Significance
In eukaryotes, meiotic recombination shuffles genetic information at each generation, making each offspring unique. In addition to its key role in heredity and evolution, this gene reshuffling is central in plant and animal breeding, which relies on the ability to combine desired alleles. However, the number of recombination events by generation is low, limiting the power of breeding programs. Here we showed that meiotic recombination can be massively increased in hybrid plants, up to almost eightfold. This opens the possibility of manipulating recombination to enhance the efficiency of plant breeding programs, which is important as we face the double challenge of ensuring food security while preserving natural resources.
Keywords: recombination, meiosis, crossovers, plant breeding, Arabidopsis
Abstract
Meiotic crossovers shuffle parental genetic information, providing novel combinations of alleles on which natural or artificial selection can act. However, crossover events are relatively rare, typically one to three exchange points per chromosome pair. Recent work has identified three pathways limiting meiotic crossovers in Arabidopsis thaliana that rely on the activity of FANCM [Crismani W, et al. (2012) Science 336:1588–1590], RECQ4 [Séguéla-Arnaud M, et al. (2015) Proc Natl Acad Sci USA 112:4713–4718], and FIGL1 [Girard C, et al. (2015) PLoS Genet 11:e1005369]. Here we analyzed recombination in plants in which one, two, or all three of these pathways were disrupted in both pure line and hybrid contexts. The greatest effect was observed when combining recq4 and figl1 mutations, which increased the hybrid genetic map length from 389 to 3,037 cM. This corresponds to an unprecedented 7.8-fold increase in crossover frequency. Disrupting the three pathways did not further increase recombination, suggesting that some upper limit had been reached. The increase in crossovers is not uniform along chromosomes and rises from centromere to telomere. Finally, although in wild type recombination is much higher in male meiosis than in female meiosis (490 cM vs. 290 cM), female recombination is higher than male recombination in recq4 figl1 (3,200 cM vs. 2,720 cM), suggesting that the factors that make wild-type female meiosis less recombinogenic than male wild-type meiosis do not apply in the mutant context. The massive increase in recombination observed in recq4 figl1 hybrids opens the possibility of manipulating recombination to enhance plant breeding efficiency.
Crossovers (COs), reciprocal exchanges between homologous chromosomes, have two consequences. First, in combination with sister chromatid cohesion, COs provide a physical link between homologs, which is required for balanced segregation of chromosomes at meiosis. Failure of the formation of at least one crossover per chromosome pair is associated with reduced fertility and aneuploidy (1). Second, COs lead to the exchange of flanking DNA, generating novel mosaics of the homologous chromatids. This translates into genetic recombination, classically measured in Morgans, or centiMorgans (cM).
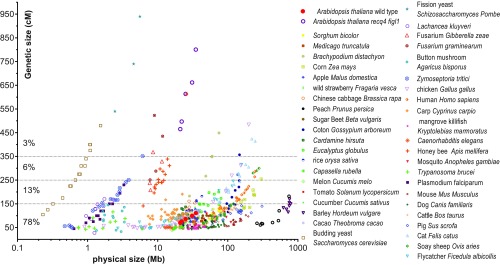
The number of COs appears to be constrained by both an upper limit and a lower limit (2). To illustrate, Fig. 1 plots the genetic size (cM) versus the physical size (DNA base pairs, log scale) of chromosomes in a diverse panel of eukaryotes. Note the large variations in physical size, while the genetic map length is much less variable. At the lower limit, chromosomes measure 50 cM, which corresponds to the requirement of at least one CO per chromosome pair (0.5 per chromatid). In some species, such as the worm Caenorhabditis elegans, the plant Capsella rubella, and the fish Kryptolebias marmoratus, all chromosomes are ∼50 cM long. Across all species, most chromosomes do not receive much more than one CO per meiosis, with 80% having fewer than three. Among the exceptional cases with more than nine COs per chromosome are the three chromosomes of the yeast Schizosaccharomyces pombe, whose meiosis has other unusual aspects, including the lack of both a synaptonemal complex and CO interference (3). Thus, the high numbers of COs per chromosome observed in a handful of species appears to be counterselected in most species, suggesting that CO rate is a trait under selection in both directions (4).
Fig. 1.
The number of meiotic crossovers is constrained in eukaryotes. Chromosomes from a range of species are plotted according to their physical (x-axis, Mb, log scale) and genetic size (y-axis, cM, linear scale). Physical size is based on genome sequence assembly, and genetic size is based on F2 or male/female average. Sex chromosomes have been excluded. An earlier version of this figure, with fewer species represented, was published by Mercier et al. (2). The data used to produce this figure are provided in Dataset S1.
Plant breeding programs rely on meiotic COs that allow the stacking of desired traits into elite lines. However, as described above, the number of COs is low. Furthermore, some regions are virtually devoid of COs, such as the regions flanking centromeres (5). This limits the genetic diversity that can be created in breeding programs and also limits the power of genetic mapping in prebreeding research. Thus, increasing recombination is a desirable trait in plant breeding (6, 7). Forward screen approaches have identified three pathways that limit recombination in Arabidopsis thaliana, relying respectively on the activity of (i) the FANCM helicase and its cofactors (8, 9), (ii) the BLM/Sgs1 helicase homologs RECQ4A and RECQ4B and the associated proteins TOP3α and RMI1 (10, 11), and (iii) the FIGL1 AAA-ATPase (12). RECQ4A and RECQ4B are duplicated genes that redundantly limit meiotic crossovers in Arabidopsis, and thus are designated RECQ4 herein for simplicity. In all mutants, recombination is largely increased compared with wild type when tested in pure lines; however, the fancm mutation, which increases meiotic recombination threefold in pure lines, has almost no effect on recombination in hybrids (12, 13). Thus, it has remained unclear whether the manipulation of these three pathways could be used to increase recombination in hybrids, the context that matters for plant breeding programs.
Results
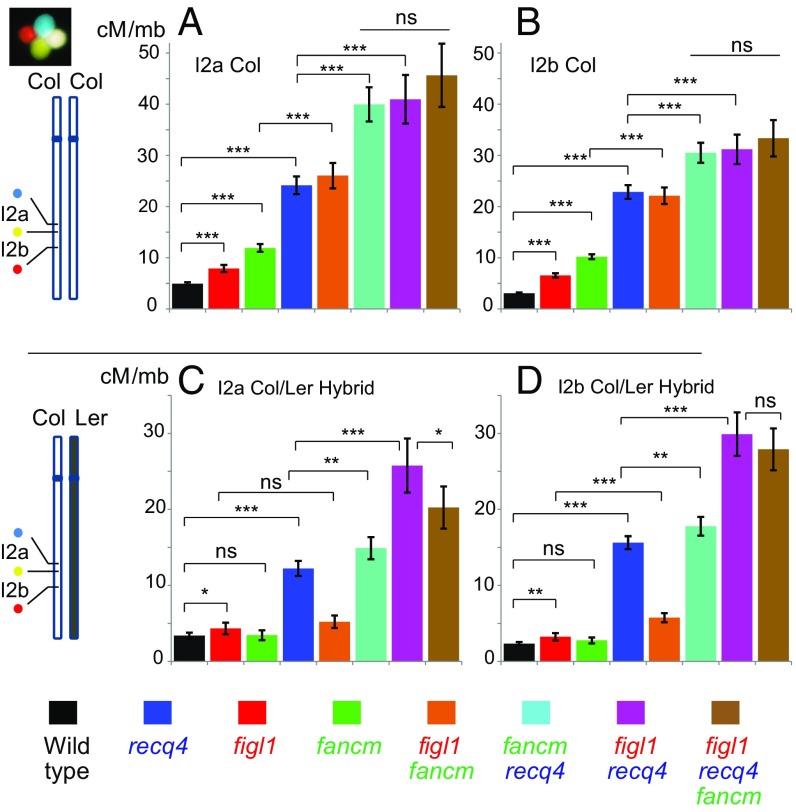
Here we analyzed recombination in single, double, and triple mutants for FANCM, RECQ4, and FIGL1 in both pure line and hybrid contexts using two complementary approaches. First, we used a fluorescent-tagged line (FTL I2ab) that relies on transgenic markers conferring fluorescence to pollen grains organized in tetrads (14) (Fig. 2). With this tool, we accurately measured recombination in two adjacent intervals in either pure line (Col) or F1 hybrid contexts (Col/Ler). Second, we used the segregation in F2 progeny of a set of 96 Col/Ler single nucleotide polymorphisms (SNPs) (12) to measure recombination genome-wide in F1 hybrid contexts (Fig. 3). In pure Col, each single mutant had higher recombination than wild type in both FTL tested intervals (Fig. 2 A and B). Each double mutant was higher than the respective single mutants, confirming that FANCM, RECQ4, and FIGL1 act in three different pathways to limit COs. The greatest increase was observed in the fancm recq4 and figl1 recq4 double mutants, both of which exhibited a ∼10-fold increase compared with wild type. The observation that each double mutant combination results in an increase in recombination compared with the single mutants (i.e., three independent pathways) predicts that the triple mutant should increase recombination even further; however, recombination in the figl1 recq4 fancm triple mutant was not higher than that in the highest double mutants, suggesting that some upper limit had been reached.
Fig. 2.
Meiotic recombination measured by FTL tetrad analysis. (A and B) Recombination in a pure Col-0 line in intervals I2a and I2b, respectively. (C and D) Recombination in the Col/Ler F1 hybrids in intervals I2a and I2b, respectively. Bars represent genetic distance calculated with the Perkins equation in cM/Mb, ± 95% confidence interval. Z-tests are indicated: ***P < 0.001; **P < 0.01; *P < 0.05; ns: P > 0.05. Raw data are provided in Dataset S2.
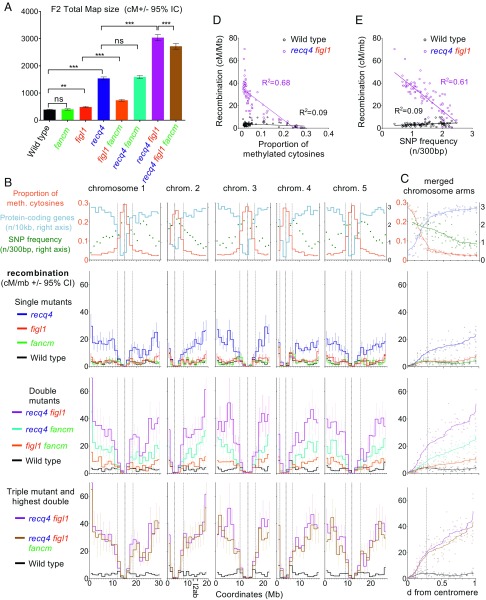
Fig. 3.
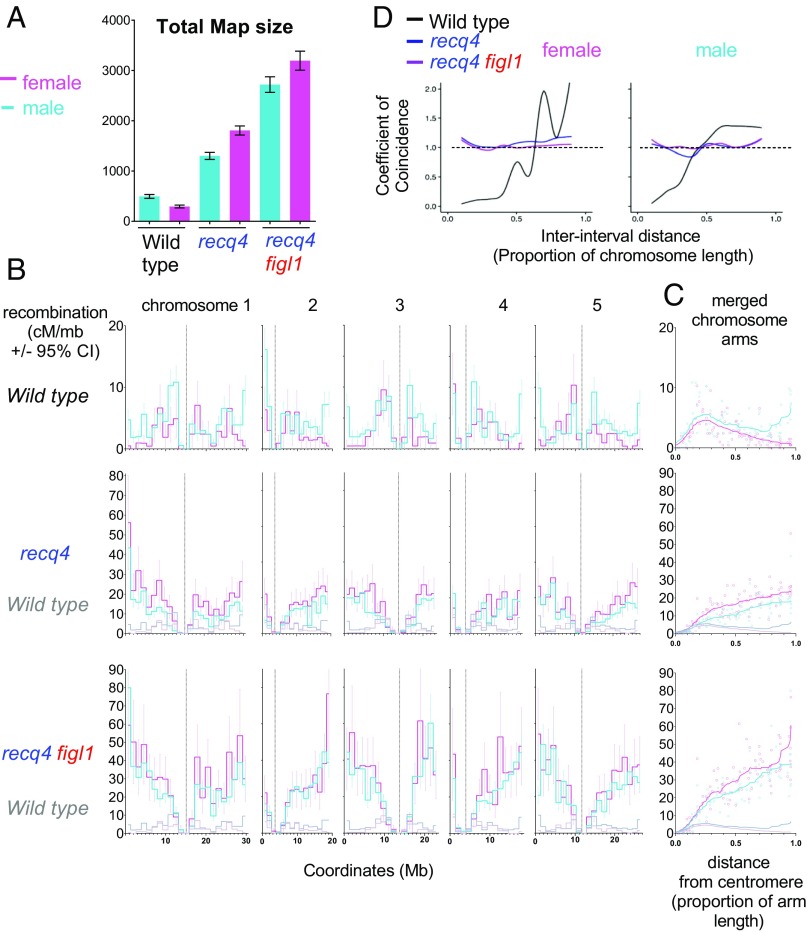
Genome-wide recombination measured by SNP segregation in F2s. (A) Total genetic maps. Tests are one-way ANOVA on the observed number of COs per F2 plant, with Dunnett’s correction. ***P < 0.001; **P < 0.01; *P < 0.05; ns: P > 0.05. (B) Plotted along each chromosomes (Mb), from top to bottom, are gene density, SNP density (19), proportion of methylated cytosines (20), and recombination in the different genotypes (cM/Mb, Haldane equation). Vertical dotted lines mark the position of the centromeres and pericentromeres. chrom, chromosome; meth, methylated. (C) The same data as in B merged for all chromosome arms (short arms of chromosomes 2 and 4 excluded), plotted along the relative distance from the centromere, and smoothed. (D) Each interval plotted according to the proportion of methylated cytosines and recombination frequency. Linear regressions and associated R2 values are shown. (E) Each interval plotted according to SNP density and recombination frequency. Intervals spanning a centromere have been excluded. Linear regressions and associated R2 values are shown. Raw genotyping data are provided in Dataset S3, and the complete dataset is shown in Dataset S4.
In the Col/Ler hybrid context (Fig. 2 C and D for FTLs; Fig. 3A for genome-wide genetic maps), both figl1 and recq4 increased recombination, but fancm had no detectable effect, as reported previously in several hybrid contexts (12, 13). Similar to observations in pure Col, double-mutant hybrids exhibited higher recombination than the corresponding singles. Notably, the fancm mutation led to a detectable increase in recombination when combined with figl1 or recq4 (Figs. 2 C and D and 3A). This finding again predicts that combining the three mutations should lead to a further increase. However, recombination was not statistically higher in the figl1 recq4 fancm triple mutant compared with the figl1 recq4 double mutant and even appeared to be reduced (Figs. 2C and 3A).
The greatest increase in the hybrid context was thus observed in the figl1 recq4 double mutant, with an ∼11-fold increase in the FTL I2ab intervals (Fig. 2 C and D) and a 7.8-fold increase genome-wide (Fig. 3A), from 389 ± 18 cM in wild type to 3,037 ± 115 cM in figl1 recq4 (translating to 7.8 ± 0.4 and 60.7 ± 2.3 COs per meiosis, respectively). This is much higher than the greatest increase in meiotic recombination observed so far in any mutant (15–17). While Arabidopsis wild type had a very typical frequency of CO, with the genetic maps of each of the five chromosomes ranging from 70 to 110 cM (an average of 1.6 CO per chromosome), the figl1 recq4 was almost the highest recombining eukaryote (Fig. 1), with the genetic maps of chromosomes ranging from 450 to 800 cM (an average of 12 COs per chromosome: Fig. 1).
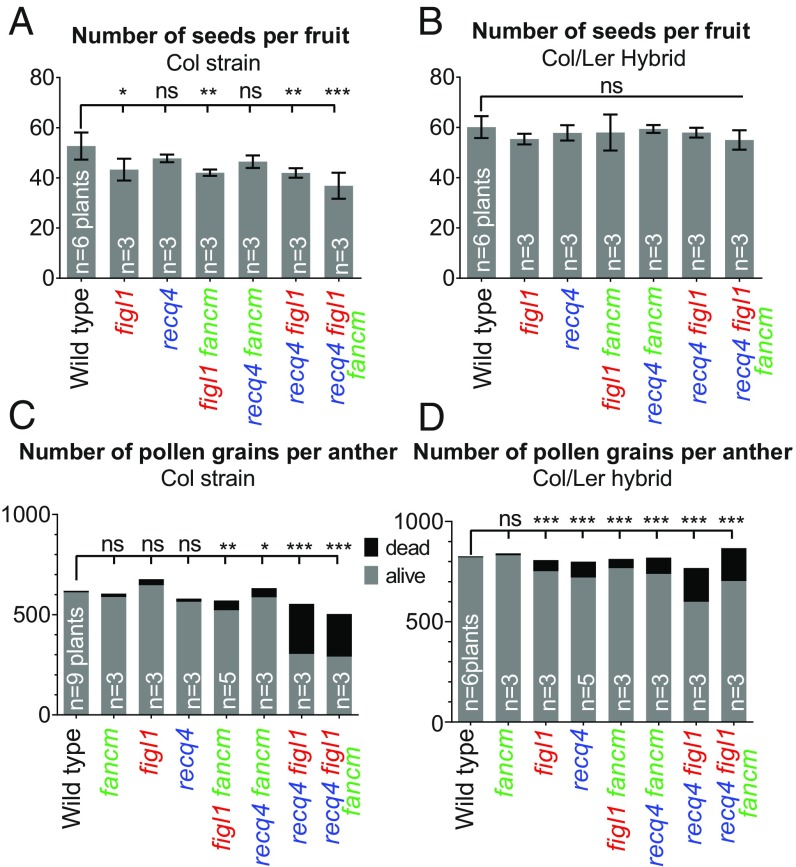
We asked whether such a large increase in recombination could reduce the fertility of plants, precluding the use of these mutant combinations in breeding programs. No growth or developmental defects were observed in any of the genotypes. In hybrids, the number of seeds per fruit was not significantly reduced in any genotype (Fig. 4B); however, a slight defect in pollen viability was observed in single mutants and multi-mutants (Fig. 4D). Furthermore, in pure Col, both reduced seed set and pollen viability defects were detected, with the highest numbers in recq4 figl1 and recq4 figl1 fancm (Fig. 4 A and C). This suggests that some fertility defects may be associated with increased recombination. The fertility defects in the different genotypes were quite poorly correlated with the increase in COs, however (compare Figs. 2 and Fig. 4); for example, Col fancm recq4 had the same large increase in recombination observed in figl1 recq4 (Fig. 2 A and B), but pollen viability was much less affected (Fig. 4C). This suggests that high levels of COs are not responsible per se for reduced fertility.
Fig. 4.
Analysis of fertility. (A and B) Number of seeds per fruit in pure Col and Col/Ler F1 hybrids, respectively. (C and D) Average numbers of viable and nonviable pollen grains per anther, following Alexander staining. n indicates for the number of plants analyzed. For each plant, the number of seeds was counted in a minimum of 10 fruits, and the number of pollen grains was counted in a minimum of three anthers. Errors bars represent SD. Tests comparing each genotype with wild type were done by ANOVA, followed by Dunnett’s correction for multiple tests. ***P < 0.001; **P < 0.01; *P < 0.05; ns: P > 0.05.
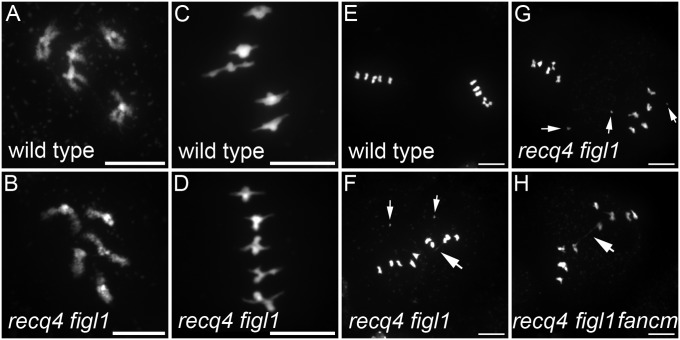
We further examined meiosis in the Col recq4 figl1 and recq4 figl1 fancm, which showed >40% pollen death (Fig. 5). At diakinesis, homologous chromosomes were associated in pairs, connected by COs. The chromosomes appeared more tightly connected in the mutants than in wild type, presumably because of increased CO numbers (Fig. 5 A and B). The shape of chromosomes at metaphase I was also suggestive of increased CO numbers (Fig. 5 C and D). At metaphase II, chromosome spreads revealed the presence of a few chromosome fragments and chromosome bridges in 50% of the cells (n = 114 metaphase II cells; Fig. 5 F–H), suggesting defective repair of a small subset of recombination intermediates.
Fig. 5.
Meiotic chromosome spreads. (A and B) Diakinesis. Five chromosome pairs are observed in both wild type (Col) and recq4 figl1 and appear to be more tightly linked in recq4 figl1. (C and D) Metaphase I. The five chromosome pairs are aligned on the metaphase plate. (E–H) Metaphase II. In wild type, five pairs of chromatids align on two metaphase plates. In recq4 figl1 and recq4 figl1fancm, chromatid bridges (large arrows) and chromosome fragments (small arrows) are observed in 50% of the cells. (Scale bars: 10 µM.)
We propose that the absence of FIGL1 and RECQ4 disturbs the double-strand break repair machinery, leading to the formation of aberrant intermediates [such as the multichromatid joint molecules observed in the yeast recq4 homolog sgs1 (18)], most of which are repaired as extra-COs and a few failing to be repaired. Ectopic intermediates and ectopic COs are other possible source of bridges and fragments. We suggest that unrepaired intermediates or aberrant products are responsible for most of the reduced fertility, but we cannot fully exclude that the extra COs themselves slightly disturb chromosome segregation.
We next explored the distribution of COs along the genome (Fig. 3 B and C) using the Col/Ler genome-wide recombination data. The first line of Fig. 3 B and C shows the gene density, SNP density (19), and proportion of methylated cytosines (20) plotted along each chromosome. The centromeres (vertical dotted line in Fig. 3B) are flanked by pericentromeric regions with low gene density and high methylated cytosine levels (>7.5%, the genome average, delimited by thin dotted lines). The three bottom lines of Fig. 3 B and C show recombination along the chromosomes in single, double, and triple mutants, respectively. At the scale used in this study (bins of ∼1.3 Mb), the wild-type curve of recombination frequency oscillates around ∼2.5–3.5 cM/Mb in chromosome arms, with the maximum observed close to pericentromeric and distal regions. The interval that spans the centromere in each chromosome has a very low level of recombination. In accordance with results for total map size (Fig. 3A), fancm is indistinguishable from wild type. The curves of all other mutant genotypes show higher recombination than wild type, with recq4 figl1 having the strongest effect (Fig. 3 B and C). In all mutants, the same tendency is observed, with no effect on recombination for intervals encompassing or immediately flanking the centromere and an increase in recombination with increasing distance from the centromere to reach a maximum close to telomeres. In figl1 recq4, the recombination frequency rises rapidly from the centromere to the first third of the arm, where it reaches ∼20 cM/Mb (approximately fivefold higher than wild-type levels). This first third of the chromosome arms correspond to the pericentromeric region, with a progressive decrease in methylation and increase in gene density with increasing distance from the centromere (Fig. 3 B and C, first row). In the remaining two-thirds of the chromosomes, methylation and gene density are relatively stable but recombination continues to increase toward the telomere, reaching an average of 45 cM/Mb (>10-fold higher than wild-type levels). Thus, knockdown of recq4 and figl1 has very different effects in different regions of the chromosome.
We propose three nonexclusive possible causes for this effect. First, the position along the chromosome itself could influence CO frequencies, as recombination occurs in the context of highly organized and dynamic chromosomes (21). Second, the accessibility of the chromatin may directly account for CO frequency, notably by influencing SPO11-dependant double-strand break formation (22). The anticorrelation of cytosine methylation and recombination (Fig. 3D) seen in recq4 figl1 (Fig. 3D; linear regression, R2 = 0.68, P < 10−4), supports this view. Third, DNA polymorphism, which decreases from centromere to telomere (Fig. 3C), may also prevent CO frequency. We observed a strong anticorrelation between recombination and SNP density in recq4 figl1 (Fig. 3E; linear regression, R2 = 0.61, P < 10−4), which is very weak and positive in wild type (R2 = 0.09, P = 0.07). The distance from the centromere, methylation, and polymorphisms are correlated with one another, but multiple linear regression suggests that each of these three parameters influences recombination in recq4 figl1 (P = 0.0012, 3.5 10−6, and 6.8 10−6, respectively). Interestingly, decreased recombination is observed in the middle of the right arm of chromosome 1 in recq4 figl1 and recq4 figl1 fancm (Fig. 3B), corresponding to a region of high polymorphism associated with a cluster of NBS-LRR disease-resistance genes (23). This supports the hypothesis that DNA polymorphisms discourage extra COs in the mutants. In some crops, such as wheat (24), barley (25), and tomato (26), large portions of chromosomes surrounding centromeres receive virtually no COs, albeit containing genes that thus are out of reach for breeding. It would be of particular interest to test the effect of recq4 figl1 in such crops to see how the CO increase seen in the short pericentromeres of Arabidopsis translates to their large pericentromeres.
We next asked whether the increase in recombination that we observed in F2s affects male and female meiosis equally. We did so by backcrossing the F1 hybrids (wild type, recq4, or recq4 figl1) by Col-0, as either male or female, and analyzing SNP segregation in the progeny (Fig. 6). In wild type, the genetic map was ∼70% larger in males than in females (495 ± 38 cM vs. 295 ± 28 cM; P < 10−4), with the difference particularly marked in the vicinity of telomeres, where male recombination is at its highest and female recombination is at its lowest (Fig. 6 B and C), in accordance with previously reported data (27). In sharp contrast, in recq4 and recq4 figl1, the female maps were larger than the male maps (P < 10−4; Fig. 6A), and the CO distributions were similar in the two sexes (Fig. 6 B and C). The total female map was increased from 295 ± 28 cM in wild type to 3,176 ± 190 cM in recq4 figl1, a >10-fold increase. The most spectacular increase was observed in the vicinity of telomeres, where the average recombination rate was 0.8 cM/Mb in female wild type and 47 cM/Mb in female recq4 figl1. In wild type, the vast majority of COs were of class I (ZMM-dependent and subject to CO interference, which limits the occurrence of two adjacent COs) while recq4 and figl1 mutations, which have been isolated as suppressors of zmm mutants, showed an increase in class II COs exclusively (2, 10, 12). Accordingly, CO interference, as measured genetically with tetrad analysis (Dataset S2) or genome-wide data (Fig. 6D), was present in the wild type but not detectable in the mutants. Thus, it appears that regulation of class I COs shapes their distribution along chromosomes and differentiates male and female meiosis, but this regulation does not apply to the class II COs up-regulated in the recq4 figl1 double mutant.
Fig. 6.
Male and female genome-wide recombination. (A) Total male (blue) and female (pink) genetic maps in the wild type, recq4, and recq4 figl1. (B) Male and female recombination in the different genotypes (cM/Mb, Haldane equation) plotted along each of the five chromosomes (Mb). Errors bars represent 95% confidence intervals. Vertical dotted lines mark the position of the centromeres. (C) The same data as in B, merged for all chromosome arms (short arms of chromosome 2 and 4 excluded), plotted along the relative distance from the centromere, and smoothed. Raw genotyping data are provided in Dataset S3, and the complete dataset is shown in Dataset S4. (D) Coefficient of coincidence (CoC) curves. The CoC is the ratio of the frequency of observed double COs to the frequency of expected double COs (the product of the CO frequencies in each of the two intervals). A CoC curve (33) plots this ratio for each pair of intervals, as a function of interinterval distance. In the presence of CO interference, CoC is around 0 for small interinterval distances, and increases to 1 with fluctuations around that value for larger interinterval distances, as seen here for wild type. A flat line at 1, as observed for the mutants, suggests an absence of interference.
Discussion
We have shown that a high CO frequency is obtained when combining recq4 and figl1 mutations, resulting in a 7.8-fold increase in total map size with only a limited effect on plant fertility and no noticeable growth or developmental defects, at least in the first two generations. This opens the possibility of manipulating recombination in plant breeding programs to increase the shuffling of genetic information, break undesirable linkages, combine desirable traits, or increase the power of genetic mapping in prebreeding research. It is intriguing that, although it is possible (as observed in a few exceptional eukaryotes and here in Arabidopsis mutants), very few eukaryotes have high CO levels per chromosome. One possibility is that a high level of COs is associated with segregation problems at meiosis and thus with fertility defects. Our data do not strongly support this possibility, however. Alternatively, it is possible that selection for a low level of recombination limits COs in eukaryotes, which could be optimal for adaptation in most contexts. An attractive idea is that in a stable environment, high recombination levels would break favorable allelic combinations that have been selected in previous generations (28). Thus, the ZMM pathway, which accounts for most COs in plants, mammals, and fungi, may have arisen in early eukaryotes to address the opposing constraints of ensuring at least one CO per chromosome while being able to adjust their numbers to low levels.
Methods
The following mutations were used in this study: Col, fancm-1 (8), figl1-1 (12), recq4a-4 (N419423) (29), and recq4b-2 (N511130) (29); Ler, fancm-10 (12), figl1-12 (12), and recq4a-W387X (10). The tetrad analysis line was I2ab (FTL1506/FTL1524/FTL965/qrt1-2) (14). Hybrid lines were obtained through the crossing of Col plants bearing the fancm, recq4a, recq4b, figl1, and qrt mutations and the FTL transgenes I2ab to Ler plants bearing the fancm, recq4a, figl1, and qrt mutations. F1 plants were grown in growth chambers (16 h/day at 21 °C, 8 h/night at 18 °C, 65% humidity) and genotyped twice for each mutation (Dataset S5). F1 sibling plants of the desired genotypes were used for tetrad analysis and fertility measures, selfed to produce the F2 population and crossed as male or female to Col-0 to produce BC1 populations. Tetrad analysis (Fig. 2), including data collection, measures of recombination (Perkins equation) and interference (interference ratio), and statistical tests, were performed as described by Girard et al. (12). F2 and BC1 populations were grown for 3 wk, and 100–150 mg of leaf material was collected from rosettes. DNA extraction and genotyping for Col/Ler polymorphisms was performed using the KASPAR technology at Plateforme Gentyane, INRA Clermont-Ferrand, France. The set of 96 KASPAR markers (Dataset S4) that are uniformly distributed on the physical map (∼every 1.3 Mb) has been described by Girard et al. (12). Genotyping data were analyzed with Fluidigm software (www.fluidigm.com) with manual corrections. The raw genotyping dataset is shown in Dataset S3. Recombination data were analyzed with MapDisto 1.7.7.0.1.1 (30). Genotyping errors were filtered using the iterative error removal function (iterations = 6, start threshold = 0.001, increase = 0.001).
Recombination (cM ± SEM) was calculated using classical fraction estimation and the Haldane mapping function. The Haldane function is preferred over the Kosambi function because the latter incorporates crossover interference, the effect of which is absent in the mutants analyzed in this study (Fig. 6D and Dataset S2), and thus likely would have underestimated the genetic distances. The obtained recombination frequencies per interval and corresponding genomic data are shown in Dataset S4. Crossover interference patterns (Fig. 6D) were analyzed using MADpatterns (31). Graphical representations were prepared with GraphPad Prism 6. The tests shown in Fig. 3A are one-way ANOVA with Dunnett’s correction on the observed number of COs (genotype transitions) per F2 plant in the genotyping data after error filtering. Linear regression analyses were performed with GraphPad Prism 6, except for the multilinear regression, which was done with R version 3.4.1. Male meiotic chromosome spreads were performed as described previously (32).
Supplementary Material
Acknowledgments
We thank Ian Henderson for discussions and sharing data before publication and Gregory Copenhaver for providing the FTL lines. We are grateful to Christine Mézard, Mathilde Grelon, and Eric Jenczewski for fruitful discussions and a critical reading of the manuscript. This work was funded by the European Research Council (Grant ERC 2011 StG 281659 MeioSight) and the Fondation Simone et Cino Del Duca/Institut de France. The Institut Jean-Pierre Bourgin benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). A.H.L. was supported by the International Outgoing Fellowships (Grant PIOF-GA-2013-628128 POLYMEIO).
Footnotes
Conflict of interest statement: Patents have been deposited by INRA on the use of RECQ4, FIGL1, and FANCM to manipulate meiotic recombination (EP3149027, EP3016506, and EP2755995).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713078114/-/DCSupplemental.
References
- 1.Hunter N. Meiotic recombination: The essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:1–35. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- 3.Berchowitz LE, Copenhaver GP. Genetic interference: Don’t stand so close to me. Curr Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritz KR, Noor MAF, Singh ND. Variation in recombination rate: Adaptive or not? Trends Genet. 2017;33:364–374. doi: 10.1016/j.tig.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Nambiar M, Smith GR. Repression of harmful meiotic recombination in centromeric regions. Semin Cell Dev Biol. 2016;54:188–197. doi: 10.1016/j.semcdb.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crismani W, Girard C, Mercier R. Tinkering with meiosis. J Exp Bot. 2013;64:55–65. doi: 10.1093/jxb/ers314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijnker E, de Jong H. Managing meiotic recombination in plant breeding. Trends Plant Sci. 2008;13:640–646. doi: 10.1016/j.tplants.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Crismani W, et al. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 9.Girard C, et al. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014;42:9087–9095. doi: 10.1093/nar/gku614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Séguéla-Arnaud M, et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA. 2015;112:4713–4718. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Séguéla-Arnaud M, et al. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017;45:1860–1871. doi: 10.1093/nar/gkw1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard C, et al. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 2015;11:e1005369, and erratum (2015) 11:e1005448. doi: 10.1371/journal.pgen.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziolkowski PA, et al. Juxtaposition of heterozygosity and homozygosity during meiosis causes reciprocal crossover remodeling via interference. Elife. 2015;4:1–29. doi: 10.7554/eLife.03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: A visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc. 2008;3:41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 15.Ziolkowski PA, et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 2017;31:306–317. doi: 10.1101/gad.295501.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Wang C, Liu Q, Liu W, Fu Y. Increasing the genetic recombination frequency by partial loss of function of the synaptonemal complex in rice. Mol Plant. 2015;8:1295–1298. doi: 10.1016/j.molp.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Youds JL, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zapata L, et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc Natl Acad Sci USA. 2016;113:E4052–E4060. doi: 10.1073/pnas.1607532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 22.de Massy B. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 23.Choi K, et al. Recombination rate heterogeneity within Arabidopsis disease resistance genes. PLoS Genet. 2016;12:e1006179. doi: 10.1371/journal.pgen.1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choulet F, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345:1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 25.Mascher M, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 26.Sim S-C, et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One. 2012;7:e40563. doi: 10.1371/journal.pone.0040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7:e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto SP. The evolutionary enigma of sex. Am Nat. 2009;174(Suppl 1):S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- 29.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorieux M. MapDisto: Fast and efficient computation of genetic linkage maps. Mol Breed. 2012;30:1231–1235. [Google Scholar]
- 31.White MA, Wang S, Zhang L, Kleckner N. Quantitative modeling and automated analysis of meiotic recombination. Methods Mol Biol. 2017;1471:305–323. doi: 10.1007/978-1-4939-6340-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross KJ, Fransz P, Jones GH. A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 1996;4:507–516. doi: 10.1007/BF02261778. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–314. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.