Significance
Bacterial secondary metabolites are of great relevance to human society and the environment. To this day, investigations of secreted metabolites focus on single compounds, compound classes, or compounds with specific bioactivities. Comparing the supernatants of Streptomyces chartreusis cultivated in different media, using liquid chromatography–coupled tandem MS, we detected a great diversity of highly regulated compounds surpassing genome-based expectations. Guided by molecular networking, a new polyether ionophore was identified and subsequently purified and characterized. The approach presented here provides a basis for structure analysis for molecules produced in amounts too low for standard methods of structure elucidation. Simultaneously, it facilitates the differential analysis of secreted metabolomes, providing insights into the chemical profiles under different cultivation conditions.
Keywords: metabolomics, secondary metabolites, antibiotics, siderophores
Abstract
Actinomycetes are known for producing diverse secondary metabolites. Combining genomics with untargeted data-dependent tandem MS and molecular networking, we characterized the secreted metabolome of the tunicamycin producer Streptomyces chartreusis NRRL 3882. The genome harbors 128 predicted biosynthetic gene clusters. We detected >1,000 distinct secreted metabolites in culture supernatants, only 22 of which were identified based on standards and public spectral libraries. S. chartreusis adapts the secreted metabolome to cultivation conditions. A number of metabolites are produced iron dependently, among them 17 desferrioxamine siderophores aiding in iron acquisition. Eight previously unknown members of this long-known compound class are described. A single desferrioxamine synthesis gene cluster was detected in the genome, yet different sets of desferrioxamines are produced in different media. Additionally, a polyether ionophore, differentially produced by the calcimycin biosynthesis cluster, was discovered. This illustrates that metabolite output of a single biosynthetic machine can be exquisitely regulated not only with regard to product quantity but also with regard to product range. Compared with chemically defined medium, in complex medium, total metabolite abundance was higher, structural diversity greater, and the average molecular weight almost doubled. Tunicamycins, for example, were only produced in complex medium. Extrapolating from this study, we anticipate that the larger part of bacterial chemistry, including chemical structures, ecological functions, and pharmacological potential, is yet to be uncovered.
Bacteria produce a multitude of structurally diverse secondary metabolites. Among those with pharmacological importance are antibiotics such as tetracyclines (1) and the siderophore desferrioxamine listed by the WHO as an essential medicine for the treatment of iron intoxication (2). Other molecules are currently under investigation, such as the antibiotic teixobactin produced by Eleftheria terrae (3).
Traditionally, secondary metabolites are identified by screening crude extracts for specific biological activities, followed by subsequent purification and characterization. This approach has proven to be efficient but can be plagued by high rediscovery rates (4) and limited to the more abundant metabolites. In fact, this approach purposefully neglects most compounds present in complex starting materials and it reveals little about a metabolite’s natural function. While not essential for the survival of microbes under laboratory conditions, secondary metabolites are thought to play a crucial role in the adaptation of bacteria to changing environmental conditions (5). Bacterial growth and survival require adaptation on different time scales. While the rapid adaptation to fluctuations in environmental parameters, such as temperature, is realized (e.g., by regulation of gene expression or protein activity), the long-term adaptation to new or changing habitats is of a more permanent nature and involves genetic alterations. In the present article “adaptation” is used to describe the rapid response to environmental fluctuations. To date, natural functions of only few metabolites are known. Siderophores are a prominent example. By sequestering iron with exquisite affinities, they allow the producer to thrive under iron-limiting conditions (6). Antibacterial and antifungal secondary metabolites are frequently secreted and are thought to provide an edge in the competition for resources (7) or allow predation (8). It is debated whether inhibitory concentrations are reached in nature, and subinhibitory concentrations are hypothesized to play a role in inter- and intraspecies communication (9, 10).
Some bacterial genera such as Streptomyces are particularly well known for their biosynthetic potential (11). Significant portions of their genomes are dedicated to secondary metabolite biosynthesis (12). Products are known for only ∼10% of in silico annotated biosynthesis gene clusters (BGCs) (12). Many secondary metabolites are produced only under specific culture conditions. For most BGCs no expression has ever been observed, which has led to the assumption that they are silent or have lost the ability to produce secondary metabolites (12). To unlock the unused chemical potential of silent BGCs, various strategies are applied, such as genetic or cultivation-dependent activation, biosynthesis in heterologous hosts, or synthetic biology approaches (13–18). Great efforts are being undertaken to characterize metabolites on the global scale to uncover novel chemistry. Data analysis, for instance, has been improved by introducing self-organizing maps, which led to the identification and purification of ciromycin A and B (19). Another example is the in-depth study of the siderophore suite of Streptomyces coelicolor M145, which led to the characterization of amphiphilic siderophores (20).
In this study, we characterized the chemical potential of Streptomyces chartreusis NRRL 3882, a strain used for the production of calcimycin and tunicamycin (21). The genome was sequenced and analyzed using antiSMASH 3.0.5 (22). The extractable and ionizable metabolites in the medium, which we refer to as the “secreted metabolome,” were analyzed by untargeted data-dependent liquid chromatography–coupled tandem MS (LC-MS/MS). MS/MS-based fragmentation patterns were analyzed by molecular networking (23). This approach allows even metabolites with unknown structures to be barcoded, compared, and tracked across samples. Known antibiotics and siderophores were identified by comparing fragment spectra with libraries and standards, and related compounds of unknown structure were structurally annotated by manual spectra annotation. Using the OSMAC (one strain many compounds) approach (14), the secreted metabolome was investigated after cultivation in different media. We demonstrate that the combination of MS and molecular networking with genomic information allows differential global metabolomic investigations as well as the structural characterization of single molecules.
Results
S. chartreusis NRRL 3882 contains a single linear chromosome and no plasmids (see SI Appendix, Table S1; European Nucleotide Archive accession numbers: genome sequence: LT962942; annotation: LT963352). The analysis tool antiSMASH 3.0.5 (22) predicts 128 putative BGCs, which account for 34% of the genome. Based on homology searches, 55 of 128 BGCs show a hit against the antiSMASH database, such as the calcimycin and tunicamycin BGCs (SI Appendix, Table S2). This analysis provides an idea of the compounds this bacterium might produce.
Metabolites secreted by S. chartreusis were detected by LC-MS/MS+ from culture supernatants under three cultivation conditions: complex medium (CM) and defined salt-based minimal media (MM) with or without iron (MM+Fe and MM−Fe, respectively). After molecular networking, charge correction, dereplication, elimination of adducts, and background and media subtraction, a total of 701 metabolites with distinct parent masses and fragment spectra were detected. Additionally, to explore the sampling depth, the culture supernatant of MM−Fe was subjected to hydrophobic interaction chromatography (HIC) with HP-20 column material, a method frequently applied for siderophore enrichment (24, 25). A further 343 metabolites were exclusively identified in the MM−Fe-derived HIC fractions. The total of 1,044 distinct metabolites detected in this study exceeds the number of BGCs by approximately an order of magnitude. However, we expect to still underestimate the chemical potential of S. chartreusis, since not all BGCs will be active under the conditions tested, the analysis was limited to positive ionization mode, and sampling depth can easily be increased, as demonstrated by the HIC experiment.
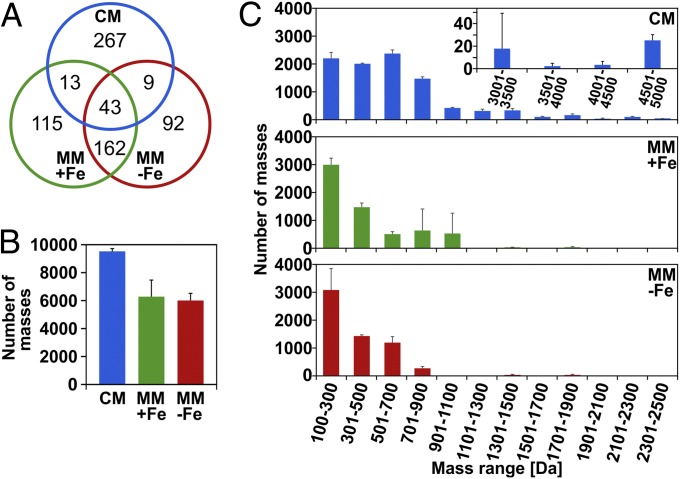
The secreted metabolome is specifically tailored to the growth conditions (Fig. 1A). Distinct sets of metabolites were detected in CM and MM, with few metabolites being produced under both conditions. In MM, iron supplementation had a profound influence. Two equally large sets of distinct metabolites were detected in MM+Fe and MM−Fe. Furthermore, global changes in the secreted metabolome correlated with nutrient availability (Fig. 1B and SI Appendix, Fig. S1). Spectral counting, based on the signal intensity of parent masses reaching the threshold for triggering fragmentation, was used to estimate compound quantities. This approach is described for quantitation of proteins and peptides (26) and also applicable for ionizable metabolites (SI Appendix, Fig. S2). In CM, metabolites with a higher molecular mass were detected more frequently (Fig. 1C). The average molecular size of all parent mass signals recorded was 621 Da, compared with 411 or 345 Da in MM+Fe or MM−Fe, respectively. Metabolites ranging from 1.9 to 5 kDa were detected only in CM. Taken together, on average, the metabolites produced in CM are larger and produced in larger quantity (Fig. 1C).
Fig. 1.
Metabolites of S. chartreusis. (A) S. chartreusis produces unique and common metabolites when cultivated in CM and MM with (MM+Fe) and without (MM−Fe) iron. A total of 701 distinct parent masses were detected. (B and C) “Number of masses” indicates the number of events of a parent mass surpassing the intensity threshold for triggering fragmentation. (B) In CM, more metabolite molecules were recorded. (C) Binning by mass range shows that size distribution varies by medium. No compounds with masses between 2,501 and 3,000 Da were detected. The experiment was performed three times independently (n = 3); averages with SDs are shown. The size distribution of distinct compounds is given in SI Appendix, Fig. S1.
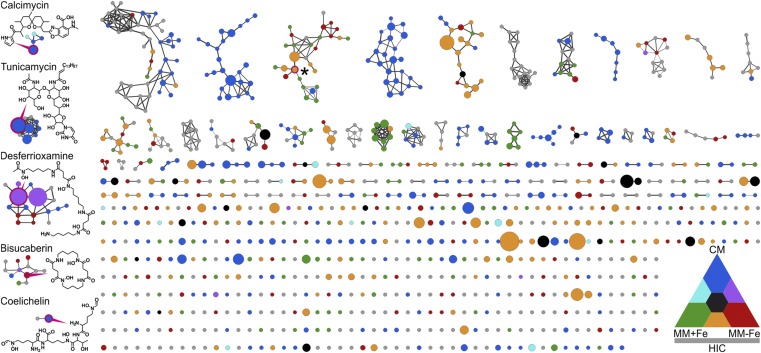
The molecular network contains 1,044 nodes, each representing one distinct metabolite [Fig. 2 and SI Appendix, Tables S3 and S4; Global Natural Products Social Molecular Networking (GNPS): https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c43717fa433e4456ac01e6cf1ce7476b, MSV000081911]. Of the nodes, 438 group in subnetworks with at least three members. Using standards and public spectral databases [GNPS (23), MetFusion (27)], members of six subnetworks highlighted in Fig. 2 were identified. For five of the subnetworks, all fragment spectra, with the exception of one bisucaberin derivative (m/z of 293.1472), were annotated and structures of previously unknown compounds predicted through comparison and annotation of fragment spectra (Fig. 3 and SI Appendix, Figs. S3–S8). Among the metabolites identified were the antibiotics tunicamycin and calcimycin, known to be produced by S. chartreusis (21). The calcimycin subnetwork contains the structurally related cezomycin (28) and N-demethyl-calcimycin (29), as well as two additional compounds (SI Appendix, Fig. S3). The calcimycin analog with an m/z of 513.2596, now named deoxacalcimycin, was purified and its structure elucidated by MS/MS, NMR, and FTIR (SI Appendix, Figs. S9–S23 and Table S5). NMR- and FTIR-based structure elucidation confirmed deoxacalcimycin to be a calcimycin analog. The final structure of deoxacalcimycin (SI Appendix, Fig. S10) was among the highest-scoring predicted structures in the MS/MS analysis but differs from the one ranked highest (SI Appendix, Fig. S3). The tunicamycin subnetwork encompasses the known tunicamycins A, B, C, D, D3, and I (30, 31) (PubChem identifiers: CID 11104835, CID 56927836, CID 56927832, CID 56927841, CID 56927833, CID 56927848), as well as four previously unknown derivatives, all of which differ in acyl-chain length (32) (SI Appendix, Fig. S4). Analytical standards were measured to aid MS/MS-based identification and structure elucidation (all spectra were added to the GNPS library; SI Appendix, Table S4). Using this approach, a subnetwork containing a molecule matching the MS/MS spectra of nalidixic acid was identified (Fig. 2 and SI Appendix, Fig. S5).
Fig. 2.
Molecular network of metabolites produced by S. chartreusis. Nodes resemble metabolites detected in the culture supernatants across a 14-d time course, with node sizes indicating cumulated abundance and colors indicating abundance in the different media (CM, blue; MM+Fe, green; MM−Fe, red). Intermediate colors indicate metabolite appearance under multiple conditions. Gray nodes resemble compounds only detected after HIC of MM−Fe. Nodes are connected if the cosine similarity of fragment spectra is ≥0.7 (line thickness reflects similarity score). Subnetworks on the left are named after one representative and contain molecules identified by database search or spectral annotation. Exemplary structures for nodes highlighted in magenta are given. *LC-MS/MS spectra match those of nalidixic acid. All structures are given in Fig. 3 and SI Appendix, Figs. S3–S8 as well as further information in SI Appendix, Table S3.
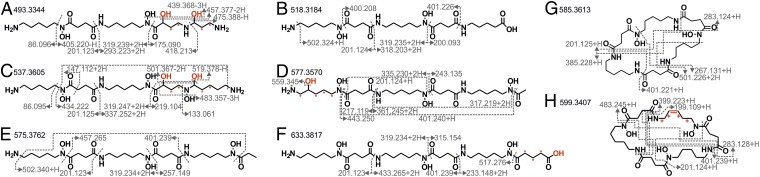
Fig. 3.
Desferrioxamine structures of S. chartreusis. (A–H) Structure prediction and annotation of fragment spectra of all previously unknown desferrioxamines. Given are the masses of protonated molecules [M+H]+ and the m/z of detected fragments. Gray dotted lines indicate fragmentation sites, while light gray dotted lines connect fragmentation sites. The “+” marks the potential positions of the red-marked double bond; asterisks “*” show alternative positions of red hydroxyl groups. Information on known desferrioxamines is provided in SI Appendix, Fig. S6 and Table S3. An overview on the procedure for structure prediction is given in SI Appendix, Fig. S25.
S. chartreusis produces a variety of hydroxamate siderophores belonging to at least two different siderophore subgroups. The bisucaberins and desferrioxamines (Fig. 2) are synthesized from two and three identical building blocks, respectively (33, 34). Shewanella putrefaciens has been shown to produce both in different stoichiometry depending on precursor availability (35). The genome of S. chartreusis harbors only one desferrioxamine BGC (SI Appendix, Table S2), which likely is responsible for the synthesis of all detected bisucaberines and desferrioxamines. The coelichelins, of which two were detected, are synthesized by a separate BGC (36). The genome of S. chartreusis harbors two further putative siderophore BGCs (SI Appendix, Table S2). Like for the antibiotics, the structures of the unknown siderophores in the described subnetworks were predicted based on mass spectrometric information (Fig. 3 and SI Appendix, Figs. S6–S8 and Table S3). Of the 17 desferrioxamines detected in total, eight were previously unknown.
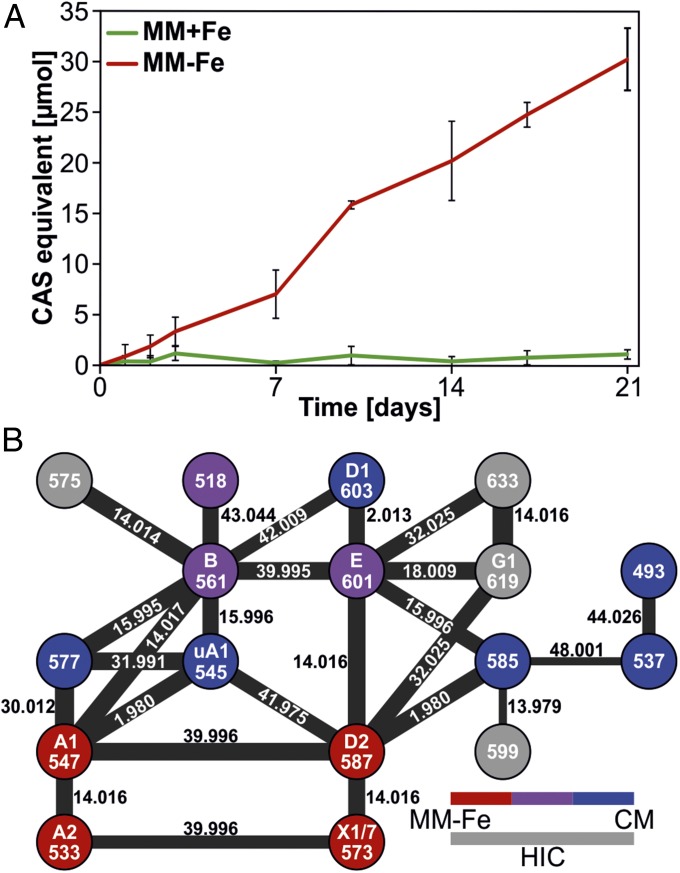
We measured total siderophore production in the supernatant of S. chartreusis over a cultivation period of 21 d in MM+Fe and MM−Fe using a colorimetric chromeazurol S (CAS) assay (Fig. 4A), in which siderophores compete with CAS for iron (37). Constituents of the CM cause dye precipitation, preventing application of the CAS assay to this medium. As expected, siderophores are produced under iron-limiting conditions. In parallel, desferrioxamine abundance was quantified using continuous MSE analysis (SI Appendix, Fig. S24). This data-independent acquisition method (38) provided better sensitivity than data-dependent acquisition of LC-MS/MS. Except for two desferrioxamines (493.3344 Da and 537.3605 Da), which are just above the detection limit, the production of all desferrioxamines parallels the siderophore levels in the CAS experiment. Notably, the LC-MS/MS experiment revealed that the desferrioxamines are not produced in equal stoichiometry when S. chartreusis is cultured in different media (Figs. 2 and 4B). The most common desferrioxamines, B and E, were also the most abundant ones in MM−Fe and in CM. The slightly smaller desferrioxamine B was the most abundant variant in MM−Fe, while E was the most abundant in CM. Both were also detected in trace amounts in MM+Fe after 14 d. A previously unknown desferrioxamine of 518.3184 Da was also detected in CM and MM−Fe. Six additional desferrioxamines were exclusively identified in CM, four in MM−Fe. The fact that four more desferrioxamines were detected in MM−Fe after HIC suggests that the product range of the biosynthetic machinery extends beyond the desferrioxamines described here. Precursor availability in the medium might contribute to the differential production of desferrioxamines. However, precursor availability is not a strict requirement, since with the more sensitive MSE analysis all desferrioxamines, including those detected only in CM by LC-MS/MS, were detected in low quantities in MM−Fe, which lacks any potential precursors (SI Appendix, Fig. S24).
Fig. 4.
Desferrioxamine levels. (A) Siderophore levels in MM+Fe and MM−Fe were measured by CAS assay (means of five biological replicates with SDs). (B) The molecular network of desferrioxamines, which are represented by their protonated mass in Da. Colors indicate appearance in media (CM, blue; MM−Fe, red; both, purple). Gray circles represent desferrioxamines only detected after HIC enrichment. The line thickness reflects cosine similarity of fragmentation spectra; factors provide the difference in parent mass in Da.
Discussion
LC-MS–Based Analysis of the Secreted Metabolome.
The bacterial genus Streptomyces is one of the richest sources of bioactive metabolites, producing ∼70% of clinically used antibiotics of natural origin (39). S. chartreusis NRRL 3882 is a typical representative regarding genome size (40) and genome-wired chemical potential. The number of predicted BGCs is close to that of the model organism S. coelicolor (RefSeq: NCC_003888.3; 128 and 118, respectively). We established an MS-based workflow that simultaneously enables structural and differential metabolome analysis (SI Appendix, Fig. S25) and employed it to study the medium dependence of metabolite secretion by S. chartreusis.
Modern LC-MS approaches allow the parallel detection of metabolites in complex mixtures and their relative quantitation across different cultivation conditions. Using our LC-MS/MS+–based workflow, in total 1,044 distinct metabolites were detected, exceeding the number of BGCs by almost an order of magnitude. Special enrichment techniques can further lower the detection limits of metabolites as demonstrated for HIC, which led to the detection of 343 additional metabolites from supernatants of MM−Fe cultures. Approaches such as the parallel use of negative-mode MS, extension of fractionation strategies, or further variation of cultivation conditions will further increase the number of detectable metabolites. It has already been shown that addition of siderophores (41) or antibiotics (42) or cocultivation (43) can alter the set of metabolites produced.
While mRNAs and proteins can be predicted from the genome sequence with suitable accuracy, metabolites cannot be predicted unequivocally from the genome directly. The gold standard for structure elucidation is NMR spectroscopy. However, it has limitations when it comes to analyzing complex mixtures of low-abundant metabolites. LC-MS and LC-MS/MS can make valuable contributions in this area, as recent works by Derewacz et al. (19) or Sidebottom et al. (20) demonstrated. In the present study, LC-MS/MS proved a useful starting point for structure elucidation of metabolites in complex mixtures. The identification of deoxacalcimycin in the molecular network and subsequent structure elucidation illustrate advantages and limitations of this method. The initial identification of deoxacalcimycin as a calcimycin analog was achieved based on MS/MS analysis, and the structural deviation from calcimycin was correctly localized to the benzoxazole region. NMR analysis revealed that deoxacalcimycin contains a 3-hydroxyanthranilic acid moiety (SI Appendix, Fig. S9) rather than a 1,3-benzoxazole moiety, which characterized the highest-scoring structure predicted by MS/MS (SI Appendix, Fig. S3). Structure elucidation using spectroscopic techniques clearly presents the main bottleneck for identification of novel structural classes. The number of annotations achievable by searching public spectral libraries is still limited. A well-characterized BGC and structural information on analogs can render future MS/MS-based structure predictions more precise. Structures can be deduced by interpreting and comparing parent and fragment mass spectra, as demonstrated here for desferrioxamines, which have been studied intensively since their discovery half a century ago (44, 45). The identification of eight previously undescribed desferrioxamines highlights the potential for the discovery of derivatives of known compound classes.
Diversity of Secreted Metabolites.
Factors contributing to the extensive metabolite diversity include, e.g., enzyme promiscuity as described for polyketide synthases, which can utilize a variety of starter and extender units (46–48), alternative cyclization or condensation reactions (49, 50), differential regulation of individual enzymes of the biosynthetic machine (51), the release of intermediates and pathway shunt products, biosynthesis mediated by gene products not detected as secondary metabolite biosynthesis genes by antiSMASH, or synthesis by cooperating biosynthetic machines (52). We cannot completely exclude that primary metabolites, and biotransformed or degraded media components, further add to metabolite diversity, although we expect those to be few. Media components of uninoculated medium were excluded from the analysis, and particularly MM contains only glucose, glutamate, and tryptophan, which could be degraded and biotransformed. Further, none of the numerous primary metabolites represented in the GNPS database were identified.
The production of structurally related derivatives offers an opportunity for fine-tuning the biological activity of metabolites to growth conditions. For example, the polyether ionophore calcimycin exerts its antibiotic effect by disturbing metal ion homeostasis in target organisms (53). The semisynthetic variant 4Br-A23187 of calcimycin differs in binding and transport capabilities for different metal ions (54–56). Similarly, the naturally occurring calcimycin derivatives detected in this study might also have divergent affinities. While cezomycin is the main product in MM, calcimycin is absent in MM but the main product in CM. It is postulated that, to produce calcimycin, cezomycin is modified by the addition of a hydroxyl group, which is converted to an amino group, which finally is methylated (29). None of these steps is expected to require complex precursors. Thus, the observed differential output of the biosynthetic machine is likely not a result of precursor availability. The biological and ecological function of these calcimycin-like ionophores is yet to be elucidated. It is possible that the derivatives have distinct biological functions or that they fulfill the same function but with different efficiencies depending on metal ion concentrations in the surroundings. It remains to be investigated if this differential production aids bacteria to prevail in their niche.
In this study, a host of siderophores was detected, some of which have very similar structures. Siderophores aid in the uptake of iron, but additional, nonclassical siderophore functions are also known. Small structural modifications can shift metal affinities and change the function from mobilization of iron to molybdenum (57). Siderophores can protect against oxidative stress, serve signaling purposes, or inhibit other organisms such as the siderophore-based antibiotic albomycin (58). Desferrioxamines can be modified to suit current living conditions (59). Traxler et al. (60) have shown that S. coelicolor, facing other Actinomycetes, produces acylated desferrioxamines. In the present study, the desferrioxamines provide another example of the different biosynthetic output of a BGC in dependence of the medium composition, which is reflected by differently colored nodes within the subnetwork (Fig. 2). In nutrient-rich CM, the more complex desferrioxamine E is most abundant (built from three N-hydroxy-N-succinylcadaverines), while the smaller desferrioxamine B (built from two N-hydroxy-N-succinylcadaverines and N-hydroxy-N-acetylcadaverine) is more abundant in MM−Fe. This differential production might be a consequence of balancing functionality and economy. Among the tested cultivation conditions, the most and largest metabolites were produced when S. chartreusis was cultivated in CM. This might reflect the availability of nutrients and precursors but can also present an adaptation to nutrient-rich environments in which secreted metabolites fulfill yet unknown functions. For most secreted metabolites, little is known about the ecological function, let alone the function of mixtures of structurally related compounds produced by a single species. Even for the comparably well-studied desferrioxamines it remains to be investigated if derivatives produced in CM fulfill different functions from those produced in MM−Fe and how the production of different products from the biosynthetic machine is regulated.
The secreted metabolome under three growth conditions revealed a high diversity and complementarity. It will be interesting to expand this LC-MS/MS–based approach by diversifying cultivation conditions as, e.g., in the OSMAC approach (14). Alternative cultivation conditions may result in the activation of additional BGCs that were silent under the three conditions tested here, an approach that has been employed successfully (18). The combination of metabolomics with global transcriptome or proteome analysis and targeted mutagenesis to match BGCs with their metabolite products will further expand our understanding of the chemical capabilities of microbes. The conditions under which compounds are produced might also provide clues as to their function. The siderophores, for instance, are produced when iron is limiting and it is tempting to speculate that among the 101 molecules detected in MM−Fe, but not MM+Fe, there are others with a role in iron acquisition.
Conclusions
We present a workflow for the global differential analysis of metabolites. Our approach provides a means to look at the diversity of molecules and at how bacteria fine-tune their chemical repertoire in response to cultivation conditions. S. chartreusis NRRL 3882 produces many metabolites in parallel with the number of distinct metabolites detectable under a single condition exceeding the number of BGCs. The set of metabolites produced is tailored specifically to the growth conditions. Further, our approach facilitates network-guided structure elucidation by tandem MS. We uncovered hitherto unknown siderophores and ionophores. Our study of the S. chartreusis NRRL 3882 secreted metabolome shows that the chemical potential of microorganisms is far from being fully characterized.
Materials and Methods
Experimental details including citations are provided in the SI Appendix.
Strains and Cultivation Conditions.
S. chartreusis NRRL 3882 was cultivated in yeast extract malt extract (39) complex medium (3 g/L yeast extract, 5 g/L peptone, 3 g/L malt extract, 55.5 mM glucose, and 73 mM saccharose) or defined MM (MM−Fe) [21 mM NaCl, 15 mM (NH4)2SO4, 8 mM MgSO4, 27 mM KCl, 50 mM Tris, 0.6 mM KH2PO4, 2 mM CaCl2, 0.01 mM MnSO4, 4.5 mM l-glutamate, 0.78 mM l-tryptophan, 11 mM d-glucose, pH 7.5], with 0.001 mM FeSO4 added when indicated (MM+Fe.)
DNA Isolation and Sequencing.
DNA was extracted after digestion of the mycelial cell wall with 5 mg/mL lysozyme in Tris-EDTA buffer (10 mM Tris, 5 mM EDTA, 0.075 mg/mL RNase A, pH 8), as described by Kieser et al. (39). The genome was sequenced on the Illumina MiSeq platform, assembled using gsAssembler 2.8 and annotated using Prokka 1.11 and GenDB 2.0. Putative BGCs were predicted using antiSMASH 3.0.5.
HIC.
Supernatant of an MM−Fe culture was incubated with 25 mg/mL of Diaion HP-20 resin. The resin was washed with water and eluted with 25%, 50%, and 100% CH3OH.
LC-MS/MS and MSE Measurements.
Over a time course of 2 wk, supernatants of cultures were harvested. After extraction with ethyl acetate, the organic and aqueous phases were dried, reconstituted in CH3OH, and subjected to LC-MS/MS and LC-MSE measurements. Separation was performed on a C18 column using an H2O/CH3CN gradient with 0.1% formic acid (FA). Mass spectra were recorded in positive mode on a Synapt G2-S HDMS with an ESI source and TOF detector. All experiments were performed three times independently.
Molecular Networking and Spectra Annotation.
After file conversion using Proteowizard (version 3.0.940), a molecular network was generated using the GNPS platform (23). Data can be accessed at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c43717fa433e4456ac01e6cf1ce7476b. The molecular network was visualized via Cytoscape (version 3.3.0). Redundancies and adducts were cleared manually. Spectra of interest were annotated with the aid of the MetFrag in silico fragmentation tool (27).
Colorimetric Siderophore Detection.
A previously described CAS assay (37) was used. Cell-free culture supernatant was mixed in equal amounts with the reaction solution (15 µM FeCl3, 150 µM CAS, 600 µM cetyl trimethylammonium bromide, 563 mM piperazin, pH 5.6). After 3 h, the absorption at 660 nm was measured.
Compound Purification and Characterization.
S. chartreusis was cultivated in international Streptomyces project medium 2 (4 g/L yeast extract, 10 g/L malt extract, 4 g/L glucose). The culture supernatant was extracted with an equal amount of ethyl acetate, which was subsequently evaporated, and the remaining residue reconstituted in CH3OH. Subsequent separation was performed by automated reversed-phase medium pressure liquid chromatography on a CombiFlash Rf using a 40-g SiliaSep C18 column, with an H2O/CH3CN gradient with 0.1% FA. Calcimycin-like compounds were detected at 360 nm. Peak fractions were combined and further analyzed. LC-MS/MS was performed on an Exactive Orbitrap mass spectrometer equipped with an Accella photodiode array detector and a SEDEX Model 80 low-temperature evaporative light-scattering detector. An H2O/CH3CN gradient with 0.1% FA was used for separation on a C18 column. All 1H and 13C NMR spectra were acquired on a 600-MHz Bruker Avance III NMR spectrometer operating at 600 and 150 MHz, respectively. Optical rotations were recorded on an Autopol III polarimeter. Infrared spectra were acquired by attenuated total reflectance using a SMART iTR accessory on a Nicolet 6700 FTIR spectrometer.
Supplementary Material
Acknowledgments
We thank Stefano Donadio (Naicons, Italy) for providing S. chartreusis NRRL 3882; Dr. Chris Kirby and Maike Fischer of Agriculture and Agri-Food Canada for providing NMR services; and Sina Schäkermann for assistance with MS and for critically reading the manuscript. M.N. thanks Ulrich Kück for support at the Department of General and Molecular Botany (RUB). The authors gratefully acknowledge financial support from the Natural Sciences and Engineering Council of Canada, the Canada Research Chair Program, the Atlantic Canada Opportunities Agency, and the Jeanne and Jean-Louis Levesque Foundation to the University of Prince Edward Island (R.G.K.), the German Research Foundation [BA 4193/6-1 (to J.E.B.), NO 407/7-1 (to M.N.)], the project “Bielefeld-Gießen Center for Microbial Bioinformatics—BiGi,” funded by the Federal Ministry of Education and Research (BMBF) (Grant 031A533) within the German Network for Bioinformatics Infrastructure (to D.W.), and the German federal state of North Rhine-Westphalia for funding the mass spectrometer (Forschungsgroßgeräte der Länder; to J.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive, https://www.ebi.ac.uk/ena/ [accession nos. LT962942 and LT963352 (genome information)], and in the Global Natural Products Social Molecular Networking (GNPS) Library, https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c43717fa433e4456ac01e6cf1ce7476b and https://gnps.ucsd.edu/ProteoSAFe/result.jsp?task=8d9039754c8f4f59bb713e7cd411fdf9&view=advanced_view (metabolome information).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715713115/-/DCSupplemental.
References
- 1.Findland M. [Clinical and laboratory study of a new antibiotic: Tetracycline] Odontoiatr Rev Iberoam Med Boca. 1954;11:381–383. Spanish. [PubMed] [Google Scholar]
- 2.WHO 2015 19th WHO list of essential medicines. Available at www.who.int/medicines/publications/essentialmedicines/EML_2015_FINAL_amended_NOV2015.pdf. Accessed February 25, 2016.
- 3.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. Retrospective analysis of natural products provides insights for future discovery trends. Proc Natl Acad Sci USA. 2017;114:5601–5606. doi: 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism: From biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 6.Miethke M. Molecular strategies of microbial iron assimilation: From high-affinity complexes to cofactor assembly systems. Metallomics. 2013;5:15–28. doi: 10.1039/c2mt20193c. [DOI] [PubMed] [Google Scholar]
- 7.Abrudan MI, et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA. 2015;112:11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumbhar C, Mudliar P, Bhatia L, Kshirsagar A, Watve M. Widespread predatory abilities in the genus Streptomyces. Arch Microbiol. 2014;196:235–248. doi: 10.1007/s00203-014-0961-7. [DOI] [PubMed] [Google Scholar]
- 9.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien J, Wright GD. An ecological perspective of microbial secondary metabolism. Curr Opin Biotechnol. 2011;22:552–558. doi: 10.1016/j.copbio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Katz L, Baltz RH. Natural product discovery: Past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 12.Baltz RH. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol. 2016;44:573–588. doi: 10.1007/s10295-016-1815-x. [DOI] [PubMed] [Google Scholar]
- 13.Thanapipatsiri A, et al. Discovery of unusual biaryl polyketides by activation of a silent Streptomyces venezuelae biosynthetic gene cluster. ChemBioChem. 2016;17:2189–2198. doi: 10.1002/cbic.201600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bode HB, Bethe B, Höfs R, Zeeck A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Reen FJ, Romano S, Dobson AD, O’Gara F. The sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar Drugs. 2015;13:4754–4783. doi: 10.3390/md13084754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand S, et al. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol Adv. 2014;32:1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Bertrand S, et al. Multi-well fungal co-culture for de novo metabolite-induction in time-series studies based on untargeted metabolomics. Mol Biosyst. 2014;10:2289–2298. doi: 10.1039/c4mb00223g. [DOI] [PubMed] [Google Scholar]
- 19.Derewacz DK, Covington BC, McLean JA, Bachmann BO. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem Biol. 2015;10:1998–2006. doi: 10.1021/acschembio.5b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chem Biol. 2013;8:2009–2016. doi: 10.1021/cb4002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doroghazi JR, et al. Genome sequences of three tunicamycin-producing Streptomyces strains, S. chartreusis NRRL 12338, S. chartreusis NRRL 3882, and S. lysosuperificus ATCC 31396. J Bacteriol. 2011;193:7021–7022. doi: 10.1128/JB.06262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber T, et al. antiSMASH 3.0:–A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M, et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han AW, et al. Turnerbactin, a novel triscatecholate siderophore from the shipworm endosymbiont Teredinibacter turnerae T7901. PLoS One. 2013;8:e76151. doi: 10.1371/journal.pone.0076151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boiteau RM, et al. Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc Natl Acad Sci USA. 2016;113:14237–14242. doi: 10.1073/pnas.1608594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: A critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 27.Ruttkies C, Gerlich M, Neumann S. Tackling CASMI 2012: Solutions from MetFrag and MetFusion. Metabolites. 2013;3:623–636. doi: 10.3390/metabo3030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Q, et al. Characterization of the biosynthesis gene cluster for the pyrrole polyether antibiotic calcimycin (A23187) in Streptomyces chartreusis NRRL 3882. Antimicrob Agents Chemother. 2011;55:974–982. doi: 10.1128/AAC.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Q, et al. Characterization of the N-methyltransferase CalM involved in calcimycin biosynthesis by Streptomyces chartreusis NRRL 3882. Biochimie. 2013;95:1487–1493. doi: 10.1016/j.biochi.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 31.Price NP, Tsvetanova B. Biosynthesis of the tunicamycins: A review. J Antibiot (Tokyo) 2007;60:485–491. doi: 10.1038/ja.2007.62. [DOI] [PubMed] [Google Scholar]
- 32.Keenan RW, Hamill RL, Occolowitz JL, Elbein AD. Biological activities of isolated tunicamycin and streptovirudin fractions. Biochemistry. 1981;20:2968–2973. doi: 10.1021/bi00513a039. [DOI] [PubMed] [Google Scholar]
- 33.Kadi N, Song L, Challis GL. Bisucaberin biosynthesis: An adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem Commun (Camb) 2008;41:5119–5121. doi: 10.1039/b813029a. [DOI] [PubMed] [Google Scholar]
- 34.Barona-Gómez F, Wong U, Giannakopulos AE, Derrick PJ, Challis GL. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J Am Chem Soc. 2004;126:16282–16283. doi: 10.1021/ja045774k. [DOI] [PubMed] [Google Scholar]
- 35.Soe CZ, Pakchung AA, Codd R. Directing the biosynthesis of putrebactin or desferrioxamine B in Shewanella putrefaciens through the upstream inhibition of ornithine decarboxylase. Chem Biodivers. 2012;9:1880–1890. doi: 10.1002/cbdv.201200014. [DOI] [PubMed] [Google Scholar]
- 36.Challis GL, Ravel J. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: Structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett. 2000;187:111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- 37.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 38.Liu S, et al. Exploring skyline for both MS(E) -based label-free proteomics and HRMS quantitation of small molecules. Proteomics. 2014;14:169–180. doi: 10.1002/pmic.201300352. [DOI] [PubMed] [Google Scholar]
- 39.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 40.Kim JN, et al. Comparative genomics reveals the core and accessory genomes of Streptomyces species. J Microbiol Biotechnol. 2015;25:1599–1605. doi: 10.4014/jmb.1504.04008. [DOI] [PubMed] [Google Scholar]
- 41.Watrous J, et al. Metabolic profiling directly from the Petri dish using nanospray desorption electrospray ionization imaging mass spectrometry. Anal Chem. 2013;85:10385–10391. doi: 10.1021/ac4023154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada BK, Wu Y, Mao D, Bushin LB, Seyedsayamdost MR. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda K, et al. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J Antibiot (Tokyo) 2000;53:979–982. doi: 10.7164/antibiotics.53.979. [DOI] [PubMed] [Google Scholar]
- 44.Keberle H. The biochemistry of desferrioxamine and its relation to iron metabolism. Ann N Y Acad Sci. 1964;119:758–768. doi: 10.1111/j.1749-6632.1965.tb54077.x. [DOI] [PubMed] [Google Scholar]
- 45.Feistner GJ, Stahl DC, Gabrik AH. Proferrioxamine siderophores of Erwinia amylovora: A capillary liquid chromatographic/electrospray tandem mass spectrometry study. J Mass Spectrom. 1993;28:163–175. [Google Scholar]
- 46.Ladner CC, Williams GJ. Harnessing natural product assembly lines: Structure, promiscuity, and engineering. J Ind Microbiol Biotechnol. 2016;43:371–387. doi: 10.1007/s10295-015-1704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandith SA, et al. Functional promiscuity of two divergent paralogs of type III plant polyketide synthases. Plant Physiol. 2016;171:2599–2619. doi: 10.1104/pp.16.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koryakina I, McArthur JB, Draelos MM, Williams GJ. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org Biomol Chem. 2013;11:4449–4458. doi: 10.1039/c3ob40633d. [DOI] [PubMed] [Google Scholar]
- 49.Austin MB, Bowman ME, Ferrer J-L, Schröder J, Noel JP. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem Biol. 2004;11:1179–1194. doi: 10.1016/j.chembiol.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 50.Beerhues L, Liu B. Biosynthesis of biphenyls and benzophenones:–Evolution of benzoic acid-specific type III polyketide synthases in plants. Phytochemistry. 2009;70:1719–1727. doi: 10.1016/j.phytochem.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 51.Vargas-Bautista C, Rahlwes K, Straight P. Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J Bacteriol. 2014;196:717–728. doi: 10.1128/JB.01022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesener SR, Potharla VY, Cheng YQ. Reconstitution of the FK228 biosynthetic pathway reveals cross talk between modular polyketide synthases and fatty acid synthase. Appl Environ Microbiol. 2011;77:1501–1507. doi: 10.1128/AEM.01513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raatschen N, et al. Extracting iron and manganese from bacteria with ionophores:–A mechanism against competitors characterized by increased potency in environments low in micronutrients. Proteomics. 2013;13:1358–1370. doi: 10.1002/pmic.201200556. [DOI] [PubMed] [Google Scholar]
- 54.Erdahl WL, Chapman CJ, Wang E, Taylor RW, Pfeiffer DR. Ionophore 4-BrA23187 transports Zn2+ and Mn2+ with high selectivity over Ca2+ Biochemistry. 1996;35:13817–13825. doi: 10.1021/bi961391q. [DOI] [PubMed] [Google Scholar]
- 55.Deber CM, Tom-Kun J, Mack E, Grinstein S. Bromo-A23187: A nonfluorescent calcium ionophore for use with fluorescent probes. Anal Biochem. 1985;146:349–352. doi: 10.1016/0003-2697(85)90550-0. [DOI] [PubMed] [Google Scholar]
- 56.Pfeiffer DR, Taylor RW, Lardy HA. Ionophore A23187: Cation binding and transport properties. Ann N Y Acad Sci. 1978;307:402–423. [Google Scholar]
- 57.Hernandez JA, George SJ, Rubio LM. Molybdenum trafficking for nitrogen fixation. Biochemistry. 2009;48:9711–9721. doi: 10.1021/bi901217p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnstone TC, Nolan EM. Beyond iron: Non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spasojevic I, Armstrong SK, Brickman TJ, Crumbliss AL. Electrochemical behavior of the Fe(III) complexes of the cyclic hydroxamate siderophores alcaligin and desferrioxamine E. Inorg Chem. 1999;38:449–454. doi: 10.1021/ic980635n. [DOI] [PubMed] [Google Scholar]
- 60.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4:e00459. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.