Significance
Activation of G protein-coupled receptors (GPCRs) initiates conformational shifts that trigger interaction with a specific G-protein subtype from a structurally homologous set. A major unsolved problem is the mechanism by which this selectivity is achieved. Structures of GPCR–G protein complexes so far fail to reveal the origin of selectivity because they all involve one G-protein subtype (Gs). In this work, we report a structural model of the activated GPCR rhodopsin in complex with another G-protein subtype (Gi) derived from intermolecular distance mapping with DEER-EPR and refinement with modeling. Comparison of the model with structures of complexes involving Gs reveals distinct GPCR–G protein-binding modes, the differences of which suggest key features of the structural selectivity filter.
Keywords: rhodopsin, GPCR, G protein, pulsed dipolar spectroscopy
Abstract
More than two decades ago, the activation mechanism for the membrane-bound photoreceptor and prototypical G protein-coupled receptor (GPCR) rhodopsin was uncovered. Upon light-induced changes in ligand–receptor interaction, movement of specific transmembrane helices within the receptor opens a crevice at the cytoplasmic surface, allowing for coupling of heterotrimeric guanine nucleotide-binding proteins (G proteins). The general features of this activation mechanism are conserved across the GPCR superfamily. Nevertheless, GPCRs have selectivity for distinct G-protein family members, but the mechanism of selectivity remains elusive. Structures of GPCRs in complex with the stimulatory G protein, Gs, and an accessory nanobody to stabilize the complex have been reported, providing information on the intermolecular interactions. However, to reveal the structural selectivity filters, it will be necessary to determine GPCR–G protein structures involving other G-protein subtypes. In addition, it is important to obtain structures in the absence of a nanobody that may influence the structure. Here, we present a model for a rhodopsin–G protein complex derived from intermolecular distance constraints between the activated receptor and the inhibitory G protein, Gi, using electron paramagnetic resonance spectroscopy and spin-labeling methodologies. Molecular dynamics simulations demonstrated the overall stability of the modeled complex. In the rhodopsin–Gi complex, Gi engages rhodopsin in a manner distinct from previous GPCR–Gs structures, providing insight into specificity determinants.
Low-light vision requires photon absorption by the G protein-coupled receptor (GPCR) rhodopsin and subsequent catalytic activation of heterotrimeric G proteins (Gαβγ) by GDP–GTP nucleotide exchange. As the photoreceptor of rod cells, rhodopsin was the first GPCR to be functionally and structurally characterized, and serves as a model for class A GPCRs. It was discovered by Franz Boll (1), subsequently studied in the late 19th century by Wilhelm Kühne (2), and has become since then a subject of intensive research (3). In the inactive state, the inverse agonist, 11-cis-retinal, is linked via a protonated Schiff base to the receptor. Absorption of light isomerizes the 11-cis-retinal to the agonist all-trans-retinal, which subsequently results in conformational changes within the receptor. Salient features of the activation mechanism of rhodopsin triggered by light were revealed via site-directed spin labeling (SDSL) and EPR spectroscopy as an outward motion of transmembrane helix (TM) 6 and a smaller inward movement of TM7 at the cytoplasmic surface (4–6). The main features of this model were subsequently confirmed by crystallography (7–9). Additional details regarding the molecular mechanism of GPCR activation have since been revealed by breakthroughs in GPCR structural biology (10–12), cryoelectron microscopy (13, 14), and spectroscopic techniques (15, 16). However, despite this wealth of information, questions remain regarding the structural origin of GPCR specificity for cognate G proteins (e.g., Gs, Gi, G12/13, Gq). For example, are all complexes between G proteins and GPCRs homologous at the level of the backbone fold, differing only by specific side-chain interactions, or is there a specificity code in the receptor involving the allowed magnitude of displacement of particular helices? Of considerable interest are GPCRs which couple to multiple G-protein subtypes and can sample diverse conformational landscapes.
In the present study, SDSL and double electron–electron resonance (DEER) spectroscopy (17) are employed to map distances between pairs of nitroxide spin labels, one in activated rhodopsin and the other in the Ras-like domain of the inhibitory G protein, Gi. The data provide structural constraints for modeling the nucleotide-free rhodopsin–Gi complex in a native-like lipid environment, free from the confines of a rigid crystalline lattice and in the absence of accessory proteins used in crystallization. In the photoreceptor cell, rhodopsin couples to transducin, Gt, which belongs to the Gi subfamily. We use the close homology between Gt and Gi to gain insight into coupling of Gi-selective GPCRs.
Results
Gt binding to rhodopsin in lipid nanodiscs was recently shown by SDSL DEER studies to select a TM6 conformation which is similar to active metarhodopsin II crystal structures but is apparently dynamic in nature (18, 19). Hence, Gt does not induce a new conformation in rhodopsin with respect to these helices but selects a preexisting conformational substate of the receptor. For the purpose of modeling based on sparse distance data, it is assumed that rhodopsin is in the metarhodopsin II crystallographic conformation.
Selection of Sites for Distance Mapping.
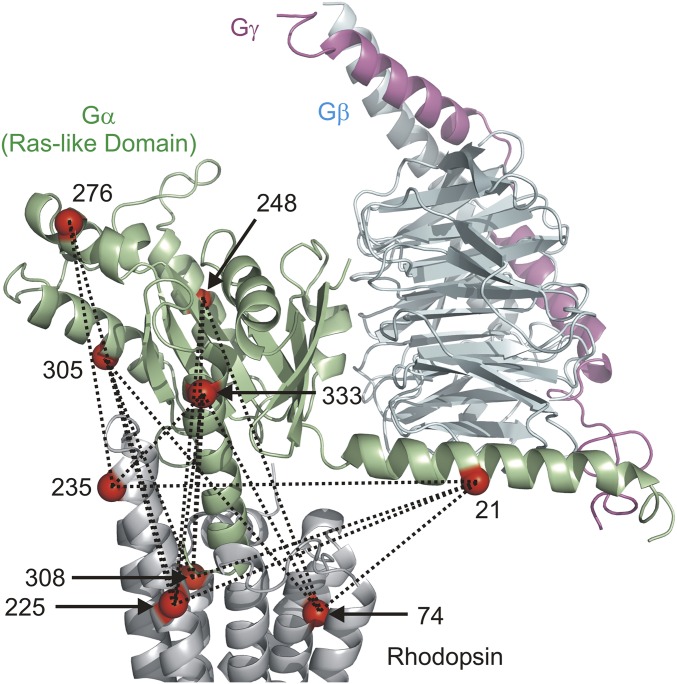
Fig. 1 shows sites on the Ras-like domain of the G protein and on rhodopsin that were mutated to single cysteines to introduce the nitroxide side chain designated R1 (20) for DEER distance measurements between the molecules in the complex. Dotted lines in the figure indicate intermolecular distances that were measured between spin pairs within the complex. Rhodopsin sites (residues 74, 308, 225, and 235) were selected based upon previous continuous-wave (CW) EPR studies (5, 21). The helical domain of the Gαi-subunit (residues 60 to 182), which acts as a lid covering the GDP-binding pocket, was omitted from Fig. 1 for clarity; no intermolecular distances involving the helical domain were collected in this study. However, previous EPR studies showed that there is disorder in the position of the helical domain with respect to the Ras-like domain in the complex (22, 23).
Fig. 1.
Rhodopsin–Gi (Gαi1Gβ1γ1) complex showing spin-labeling sites within Gi and rhodopsin. Dotted lines indicate the pairwise distances measured. The helical domain of Gαi is omitted for clarity. The gray ribbon is the rhodopsin backbone, while the green, cyan, and magenta ribbons identify the Ras-like domain, Gβ-subunit, and Gγ-subunit, respectively.
Sites for introduction of R1 in the Ras-like domain of Gαi were a subset of those from previous CW EPR studies (22–25), and include residue 21 in the Gα N-terminal helix that contacts the Gβ-subunit and residue 333 in the C-terminal α5-helix, a key interaction element with the receptor (24). Sites 248, 276, and 305 were selected to sample other helical segments in the Ras-like domain. In each case, most residues in rhodopsin and Gαi were located on the outer, solvent-exposed surfaces of the helices, where structural perturbation due to R1 is minimal. CW EPR spectra of all labeled mutants are shown in Fig. S1. The purity of the Gαi mutants used to form the complexes was typically >95% as judged by SDS/PAGE (Fig. S2).
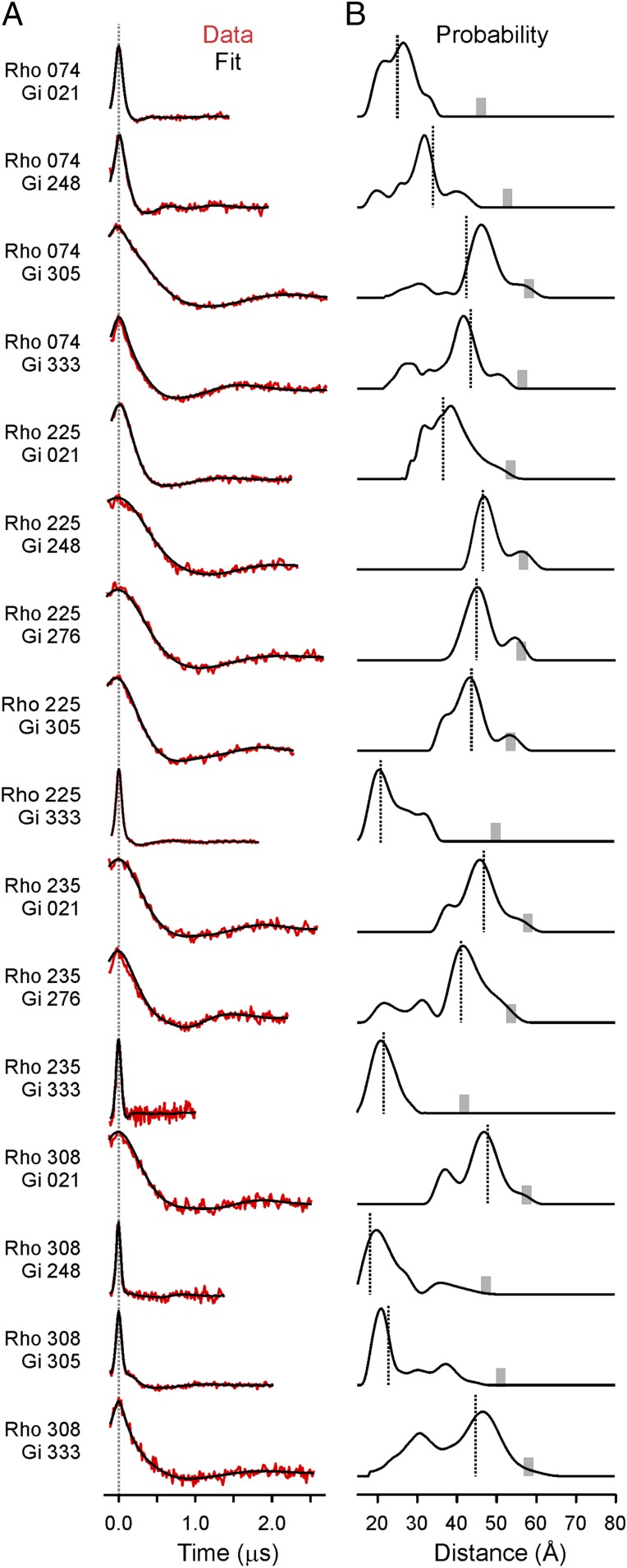
For the DEER measurements, spin-labeled rhodopsin mutants incorporated into lipid nanodiscs [70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 30% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine] were mixed with spin-labeled Gαi mutants in the presence of Gβγ-subunits (1:1:1 molar ratio of rhodopsin:Gα:Gβγ) and light-activated to initiate complex formation. Fig. 2A shows the background-corrected dipolar evolution functions (DEFs) (26) from the four-pulse DEER experiment for the 16 spin-labeled rhodopsin–Gi pairs measured. For a random distribution of spins, the background-corrected DEF is a featureless horizontal line (Fig. S3); only when discrete spin pairs are present is the oscillatory behavior of the DEF seen in Fig. 2A obtained. Thus, the appearance of the nonzero DEF signal confirms complex formation. The distance distributions directly derived from the DEFs are shown in Fig. 2B.
Fig. 2.
DEER distance measurements. (A) Background-corrected dipolar evolution functions (red traces) are offset for clarity, and fits to the data are shown (black traces). (B) Distance distributions for each of the rhodopsin–G protein pairs. The vertical dotted lines represent internitroxide distances from the final DEER model. The gray bar on the x axis of each distribution shows the upper distance limit for reliable determination given the DEF collection times.
Modeling of the Rhodopsin–Gi Complex.
Fig. 2 reveals that the distance distributions are multimodal and the complete distribution can span more than 20 Å in width, outside the range for the known rotamers of R1 (27). In simple proteins whose function does not involve changes in global backbone topology (such as holomyoglobin), the distance distributions between R1 pairs in stable helices are much narrower, on the order of 5 Å or less (28). Thus, the widths likely reflect structural heterogeneity of the complex, and hence flexibility under physiological conditions. The molecular origin of the heterogeneity and flexibility is of great interest and will be the subject of future studies, but for the purposes of modeling the most significant complex structure the most probable distances of the distributions were used as initial global constraints. The first step of modeling assumed that the activated G protein and activated rhodopsin could be approximated as rigid bodies (at the level of the backbone fold) upon which relative rotation and translation operations could be performed to minimize differences between the most probable measured and model distances.
For modeling, the Ras-like domain of Gs in the β2-adrenergic receptor–Gs complex [β2AR–Gs complex; Protein Data Bank (PDB) ID code 3SN6 (12)] was used as an initial template for the Gαi backbone fold. The backbone folds of the nucleotide domains of Gαi and Gαs are essentially identical in crystal structures, with the main difference being an extended loop in Gαs preceding the α4-helix. The Ras-like domain of Gαi [PDB ID code 1GP2 (residues 5 to 59 and 183 to 326) (29)] was overlaid on that of Gαs in the complex with β2AR. The last six amino acids in the C-terminal α5-helix of Gαi are not resolved in the 1GP2 structure (29). Therefore, the α5-helix from a different Gαi structure [PDB ID code 1AGR (residues 330 to 354) (30)] was overlaid with the corresponding helix of Gαs in the β2AR–Gs complex. This construct was the initial template for Gαi. Finally, Gβγ-subunits were taken to have the same orientation relative to the Ras-like Gα-domain as in the heterotrimeric G-protein crystal structures [i.e., PDB ID code 1GP2 (29)], although no experimental distances were measured to confirm this orientation. The metarhodopsin II crystal structure [PDB ID code 3PXO (19)] was used as a template for the activated receptor backbone fold. These templates with modeled nitroxide R1 side chains were then docked to minimize the differences between the measured (most probable) and modeled internitroxide distances. The modeling process at this point was entirely “data-driven,” with no manual alignment steps. Details of the modeling are provided in Materials and Methods. The interspin distances in the final model and the most probable experimental values are in excellent agreement, as shown in Fig. 2B as vertical dashed lines in the distance distributions.
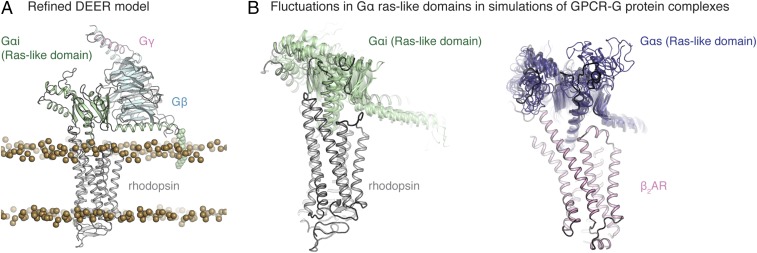
The DEER model was further refined as described in Materials and Methods and SI Materials and Methods. In particular, missing residues were added and hydrogen atoms included to fully model the hydrogen-bonding network, and known posttranslational modifications were added. The protein was placed in a bilayer using the OPM database [Orientations of Proteins in Membranes (31)]. In the complex, the N-terminal helix of the Ras-like domain lies along the surface of the polar head groups of the lipids, as expected, with the residue C3 palmitoylation acting as an anchor on the membrane surface (32). Energy minimization steps were carried out to reduce clashes at the α5-helix–rhodopsin interface. The final refined DEER model is shown in Fig. 3A (see Materials and Methods and SI Materials and Methods for details). The interspin distances in the refined model were essentially unchanged from the DEER model (Fig. 3B, Left and Fig. S4). In addition to the DEER distance constraints, earlier CW EPR studies provide support for the model in Fig. 3A (23–25, 33). For example, CW EPR line shape changes of spin-labeled Gαi upon receptor binding clearly delineate contact surfaces with the receptor (23–25, 33) (Fig. S5).
Fig. 3.
Structural models of receptor–G protein complexes. (A) Rendering of the refined rhodopsin–Gi complex embedded in the lipid bilayer based on DEER distance mapping. Lipid bilayer head group phosphates are shown as gold spheres. The refined model has posttranslational modifications on both rhodopsin and Gαi. (B) Overlaid snapshots from simulations of rhodopsin–Gi (simulation 3 of this paper) and β2AR–Gs (simulation 24, table S1 of ref. 34) complexes. In both cases, snapshots are shown every 54 ns of simulation between 0 and 500 ns, after removing frames within the first 100 ns to account for relaxation of the complex. Simulation snapshots were aligned on TM1 to TM4 of the receptor. As receptor conformation remained fairly constant across all simulations, we display multiple snapshots of only the Gα Ras-like domain. The overall orientation of the Gαi-subunit, including the α5-helix, remains fairly constant in MD simulations. Some fluctuations are observed in terms of the tightness of packing at the rhodopsin–Gαi interface and in terms of the position of the N-terminal helix of Gα. The starting DEER model is outlined in darker lines (B, Left).
Molecular dynamics (MD) simulations of the refined model were performed to verify its stability. Three simulations, each 600 ns in length, indicate the overall stability of the rhodopsin–Gαi interface and of the orientation of Gαi within the rhodopsin-binding cavity (Fig. 3B, Left). In the simulations, the interface contact area remained fairly constant (Fig. S4), while larger motions were observed for the Gβγ-subunits for which no DEER distances were measured (Fig. S6). The simulations relax to an overall orientation of the Gα-subunit and its α5-helix that remains fairly constant in rhodopsin–Gi complex simulations, lending support to the DEER model of the rhodopsin–Gi complex. Moreover, the fluctuations observed in these simulations only slightly exceed those observed in earlier simulations of the crystal structure of the β2AR–Gs complex (simulations 22 to 24 in table S1 of ref. 34; Fig. 3B, Right), with the N-terminal helix of the Gαi-subunit moving laterally across the membrane and the α5-helix of Gα shifting slightly within the β-barrel region of the Ras-like domain (Fig. 3B, Left and Fig. S4). We note that further model refinements at the receptor-binding interface might reduce the dynamic nature of individual residue–residue contacts observed in simulation. Collectively, the simulations generally support the overall orientation and conformation of the rhodopsin–Gi complex.
Discussion
Comparison with Other Ternary Complexes.
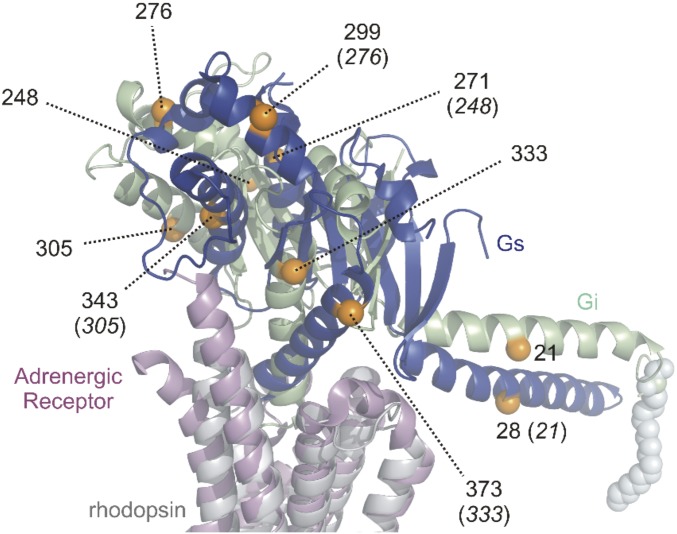
Besides the crystal structures of the β2AR–Gs complex (12) and the adenosine A2A receptor in complex with an engineered “mini-Gαs” G protein (11), the cryoelectron microscopy structures of the calcitonin receptor as well as the GLP-1 receptor both in complex with Gs have been solved (13, 14). These structures all show a nearly identical receptor–Gαs interaction on the protein backbone level (Fig. S7). Fig. 4 shows a comparison of the rhodopsin–Gi model with the crystal structure of the β2AR–Gs complex (12). Only the respective Ras-like domains and receptors are shown for clarity. A difference in binding mode of Gi versus Gs is evident. The Ras-like domain sits more upright on the receptor in the rhodopsin–Gi model, and the TM5–TM6 loop of rhodopsin makes contact with the β6-sheet of Gi. This contact is absent in the β2AR–Gs crystal structure. The inclination angle of the C-terminal α5-helix of the Ras-like domain is also different in the two models, whereas this inclination angle is similar in the present rhodopsin–Gi model and the crystal structure of metarhodopsin II in complex with a C-terminal peptide derived from Gαt (19).
Fig. 4.
Comparison of the Ras-like domain orientation in the different ternary complex structures of rhodopsin–Gi and β2AR–Gs (PDB ID code 3SN6). Sites in the Ras-like domain of Gαi used for distance mapping are shown as orange spheres of the Cα atoms. Corresponding residues in Gαs (orange) are indicated with Gαi numbering in parentheses to guide the eye in assessing the magnitude of structural differences.
Selectivity Determinants for G-Protein Coupling.
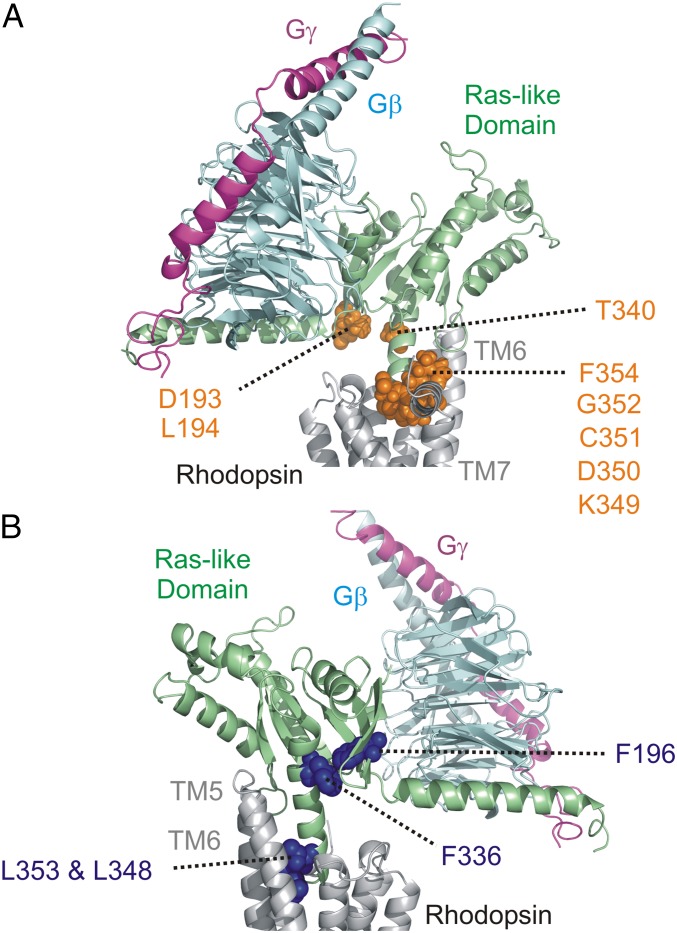
To elucidate potential roles of particular G-protein residues in receptor selectivity, residues involved in a “selectivity barcode” (35) (www.gpcrdb.org/) are highlighted on the rhodopsin–Gi model in Fig. 5A. Most of the residues lie in the extreme C terminus of the G protein (K349, D350, C351, G352, and F354), which has been predicted to modulate G protein-subtype selectivity (36, 37). Remarkably, in the refined model of Fig. 3A, they pack efficiently against the inside of rhodopsin’s cytoplasmic TM6 surface near the C-terminal portion of TM7. Importantly, a different insertion angle of the G-protein C-terminal helix (such as seen in the β2AR–Gs crystal structure) would yield steric clashes with TM6 of rhodopsin. Therefore, the identity of the C-terminal residues as well as the magnitude of TM6 movement, which is different from one receptor to another, will have a strong influence on receptor–G protein specificity. Indeed, like rhodopsin, the μ-opioid receptor displays a smaller TM6 movement in the active state than β2AR and couples to Gi (10). Receptors which can couple to multiple G-protein subtypes show interesting behaviors where the inversion of residues within TM6 can alter specificity toward Gs coupling (38, 39). This suggests that TM6 flexibility in the active state may also be important along with the C-terminal G-protein sequence. Additional residues predicted to be specificity determinants are L194 and D193 in the β2–β3 loop of the Ras-like domain (35) (Fig. 5A). Residues in this loop are in proximity to the TM3–TM4 loop of GPCRs. Local interactions in this region may yield additional contributions to selectivity. Indeed, V217 in the β2–β3 loop of Gs (analogous to L194 in Gi) makes contact with the TM3–TM4 loop of the receptor (residue F139) in the β2AR–Gs structure (12). Mutation of F139 to alanine disrupted receptor–G protein coupling (40).
Fig. 5.
Conserved G-protein residues and selectivity determinants. (A) Residues across G-protein subtypes that are involved in G-protein selectivity. Side chains are colored orange on the rhodopsin–Gi complex model. The residue numbers for Gαi are indicated. (B) Conserved residues across G-protein subtypes involved in stabilizing the receptor-bound state. Side chains are indicated in dark blue on the rhodopsin–Gi complex model.
Conserved Residues Across G-Protein Subtypes.
Fig. 5B shows a subset of residues in Gαi which are conserved across G-protein subtypes (35). Site F336 has been shown to increase basal nucleotide exchange rates upon mutation to alanine (41) by creating a cavity between the Ras-like domain body and the C-terminal helix of Gαi. This residue is displaced upon Gαi binding to rhodopsin with corresponding α5-helix movements. A translation and rotation of the α5-helix in the nucleotide-free model is compatible with a similar motion reported earlier (24). The F336 site comes into contact with another conserved phenylalanine (F196) of Gαi in the refined model. The F196 sequence region of the β3-sheet of Gαi is just upstream of the well-known “switch II” helix, and perturbations in the β3-sheet may propagate to this helix. Indeed, switch II conformational rearrangements have been detected in G proteins upon receptor binding (33). Some additional conserved residues shown in Fig. 5B include two leucines (L348 and L353), which in the refined model pack against the interior surface of TM6 in rhodopsin but do not influence selectivity.
In conclusion, the structural model presented reveals an alternative docking mode of heterotrimeric G proteins bound to GPCRs. We believe that the specificity of GPCR interactions is related to the binding orientation of the Ras-like domain on the cell-surface receptors and the corresponding differences in the C-terminal helix insertion angle. Comparing the rhodopsin–Gi and β2AR–Gs complexes shows that the bulkiness of the receptor-interacting part of the Gα α5-helix also determines the degree of TM6 movement needed for coupling, which is in agreement with earlier MD simulations on β2AR with C-terminal peptides derived from Gαi and Gαs (42). In regard to the proposed specificity barcodes for heterotrimeric G proteins (35), it appears that residues within the β6-sheet of the Ras-like domain of Gαi contribute to the contact interface with the TM5–TM6 loop of rhodopsin [as evidenced by the data-driven model and earlier CW EPR studies (24)]. Hence, it is clear that this region of Gαi is also crucial for receptor engagement. This is consistent with peptide competition studies which showed transducin peptides covering this sequence region competed against the heterotrimeric G protein for rhodopsin engagement (43). MD simulations of the rhodopsin–Gi complex together with DEER distance measurements are beginning to uncover dynamic elements of the nucleotide-free GPCR–G protein complex that are crucial for guanine nucleotide exchange.
Materials and Methods
Protein Expression and Spin Labeling of Rhodopsin Mutants.
Expression, purification, and spin labeling of bovine rhodopsin mutants were performed as previously described (18, 44). For the present study, single rhodopsin cysteine mutants Y74C, Q225C, A235C, and M308C in a C140S/C316S base mutant were produced in stable HEK293S GnTI− cell cultures. All buffers used for purification were filtered and degassed. Rhodopsin cysteine mutants were immobilized on a 1D4 resin and spin-labeled with 100 μM S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (Toronto Research Chemicals) for 2.5 h at room temperature. The excess of spin label was removed by successive washes (18, 44). The spin-labeled mutants were eluted from a 1D4 antibody resin in a buffer containing 90 mM octylglucoside at 4 °C (for incorporation into nanodiscs) using a peptide with sequence identity to the last 18 amino acids of rhodopsin’s C terminus. Each spin-labeled rhodopsin mutant showed wild-type UV-visible absorption characteristics consistent with previous reports (5, 18, 21).
Purification of the MSP1E3D1 Scaffold Protein and Reconstitution of Rhodopsin Nanodiscs.
The MSP1E3D1 scaffold protein was expressed in Escherichia coli and purified as described (18). Nanodiscs containing mutant rhodopsin were made using published protocols (45). The phospholipid composition of the prepared nanodiscs was 70% 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 30% 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine.
G-Protein Expression, Purification, and Spin Labeling.
A rat Gαi1 construct with six cysteines replaced [C3S-C66A-C214S-C305S-C325A-C351I (25)] was cloned into a modified pET15b vector, generously provided by David Eisenberg, University of California, Los Angeles, which produced a fusion construct coding for an N-terminal hexahistidine tag, maltose-binding protein (MBP), and tobacco etch virus (TEV) protease-specific cleavage site merged to Gαi (with its start methionine). Expression of the protein in E. coli BL21Gold(DE3) cells was performed as previously described (22). The purity of the cleaved Gαi mutants was >95% as judged by SDS/PAGE. Purified Gαi mutants were spin-labeled by mixing equimolar amounts of protein with S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate for 10 min followed by extensive washes in Amicon filters to remove noncovalently attached label. Gβγ (β1γ1-isoform) was purified from bovine retinae essentially as described (23, 46). The functionality of each Gαi cysteine mutant was assessed by aluminum fluoride activity tests as previously described (25). The excitation and emission wavelengths of tryptophan fluorescence assays were 290 and 340 nm, respectively.
Double Electron–Electron Resonance and Continuous-Wave EPR Spectroscopy.
For DEER measurements, spin-labeled rhodopsin in nanodiscs (90 μM) was mixed with an equivalent concentration (90 μM) of reconstituted G-protein heterotrimer (i.e., Gαβγ). The G-protein heterotrimer was reconstituted by mixing spin-labeled Gαi with Gβγ in a 1:1 molar ratio. DEER experiments were performed in 20 mM Mops, 100 mM NaCl, 2 mM MgCl2, 20 μM GDP, and 20% deuterated glycerol. Samples were illuminated within quartz capillaries (1.5 mm i.d. and 1.8 mm o.d.) before freezing using a >515-nm long-pass filter and loaded into an EN 5107D2 resonator (Bruker). Q-band measurements were performed at 80 K on a Bruker Elexsys 580 spectrometer with a Super Q-FTu bridge. A 32-ns π-pump pulse was applied to the low-field peak of the nitroxide field-swept spectrum, and the observer π/2 (16 ns) and π (32 ns) -pulses were positioned 50 MHz (17.8 G) upfield, which corresponds to the nitroxide center line. Distance distributions were obtained from the raw dipolar evolution data using the program LongDistances (developed in-house using National Instruments LabVIEW). It can be downloaded from www.biochemistry.ucla.edu/Faculty/Hubbell/.
As a control, a few DEER experiments were collected in the dark versus light. Data collected in the dark showed only background signal and were consistent with no ternary complex formation. All samples in the light, however, showed clear dipolar oscillations consistent with rhodopsin–G protein interactions.
Modeling of the Rhodopsin–G Protein Complex Using Sparse Distance Constraints.
R1 rotamers for the starting positions in each molecule corresponded to one of the three most probable as determined from crystal structures (27). For rhodopsin, the rotamers were those determined in earlier DEER distance mapping of rhodopsin (6). In those experiments, the rotamers were chosen to best match the experimental distances in dark rhodopsin, assuming that the crystal structure of the inactive molecule represented the structure in solution. For Gi, rotamer selection [from the most probable set (27)] was based on lack of steric clash in the structure and best match to experimental intramolecular distances from earlier DEER distance mapping (22, 24, 25).
Starting with the initial coordinates, two sets of constraints were simultaneously imposed for the geometry optimization of the nitroxide spin positions: All measured intermolecular nitroxide spin distances (between rhodopsin and Gi) were assigned from the DEER results. All intramolecular nitroxide spin distances between the investigated sites (within rhodopsin or within Gi) were assigned from the existing starting positions of the modeled R1 side chains. The intramolecular distances ensured approximate rigid-body movements of the two molecules, while still allowing some local flexibility when trying to accommodate the intramolecular distance constraints from DEER.
The DEER data only reflect pairwise distances and not absolute locations, so the molecular coordinates of the proteins need to be realigned with the optimized geometry. This was done in two steps. First, the optimized geometry (of all nitroxide spin positions) was realigned to best match the respective starting positions on rhodopsin in terms of least squares residual distances. Second, the entire Gi was realigned to best match the best fit Gi positions, and the PDB file was rewritten with the transformed new coordinates. These steps result in two aligned PDB files (rhodopsin and Gi aligned to satisfy the distance data) that can be used as a starting point for further modeling. Note that the process was entirely data-driven to avoid any possible bias from manual alignment. The relative weighting of intra- vs. intermolecular distance constraints was varied over two orders of magnitude giving very similar results, indicating that the set of distances is consistent. There is some amount of uncertainty, because we do not exactly know the spin positions relative to the molecules, and each molecule can be moved or rotated within (narrow) limits without changing the constraints considerably. While this method is purely geometrical and does not account for molecular overlap, the resulting complex showed a near-perfect fit.
Model Refinement and Molecular Dynamics Simulation.
Missing residues in the DEER-based model were added, including residues at the Gαi N terminus, Gγ C terminus, and rhodopsin C terminus, to add lipid modifications important for membrane anchoring. In total, the final system contained three palmitoylcysteines. All spin-labeled cysteines were reverted to their wild-type identities, several acidic residues known to be protonated in the active state of rhodopsin were protonated (47), and the stabilizing disulfide between mutated residues N2C/D28C on the extracellular face of rhodopsin was introduced to stabilize the active binding pocket (48, 49). Finally, the Minimize tool in Maestro (Schrödinger) was used to optimize intermolecular contacts formed at the rhodopsin–Gi interface, followed by an additional minimization performed on the entire complex.
The refined and prepared rhodopsin–Gi complex was inserted into a hydrated POPC bilayer, creating a final system of 231,641 atoms. We performed three unbiased simulations of the prepared rhodopsin–Gi complex with the CHARMM36 force field (with CHARMM36m protein parameters) using the AMBER 16 software suite on graphics processing units (GPUs) (50, 51). Each of the three simulations was ∼600 ns in length. See SI Materials and Methods for a complete description of structure refinement and simulation protocols.
Supplementary Material
Acknowledgments
We thank Lu Chen and Yang Shen for technical assistance. This work was supported by NIH Grant R01EY05216 (to W.L.H.), the Canada Excellence Research Chairs program (O.P.E.), Canadian Institute for Advanced Research (O.P.E.), Anne and Max Tanenbaum Chair in Neuroscience (O.P.E.), Jules Stein Professor Endowment (W.L.H.), an institutional Biomedical Informatics Training Grant postdoctoral fellowship (to S.A.H.) (T15-LM007033-33), and National Eye Institute Core Grant P30EY00331. N.R.L. is a recipient of a National Science Foundation Graduate Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721896115/-/DCSupplemental.
References
- 1.Baumann C. Franz Boll. Vision Res. 1977;17:1267–1268. doi: 10.1016/0042-6989(77)90113-4. [DOI] [PubMed] [Google Scholar]
- 2.Crescitelli F. Friedrich Wilhelm Kühne. The centennial of rhodopsin. Arch Ophthalmol. 1977;95:1766. doi: 10.1001/archopht.1977.04450100068003. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 5.Altenbach C, et al. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: A site-directed spin-labeling study. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 6.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 8.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 9.Zhou XE, Melcher K, Xu HE. Structure and activation of rhodopsin. Acta Pharmacol Sin. 2012;33:291–299. doi: 10.1038/aps.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, et al. Structural insights into µ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpenter B, Nehmé R, Warne T, Leslie AG, Tate CG. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang YL, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L, Van Eps N, Zimmer M, Ernst OP, Prosser RS. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature. 2016;533:265–268. doi: 10.1038/nature17668. [DOI] [PubMed] [Google Scholar]
- 16.Manglik A, et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 18.Van Eps N, et al. Conformational equilibria of light-activated rhodopsin in nanodiscs. Proc Natl Acad Sci USA. 2017;114:E3268–E3275. doi: 10.1073/pnas.1620405114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 20.Todd AP, Cong J, Levinthal F, Levinthal C, Hubbell WL. Site-directed mutagenesis of colicin E1 provides specific attachment sites for spin labels whose spectra are sensitive to local conformation. Proteins. 1989;6:294–305. doi: 10.1002/prot.340060312. [DOI] [PubMed] [Google Scholar]
- 21.Altenbach C, Cai K, Khorana HG, Hubbell WL. Structural features and light-dependent changes in the sequence 306–322 extending from helix VII to the palmitoylation sites in rhodopsin: A site-directed spin-labeling study. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 22.Van Eps N, Thomas CJ, Hubbell WL, Sprang SR. The guanine nucleotide exchange factor Ric-8A induces domain separation and Ras domain plasticity in Gαi1. Proc Natl Acad Sci USA. 2015;112:1404–1409. doi: 10.1073/pnas.1423878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Eps N, et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci USA. 2011;108:9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 25.Medkova M, Preininger AM, Yu NJ, Hubbell WL, Hamm HE. Conformational changes in the amino-terminal helix of the G protein alpha(i1) following dissociation from Gbetagamma subunit and activation. Biochemistry. 2002;41:9962–9972. doi: 10.1021/bi0255726. [DOI] [PubMed] [Google Scholar]
- 26.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 27.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerch MT, Yang Z, Brooks EK, Hubbell WL. Mapping protein conformational heterogeneity under pressure with site-directed spin labeling and double electron-electron resonance. Proc Natl Acad Sci USA. 2014;111:E1201–E1210. doi: 10.1073/pnas.1403179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 30.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4−-activated G(i alpha1): Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 31.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 32.Wedegaertner PB, Wilson PT, Bourne HR. Lipid modifications of trimeric G proteins. J Biol Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- 33.Van Eps N, Oldham WM, Hamm HE, Hubbell WL. Structural and dynamical changes in an alpha-subunit of a heterotrimeric G protein along the activation pathway. Proc Natl Acad Sci USA. 2006;103:16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dror RO, et al. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flock T, et al. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature. 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 37.Conklin BR, et al. Carboxyl-terminal mutations of Gq alpha and Gs alpha that alter the fidelity of receptor activation. Mol Pharmacol. 1996;50:885–890. [PubMed] [Google Scholar]
- 38.Abadji V, Lucas-Lenard JM, Chin C, Kendall DA. Involvement of the carboxyl terminus of the third intracellular loop of the cannabinoid CB1 receptor in constitutive activation of Gs. J Neurochem. 1999;72:2032–2038. doi: 10.1046/j.1471-4159.1999.0722032.x. [DOI] [PubMed] [Google Scholar]
- 39.Ulfers AL, et al. Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity. Protein Sci. 2002;11:2526–2531. doi: 10.1110/ps.0218402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moro O, Lameh J, Högger P, Sadée W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J Biol Chem. 1993;268:22273–22276. [PubMed] [Google Scholar]
- 41.Kapoor N, Menon ST, Chauhan R, Sachdev P, Sakmar TP. Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J Mol Biol. 2009;393:882–897. doi: 10.1016/j.jmb.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 42.Rose AS, et al. Position of transmembrane helix 6 determines receptor G protein coupling specificity. J Am Chem Soc. 2014;136:11244–11247. doi: 10.1021/ja5055109. [DOI] [PubMed] [Google Scholar]
- 43.Hamm HE, et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 44.Caro LN, et al. Rapid and facile recombinant expression of bovine rhodopsin in HEK293S GnTI(−) cells using a PiggyBac inducible system. Methods Enzymol. 2015;556:307–330. doi: 10.1016/bs.mie.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 46.Kleuss C, Pallast M, Brendel S, Rosenthal W, Schultz G. Resolution of transducin subunits by chromatography on Blue Sepharose. J Chromatogr A. 1987;407:281–289. doi: 10.1016/s0021-9673(01)92625-1. [DOI] [PubMed] [Google Scholar]
- 47.Mahalingam M, Martínez-Mayorga K, Brown MF, Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc Natl Acad Sci USA. 2008;105:17795–17800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie G, Gross AK, Oprian DD. An opsin mutant with increased thermal stability. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 49.Standfuss J, et al. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J, et al. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 52.Degtyarev MY, Spiegel AM, Jones TL. Palmitoylation of a G protein alpha i subunit requires membrane localization not myristoylation. J Biol Chem. 1994;269:30898–30903. [PubMed] [Google Scholar]
- 53.Betz RM. 2017 Dabble. Available at https://zenodo.org/record/836914#.WoG9GCXwbcs. Accessed July 1, 2017.
- 54.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanommeslaeghe K, MacKerell AD., Jr Automation of the CHARMM general force field (CGenFF) I: Bond perception and atom typing. J Chem Inf Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr Automation of the CHARMM general force field (CGenFF) II: Assignment of bonded parameters and partial atomic charges. J Chem Inf Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Case DA, et al. 2017 AMBER 2017 (University of California, San Francisco). Available at ambermd.org.
- 59.Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J Chem Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 60.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 61.Roe DR, Cheatham TE., III PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 62.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.