In contrast to other biopolymers such as RNA, most foldable protein sequences have a single, unique native structure, which is the most stable conformation. Understanding this remarkable property has been the long-held goal of protein folding research (1, 2). Once folded, proteins will undergo transitions between different conformational states within the native free-energy basin, often associated with their function; however, in most cases, these can be thought of as only local changes or deformations of structure that do not change the overall fold. On the other hand, a small number of proteins have been identified in which a more profound rearrangement of the fold can occur, with the most extreme examples being protein sequences that can switch to a completely different folded state as a function of environmental conditions or ligand binding (3). An example in which a subtle change of fold is used for functional regulation is found in the serine protease inhibitor family known as serpins. In this case, it appears that the initially formed “native” structure is not the thermodynamically most stable fold. In PNAS, Rao and Gosavi (4) investigate the origin of this kinetically controlled folding for a prototypical serpin, α1-antitrypsin.
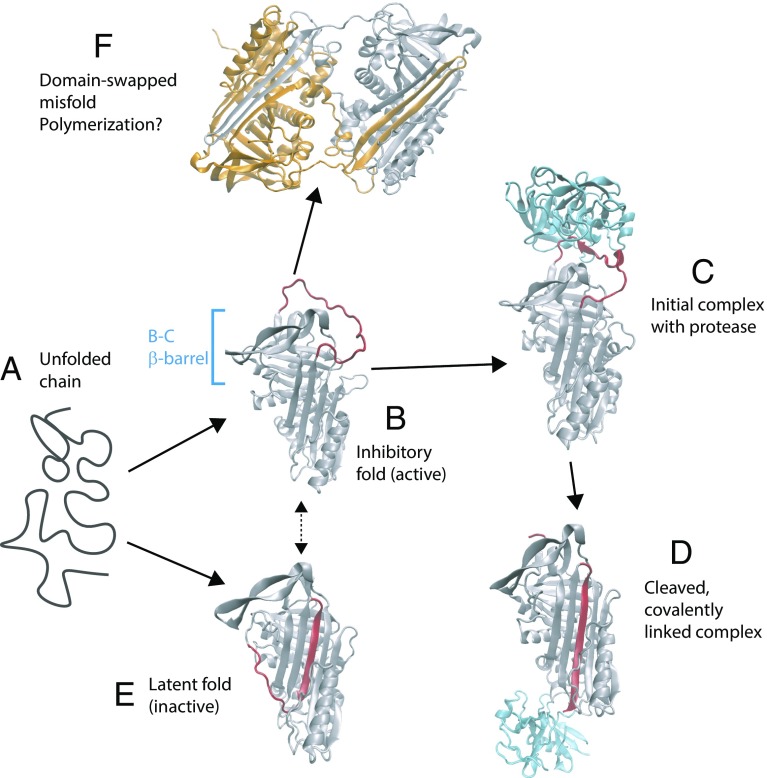
Serpins are an abundant type of protease inhibitor, most commonly inhibiting chymotrypsin-like serine proteases (5) by a suicide mechanism: The protein initially folds to an active, inhibitory fold (Fig. 1, Top Left) in which the exposed reactive center loop (RCL) serves as a recognition motif for proteases. Binding of the protease results in cleavage of the RCL loop and covalent linkage of the serpin to the protease. At the same time, the remainder of the RCL loop inserts into a β-sheet in the serpin structure. Interestingly, a similar uncleaved “latent” fold with an inserted RCL sequence also exists (Fig. 1, Bottom Left) and, moreover, is more thermodynamically stable than the inhibitory fold. However, the inhibitory structure is formed first, presumably because it is under kinetic control. Some serpins, such as plasminogen activator inhibitor-1, can also spontaneously convert from the inhibitory form to the latent form (dashed arrow in Fig. 1), providing yet another mode of regulation. The fascinating complexity of the serpin inhibition mechanism has been illuminated by the success of experimental structural biology in identifying many of the stable states populated by these proteins (including the examples shown in Fig. 1 and more) (5). In addition, equilibrium and kinetic folding studies have indicated the formation of a stable folding intermediate on the folding pathways of serpins, including α1-antitrypsin (6). Nonetheless, ensemble experiments can only probe stable or metastable states, and not the detailed mechanisms for their interconversion. Filling in these gaps is the natural domain of theory and simulation, although the problem is still challenging owing to the time scales involved. An excellent example is the above-mentioned spontaneous conversion of plasminogen activator inhibitor-1 from the inhibitory form to the latent form, a process that occurs on a time scale of hours. By using a recently developed path-sampling algorithm, the mechanism of this transition could be studied in recent all-atom simulations, with results very consistent with experimental changes of rate obtained for mutants (7).
Fig. 1.
Stable states populated by the serpin α1-antitrypsin. The unfolded protein (a) initially folds to an active, inhibitory conformation (b) in which the RCL (red) is exposed [Protein Data Bank (PDB) ID code 1QLP (19)]. (c) Protease (in this case, trypsin, shown in blue) binds to the RCL [PDB ID code 1OPH (20)]. (d) Loop is cleaved by the protease, resulting in the enzyme being covalently linked to the serpin and insertion of the RCL sequence into a β-sheet [PDB ID code 1EZX (21)]. (e) Serpin can also adopt a latent fold, similar to the cleaved structure, which is more thermodynamically stable than the initially formed inhibitory fold [PDB ID code 1DVN (22)]. (f) Domain-swapped dimer structure [PDB ID code 2ZNH (16)] hints at a possible mechanism of polymerization.
Many mechanistic questions about serpin folding and conformational transitions remain. Foremost among these is why the active, inhibitory form wins the initial race to the native (inhibitory) state over the thermodynamically more stable latent form. Rao and Gosavi (4) now address this question using a structure-based, coarse-grained model in which the only residue pairs with attractive interactions are those formed in a given target structure. Such models are motivated by the energy landscape theory of protein folding, which posits that the folding free-energy landscape is funneled toward a single native state (8–11). While such a model may at first seem inconsistent with the folding scenario of serpins, it allows direct folding to the alternative latent structure to be studied, without the competing inhibitory fold. By computing separate folding free-energy landscapes for the inhibitory and latent structures, the authors found a free-energy barrier for folding to the latent form that was significantly higher (by ∼4 kBT). An important consideration in comparing free-energy barriers is relative stabilities, considering that the latent form is more stable and stabilizing a protein tends to lower its folding barrier. In the context of the simulation model, it could be shown that the latent form would have to be ∼13 kBT more stable than the inhibitory form for the folding barriers to be comparable. The lower barrier for folding to the inhibitory form appears to be the consequence of a metastable on-pathway intermediate in which a domain consisting of the B-C β-barrel is folded, while the remainder of the protein is not. This intermediate is in accord with most experimental data reflecting the folding equilibrium and kinetics for this protein and other serpins (5). The latent form folds instead via a single transition state, with some similarities to the above intermediate, but in which key β-strand contacts with the RCL are already formed very early. The authors have proposed that this mechanism could be tested by engineering disulfide bonds that lock in these contacts, and thus favor formation of the latent state.
The simulations described above have addressed the left-hand side of the diagram in Fig. 1, namely, folding of and spontaneous conversion between the inhibitory and latent forms. From a folding perspective, this is elegant because all these steps involve the energy landscape of the same unbroken protein chain. However, the functional energy landscape of a serpin includes additional steps: interaction with a protease, cleavage of the RCL, covalent linkage of the serpin to the protease, and conversion of the cleaved protease to the inactive form, with a structure similar to the latent form. An interesting future mechanistic question for simulation is how the cleavage of the RCL is coupled to the structural conversion of the serpin fold to the inactive state. While such a problem would be difficult to study with standard topology-based models, so-called dual-topology models can accommodate alternative stable states (12–14), allowing the coupling between bond cleavage and conformational transitions to be studied directly in a simulation model. Another interesting aspect of serpins are serinopathy diseases, characterized by formation of pathological polymers of these proteins (ref. 15, also hinted at in ref. 4). The identification of a domain-swapped serpin dimer structure suggests a potential mechanism for head-to-tail polymerization (16) (Fig. 1F). In this structure, each protein swaps one β-hairpin (involving the RCL) with the other; were the exchange not mutual, such interactions could be used to form a chain of serpin molecules. If polymerization indeed occurs by domain swapping, it should also be possible to study the mechanism using simulations with structure-based models (17, 18). There is thus rich potential for theory and simulation to provide further insights into the mechanisms of serpin folding, inhibitory function, misfolding, and polymerization.
Acknowledgments
R.B.B. is supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
Footnotes
The author declares no conflict of interest.
See companion article on page 1998 in issue 9 of volume 115.
References
- 1.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 2.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 3.Bryan PN, Orban J. Proteins that switch folds. Curr Opin Struct Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giri Rao VVH, Gosavi S. On the folding of a structurally complex protein to its metastable active state. Proc Natl Acad Sci USA. 2018;115:1998–2003. doi: 10.1073/pnas.1708173115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whisstock JC, Bottomley SP. Molecular gymnastics: Serpin structure, folding and misfolding. Curr Opin Struct Biol. 2006;16:761–768. doi: 10.1016/j.sbi.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Tsutsui Y, Dela Cruz R, Wintrode PL. Folding mechanism of the metastable serpin α1-antitrypsin. Proc Natl Acad Sci USA. 2012;109:4467–4472. doi: 10.1073/pnas.1109125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cazzolli G, et al. Serpin latency transition at atomic resolution. Proc Natl Acad Sci USA. 2014;111:15414–15419. doi: 10.1073/pnas.1407528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryngelson JD, Wolynes PG. Intermediates and barrier crossing in a random energy model (with applications to protein folding) J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 9.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zwanzig R. Simple model of protein folding kinetics. Proc Natl Acad Sci USA. 1995;92:9801–9804. doi: 10.1073/pnas.92.21.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bicout DJ, Szabo A. Entropic barriers, transition states, funnels, and exponential protein folding kinetics: A simple model. Protein Sci. 2000;9:452–465. doi: 10.1110/ps.9.3.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Best RB, Chen Y-G, Hummer G. Slow protein conformational dynamics from multiple experimental structures: The helix/sheet transition of arc repressor. Structure. 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Maragakis P, Karplus M. Large amplitude conformational change in proteins explored with a plastic network model: Adenylate kinase. J Mol Biol. 2005;352:807–822. doi: 10.1016/j.jmb.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Miyashita O, Wolynes PG, Onuchic JN. Simple energy landscape model for the kinetics of functional transitions in proteins. J Phys Chem B. 2005;109:1959–1969. doi: 10.1021/jp046736q. [DOI] [PubMed] [Google Scholar]
- 15.Gettins PGW, Olson ST. Inhibitory serpins. New insights into their folding, polymerization, regulation and clearance. Biochem J. 2016;473:2273–2293. doi: 10.1042/BCJ20160014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamasaki M, Li W, Johnson DJD, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 17.Yang S, et al. Domain swapping is a consequence of minimal frustration. Proc Natl Acad Sci USA. 2004;101:13786–13791. doi: 10.1073/pnas.0403724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borgia MB, et al. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott PR, Pei XY, Dafforn TR, Lomas DA. Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000;9:1274–1281. doi: 10.1110/ps.9.7.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dementiev A, Simonovic M, Volz K, Gettins PG. Canonical inhibitor-like interactions explain reactivity of alpha1-proteinase inhibitor Pittsburgh and antithrombin with proteinases. J Biol Chem. 2003;278:37881–37887. doi: 10.1074/jbc.M305195200. [DOI] [PubMed] [Google Scholar]
- 21.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 22.Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: A possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]