinositol 4,5-bisphosphate rescues TRPM4 channels from desensitization. J Biol Chem. 2005;280:39185–39192. doi: 10.1074/jbc.M506965200. [DOI] [PubMed] [Google Scholar]
- 8.Nilius B, Prenen J, Voets T, Droogmans G. Intracellular nucleotides and polyamines inhibit the Ca2+-activated cation channel TRPM4b. Pflugers Arch. 2004;448:70–75. doi: 10.1007/s00424-003-1221-x. [DOI] [PubMed] [Google Scholar]
- 9.Barbet G, et al. The calcium-activated nonselective cation channel TRPM4 is essential for the migration but not the maturation of dendritic cells. Nat Immunol. 2008;9:1148–1156. doi: 10.1038/ni.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earley S. TRPM4 channels in smooth muscle function. Pflugers Arch. 2013;465:1223–1231. doi: 10.1007/s00424-013-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H, et al. TRPM4 controls insulin secretion in pancreatic beta-cells. Cell Calcium. 2007;41:51–61. doi: 10.1016/j.ceca.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schattling B, et al. TRPM4 cation channel mediates axonal and neuronal degeneration in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat Med. 2012;18:1805–1811. doi: 10.1038/nm.3015. [DOI] [PubMed] [Google Scholar]
- 13.Vennekens R, et al. Increased IgE-dependent mast cell activation and anaphylactic responses in mice lacking the calcium-activated nonselective cation channel TRPM4. Nat Immunol. 2007;8:312–320. doi: 10.1038/ni1441. [DOI] [PubMed] [Google Scholar]
- 14.Kruse M, et al. Impaired endocytosis of the ion channel TRPM4 is associated with human progressive familial heart block type I. J Clin Invest. 2009;119:2737–2744. doi: 10.1172/JCI38292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, et al. Gain-of-function mutations in TRPM4 cause autosomal dominant isolated cardiac conduction disease. Circ Cardiovasc Genet. 2010;3:374–385. doi: 10.1161/CIRCGENETICS.109.930867. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs G, et al. Enhanced β-adrenergic cardiac reserve in Trpm4−/− mice with ischaemic heart failure. Cardiovasc Res. 2015;105:330–339. doi: 10.1093/cvr/cvv009. [DOI] [PubMed] [Google Scholar]
- 17.Mathar I, et al. Increased β-adrenergic inotropy in ventricular myocardium from Trpm4−/− mice. Circ Res. 2014;114:283–294. doi: 10.1161/CIRCRESAHA.114.302835. [DOI] [PubMed] [Google Scholar]
- 18.Stallmeyer B, et al. Mutational spectrum in the Ca(2+)-activated cation channel gene TRPM4 in patients with cardiac conductance disturbances. Hum Mutat. 2012;33:109–117. doi: 10.1002/humu.21599. [DOI] [PubMed] [Google Scholar]
- 19.Demion M, et al. Trpm4 gene invalidation leads to cardiac hypertrophy and electrophysiological alterations. PLoS One. 2014;9:e115256. doi: 10.1371/journal.pone.0115256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse M, Pongs O. TRPM4 channels in the cardiovascular system. Curr Opin Pharmacol. 2014;15:68–73. doi: 10.1016/j.coph.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Abriel H, Syam N, Sottas V, Amarouch MY, Rougier JS. TRPM4 channels in the cardiovascular system: Physiology, pathophysiology, and pharmacology. Biochem Pharmacol. 2012;84:873–881. doi: 10.1016/j.bcp.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Schott JJ, et al. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 23.Winkler PA, Huang Y, Sun W, Du J, Lü W. Electron cryo-microscopy structure of a human TRPM4 channel. Nature. 2017;552:200–204. doi: 10.1038/nature24674. [DOI] [PubMed] [Google Scholar]
- 24.Guo J, et al. Structures of the calcium-activated, non-selective cation channel TRPM4. Nature. 2017;552:205–209. doi: 10.1038/nature24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Autzen HE, et al. Structure of the human TRPM4 ion channel in a lipid nanodisc. Science. 2018;359:228–232. doi: 10.1126/science.aar4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 27.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 28.García-Sanz N, et al. A role of the transient receptor potential domain of vanilloid receptor I in channel gating. J Neurosci. 2007;27:11641–11650. doi: 10.1523/JNEUROSCI.2457-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohács T, Lopes CMB, Michailidis I, Logothetis DE. PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat Neurosci. 2005;8:626–634. doi: 10.1038/nn1451. [DOI] [PubMed] [Google Scholar]
- 30.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 34.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhl S, Mathar I, Vennekens R, Freichel M. Adenylyl cyclase-mediated effects contribute to increased isoprenaline-induced cardiac contractility in TRPM4-deficient mice. J Mol Cell Cardiol. 2014;74:307–317. doi: 10.1016/j.yjmcc.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Mathar I, et al. Increased catecholamine secretion contributes to hypertension in TRPM4-deficient mice. J Clin Invest. 2010;120:3267–3279. doi: 10.1172/JCI41348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerzanich V, et al. De novo expression of Trpm4 initiates secondary hemorrhage in spinal cord injury. Nat Med. 2009;15:185–191. doi: 10.1038/nm.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ru H, et al. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell. 2015;163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Comput Crystallogr Newsl. 2013;4:43–44. [Google Scholar]
- 48.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.