Significance
The Hsp90 molecular chaperone remodels and activates a large variety of client proteins. Its activity requires collaboration with the more than 25 Hsp90 cochaperone proteins, including the Hsp70 chaperone. In higher eukaryotes, Hsp90 collaborates with Hsp70 through a bridging protein, the Hop cochaperone. We show that yeast Hsp90 (Hsp82) and Hsp70 (Ssa1) directly interact in vitro in the absence of yeast Hop (Sti1) and identify a region in the middle domain of yeast Hsp90 that is important for interaction with Hsp70. This region of Hsp90 is also important for interactions with several cochaperones and client proteins, suggesting that collaboration between Hsp70 and Hsp90 in protein remodeling may be modulated through competition between Hsp70 and Hsp90 cochaperones for the interaction surface.
Keywords: Ssa1, Hsp82, molecular chaperones, Sti1, Ydj1
Abstract
Heat shock protein 90 (Hsp90) is a highly conserved ATP-dependent molecular chaperone that is essential in eukaryotes. It is required for the activation and stabilization of more than 200 client proteins, including many kinases and steroid hormone receptors involved in cell-signaling pathways. Hsp90 chaperone activity requires collaboration with a subset of the many Hsp90 cochaperones, including the Hsp70 chaperone. In higher eukaryotes, the collaboration between Hsp90 and Hsp70 is indirect and involves Hop, a cochaperone that interacts with both Hsp90 and Hsp70. Here we show that yeast Hsp90 (Hsp82) and yeast Hsp70 (Ssa1), directly interact in vitro in the absence of the yeast Hop homolog (Sti1), and identify a region in the middle domain of yeast Hsp90 that is required for the interaction. In vivo results using Hsp90 substitution mutants showed that several residues in this region were important or essential for growth at high temperature. Moreover, mutants in this region were defective in interaction with Hsp70 in cell lysates. In vitro, the purified Hsp82 mutant proteins were defective in direct physical interaction with Ssa1 and in protein remodeling in collaboration with Ssa1 and cochaperones. This region of Hsp90 is also important for interactions with several Hsp90 cochaperones and client proteins, suggesting that collaboration between Hsp70 and Hsp90 in protein remodeling may be modulated through competition between Hsp70 and Hsp90 cochaperones for the interaction surface.
Heat shock protein 90, Hsp90, is a highly conserved and abundant ATP-dependent molecular chaperone (1–3). It is found in most organisms and is essential in eukaryotes. Hsp90 functions in a wide variety of cellular processes by facilitating protein remodeling and activation of more than 200 client proteins (1–4). In yeast, as in many eukaryotes, there are two cytosolic Hsp90 isoforms, Hsp82 and Hsc82. Hsp82 and Hsc82 are 97% identical in sequence (Fig. S1), with differences in 16 amino acids. In addition, Hsp82 has a five amino acid insertion and a single amino acid deletion, compared with Hsc82. Cells expressing Hsc82 or Hsp82 as the sole source of Hsp90 are viable under both optimal and stress conditions (5). Hsp82 is up-regulated ∼20-fold under heat stress and other stress conditions, while Hsc82 is constitutively expressed and makes up 1–2% of the total cellular protein (5).
Protein remodeling and activation by Hsp90 requires the participation of various Hsp90 cochaperones and the Hsp70 chaperone (3, 4, 6). Various clients require different subsets of cochaperones for remodeling, and the specific functions of many of the cochaperones are not fully understood (3, 7, 8). Structurally, Hsp90 is a homodimer with each protomer consisting of three domains (1, 3). The N-terminal domain (N-domain) is involved in ATP binding and hydrolysis (9). A flexible linker of unknown function and variable length, referred to as the “charged linker,” connects the N-domain to the middle domain (M-domain) (10, 11). The M-domain plays a role in client and cochaperone binding (3, 12, 13). The C-terminal domain (C-domain) is required for dimerization and also participates in client binding (1–3, 12). Hsp90 undergoes conformational changes in response to ATP binding, hydrolysis, and ADP release (1, 3, 6, 14–16). In the absence of ATP, the Hsp90 dimer acquires an open, V-shaped structure such that the protomers interact via the C-terminal dimerization domain (16). When ATP is bound, the protein takes on a closed conformation with the two N-domains of the dimer interacting and a portion of the N-domain, the “lid,” closing over the nucleotide in each protomer (16, 17). Additional conformational changes occur upon ATP hydrolysis, resulting in a distinct compact ADP-bound conformation (14–16). After ADP release, Hsp90 reverts to the open conformation (1, 3). However, client protein binding and cochaperone interactions bias the conformational dynamics of Hsp90 and affect the residence time in the various conformations (1, 3, 4, 18).
In the absence of cochaperones and Hsp70, Hsp90 functions in vitro in an ATP-independent manner as a holdase to prevent aggregation of some client proteins (19–21). However, both in vivo and in vitro protein activation and reactivation by Hsp90 requires ATP hydrolysis by Hsp90. In addition, protein remodeling of some clients requires collaboration with the Hsp70 chaperone, Hsp90 cochaperones, and Hsp70 cochaperones (3, 7, 22–24). In higher eukaryotes, the collaboration between Hsp90 and Hsp70 requires the participation of a bridging protein, Hop (Hsp70–Hsp90 organizing protein) (1, 3, 4). Although yeast possess a Hop cochaperone homolog, Sti1, previous work from Lindquist and coworkers (25) and from Johnson and coworkers (26) showed that yeast Hsp90 associates with Hsp70 both in wild-type yeast cell lysates and in lysates of cells deleted for sti1. The results show that Sti1 is not required for the interaction between Hsp90 and Hsp70 and suggest that the two proteins interact directly. Unlike yeast, bacteria do not possess a Hop/Sti1 homolog, and bacterial Hsp90 and Hsp70 interact directly (27–29). In Escherichia coli a region of the M-domain of Hsp90 is required for physical and functional interaction with the nucleotide-binding domain of the E. coli Hsp70, DnaK (28, 30).
In this work, we tested if yeast Hsp90 interacts directly with yeast Hsp70 in vitro and found that the two chaperones physically interact in the absence of Hop/Sti1. Additionally, we identified a region in the M-domain of yeast Hsp90, homologous to the DnaK-interacting region of E. coli Hsp90 (Hsp90Ec) that is important for the interaction and for the functional collaboration between yeast Hsp90 and Hsp70.
Results
Yeast Hsp90 and Hsp70 Interact in Vitro.
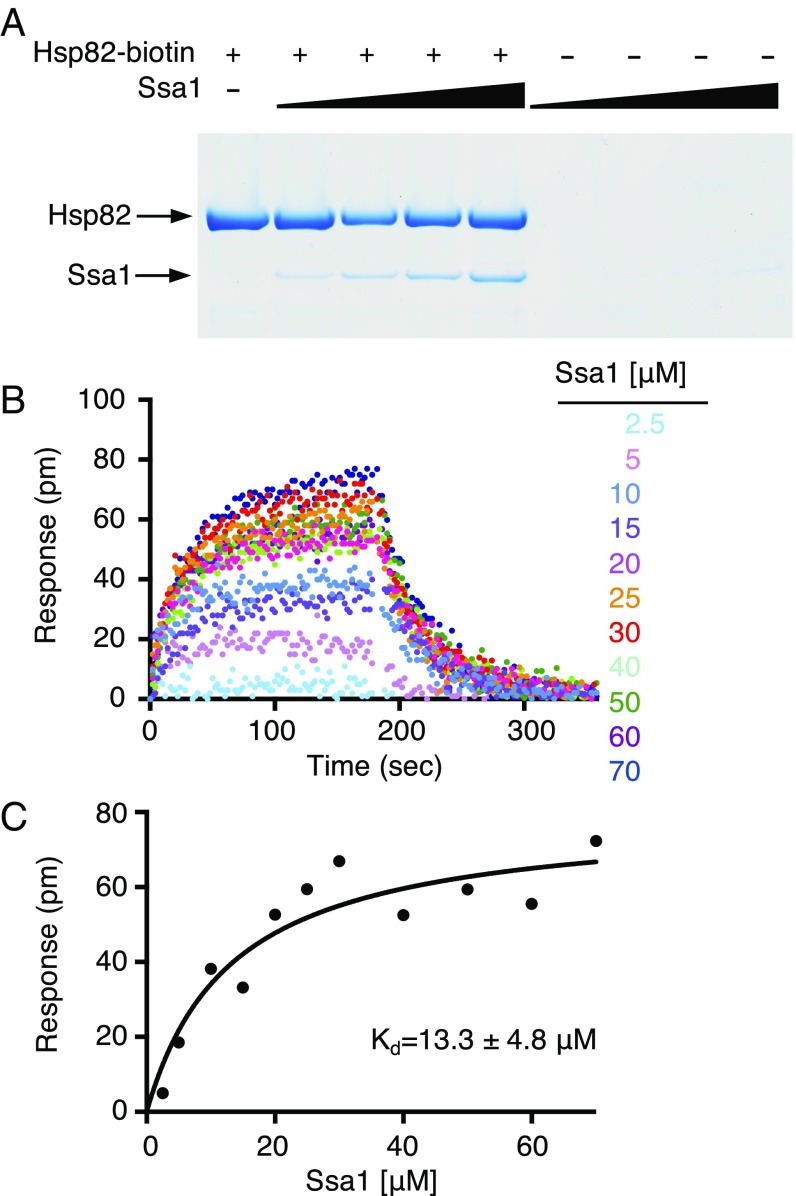
To test for a direct interaction between yeast Hsp90 and yeast Hsp70, we utilized an in vitro protein–protein interaction assay (pull-down assay). For these experiments, we used Hsp82, which is the yeast heat shock-inducible Hsp90 homolog, and Ssa1, one of the several cytoplasmic yeast Hsp70 homologs. Ssa1 was chosen since it has been previously shown to function in vitro with Hsp82 (31, 32). Biotin-labeled Hsp82 was incubated with Ssa1; protein complexes that bound NeutrAvidin agarose beads were eluted, separated by SDS/PAGE, and visualized by Coomassie blue staining (Fig. 1A). We observed Ssa1 associated with Hsp82, suggesting a direct physical interaction. As the concentration of Ssa1 was increased, more Ssa1 associated with Hsp82-biotin. In the absence of Hsp82, Ssa1 did not associate with the NeutrAvidin beads (Fig. 1A).
Fig. 1.
Direct interaction between Hsp82 and Ssa1 in vitro. (A) The interaction between Hsp82 and Ssa1 was monitored using a pull-down assay. Various concentrations of Ssa1 (1.6, 3.2, 6.3, and 12.6 μM) were incubated in the presence and absence of Hsp82-biotin (2 μM). Proteins were captured on NeutrAvidin agarose and analyzed by Coomassie blue staining following SDS/PAGE as described in Materials and Methods. A representative gel from three independent experiments is shown. (B) The interaction between Hsp82 and Ssa1 was measured by BLI. The kinetics of association and dissociation were monitored at various concentrations of Ssa1 (2.5, 5, 10, 15, 20, 25, 30, 40, 50, 60, and 70 μM) with Hsp82-biotin in the presence of ATP as described in Materials and Methods. The single exponential fit of the association at each concentration of Ssa1 was used to obtain the response plateau value. (C) The response value for each concentration of Ssa1 from B is plotted as a function of the concentration and fit using a one-site binding model in GraphPad Prism. The steady-state Kd and Bmax were determined from the fit and are 13.3 ± 4.8 μM and 79 ± 9 pm, respectively.
To obtain an apparent binding constant for the Hsp82 and Ssa1 interaction, we used bio-layer interferometry (BLI). Biotin-labeled Hsp82 was immobilized on a streptavidin-coated biosensor, and Ssa1 binding was monitored at various concentrations of Ssa1 (2.5–70 μM) in the presence of ATP as described in Materials and Methods (Fig. 1B). At high Ssa1 concentrations the binding curves were complex. Therefore, we used a steady-state analysis method and calculated a Kd of 13 ± 5 μM (Fig. 1C). The observed Kd is consistent with the apparent Kd of ∼13 μM previously determined for the Hsp90Ec–DnaK interaction (30). These results showing that Hsp82 and Ssa1 form a complex independent of other cochaperones are also consistent with weak interactions observed between Synechococcus elongates Hsp90 and Hsp70 (29) and mitochondrial Hsp90 (TRAP) and Hsp70 (Mortalin) (33).
Predicted Yeast Hsp90–Hsp70 Interaction Region.
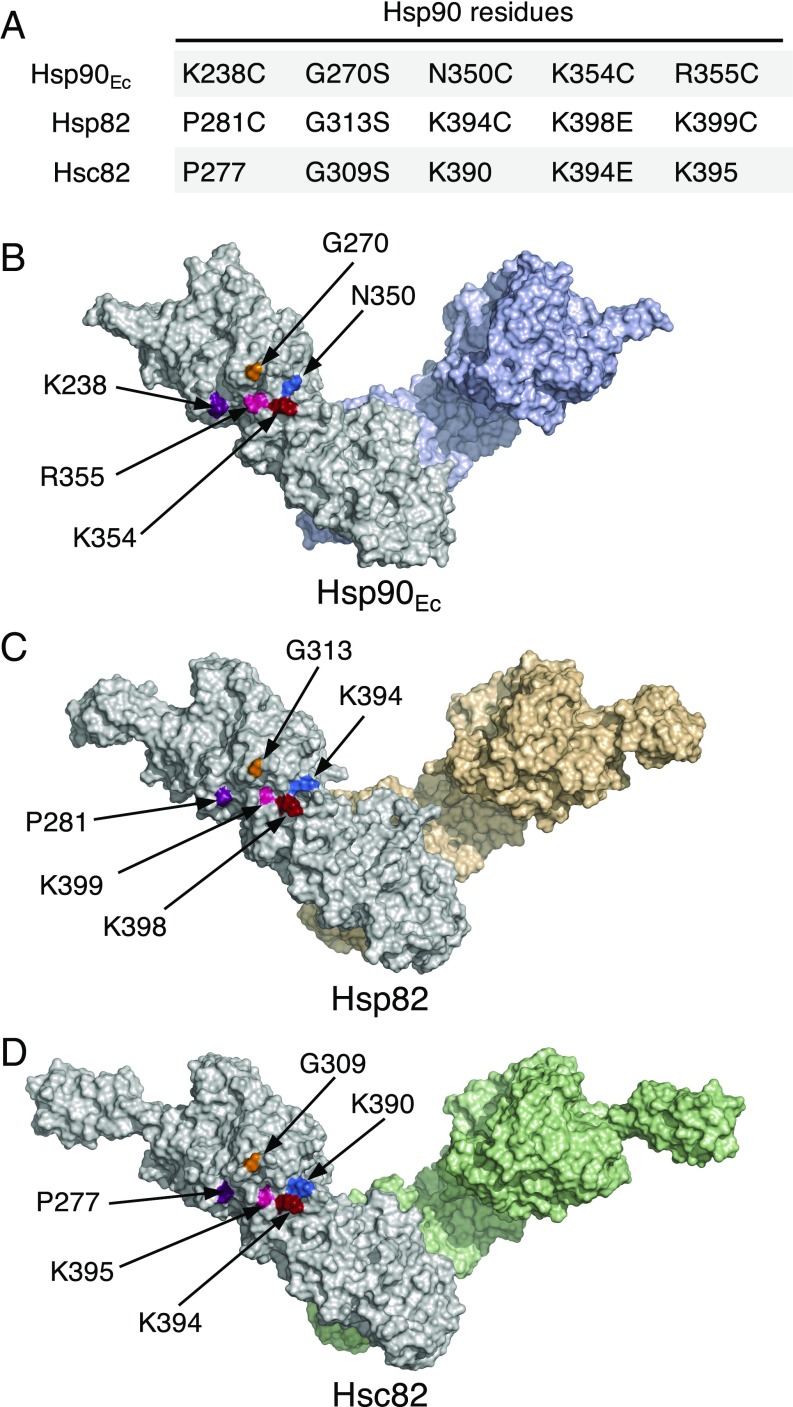
We next wanted to determine the region of yeast Hsp90 important for the interaction with Hsp70. We had previously identified a region in the M-domain of Hsp90Ec that was essential for interaction with DnaK (28). A bacterial two-hybrid assay had been used as a screen for Hsp90Ec mutants defective in interaction with DnaK in vivo. The mutant Hsp90Ec proteins were then tested in vitro for the ability to interact with DnaK and were shown to be defective in direct interaction with DnaK. The four most defective Hsp90Ec mutants contained substitutions in residues K238, G270, K354, and R355 (Fig. 2A and shown on a model of Hsp90Ec in Fig. 2B) (28). We postulated that the homologous region of yeast Hsp90 could also be important for interaction with yeast Hsp70 and therefore focused our attention on the residues in yeast Hsp90 that aligned with these Hsp90Ec residues, Hsp82-P281, -G313, -K398, and -K399 (Fig. 2A). In addition, Hsp82-K394 was included in this study due to its surface-exposed position on the model of Hsp82 near K398 and K399 (Fig. 2 A and C). These five residues are in the M-domain of Hsp82 and are on the surface in both the open model of the Hsp82 dimer and the closed structure of the dimer (Fig. 2C) (16, 17, 28). Three of these residues are identical or conserved in Hsp90Ec and Hsp82; the other two are nonconserved (Fig. 2A and Fig. S1).
Fig. 2.
Yeast Hsp90 residues potentially involved in Ssa1 interactions. (A) Chart showing residues in Saccharomyces cerevisiae Hsp82 and Hsc82 that align with five residues in E. coli Hsp90 (Hsp90Ec) that were previously shown to be important for the Hsp90Ec–DnaK interaction (28). The substitution mutants previously made in Hsp90Ec and those made in Hsp82 and Hsc82 in this study are also shown. (B–D) Location of the five residues of interest in A shown, in color, on surface-rendered models of dimeric Hsp90Ec (B), Hsp82 (C), and Hsc82 (D). In B, a surface-rendered model of the crystal structure of the Hsp90Ec dimer in the apo form (PDB ID code 2IOQ) (16) with the C-terminal domains aligned to the crystal structure of the isolated C-domain (PDB ID code 1SF8) (64) generated using PyMOL (https://pymol.org/2/). In C and D, models were constructed as described in Materials and Methods. Several C-terminal amino acid residues of Hsp82 (676–709) and Hsc82 (670–705) were excluded due to the uncertainty in the predicted structure (see Materials and Methods). In B–D, one protomer of each dimer is gray, and the other is colored.
hsp82 and hsc82 Mutants in the Potential Ssa1 Interaction Region Exhibit Physiological Defects.
Previously, mutations in yeast Hsp82-G313 (Fig. 2 A and C), one of the five Hsp82 residues we postulated to be important for the interaction between Hsp82 and Hsp70, had been identified in two independent screens for hsp82 mutants that were temperature sensitive for growth when expressed as the sole source of Hsp90 (34, 35). Moreover, a mutation of the Hsc82 residue homologous to Hsp82-G313S, Hsc82-G309S (Fig. 2 A and D), caused temperature sensitivity when expressed as the only Hsp90 source (26). Since Hsp82 and its isoform Hsc82 are very similar in sequence (Fig. S1) and function, these findings support the hypothesis that this region of Hsp82 is important for Hsp90 function.
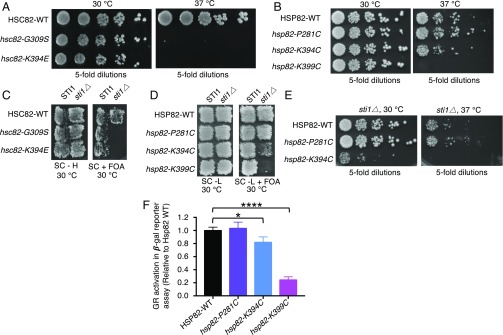
Further support for the importance of this region of Hsp90 was gained when a screen for hsc82 temperature-sensitive mutants identified a substitution mutant in another residue predicted above to be involved in the Hsp90–Hsp70 interaction. An hsc82 library that was generated using error-prone PCR mutagenesis was screened for temperature-sensitive mutants; one of the 10 hsc82 mutants we obtained was in K394, which is homologous to Hsp82 residue K398 (Fig. 2 A and D). Cells expressing this mutant protein, Hsc82-K394E, as the sole source of Hsp90, like those expressing Hsc82-G309S (Fig. 3A) (26), grew similarly to the wild type at 30 °C but were unable to grow at 37 °C (Fig. 3A and Table S1). In control experiments, Hsc82-G309S and Hsc82-K394E were expressed at similar levels as the wild type at 30 and 37 °C (Fig. S2A).
Fig. 3.
In vivo phenotypes of hsc82 and hsp82 mutant alleles. (A) Plasmids expressing wild-type or mutant Hsc82 were expressed as the sole Hsp90 in JJ816 cells (hsc82hsp82). After overnight growth, colonies were serially diluted fivefold and grown for 2 d at the indicated temperature. (B) Plasmids expressing wild-type or mutant Hsp82 were expressed as the sole Hsp90 in strain MR318 (hsc82hsp82). After overnight growth, colonies were serially diluted fivefold and grown for 2 d at the indicated temperature. (C) Plasmids expressing wild-type HSC82, hsc82-G309S, or hsc82-K394E were transformed into JJ816 cells (hsc82hsp82/YEp-HSP82) or JJ832 cells (hsc82hsp82sti1/YEp24-HSP82). Cells were replica-plated onto medium containing 5-FOA, which prevents the growth of cells carrying the URA3 plasmid, and then were grown for 2 d at 30 °C. Growth on 5-FOA indicates the Hsc82 expressed from the plasmid can support growth as the only Hsp90 in the cell. (D) Strain MR318 (STI1 hsc82hsp82) or 1670 (sti1∆ hsc82hsp82) carrying a URA3 plasmid encoding wild-type Hsp82 and the indicated Hsp82 alleles on a LEU2 plasmid were grown on medium allowing loss of the URA3 plasmid and then were replica-plated onto medium containing 5-FOA, as in C. (E) Strain 1670 (sti1∆ hsc82hsp82) expressing wild-type or mutant Hsp82 was grown in liquid rich medium, serially diluted fivefold, and incubated for 2 d at 30 or 37 °C on rich medium. (F) GR maturation. Strains expressing wild-type or mutant Hsp82 were transformed with plasmids encoding GR and a downstream LacZ reporter. After growth at 30 °C in the presence of deoxycorticosteroid, β-galactosidase activity was measured. Data from three replicates are presented as mean ± SD. *P < 0.05, ****P ≤ 0.0001 (ANOVA; see Materials and Methods). In A–E, representatives of at least three independent experiments are shown.
We next constructed substitution mutations in the other yeast Hsp90 residues predicted to be important for Hsp70 interaction (Fig. 2 A and C and Fig. S1), including hsp82-P281C, hsp82-K394C (not to be confused with Hsc82-K394E), and hsp82-K399C. These substitutions were made in Hsp82 since we had used Hsp82 to establish the direct interaction between yeast Hsp90 and Hsp70 (Fig. 1). Cysteine substitutions were chosen because Hsp82 lacks cysteine residues, and it was anticipated that they would be useful in future experiments. The strains expressing the Hsp82 mutants grew similarly to the wild type at 30 °C (Fig. 3B and Table S1). However, at 37 °C, cells expressing Hsp82-K399C as the only Hsp90 source were unable to grow (Fig. 3B). Cells expressing Hsp82-K394C were able to grow at 37 °C, although they grew more slowly than the wild type, suggesting a partial defect (Fig. 3B). Cells expressing Hsp82-P281C grew similarly to the wild type at 37 °C (Fig. 3B). In control experiments, the three Hsp82 mutant strains expressed similar levels of Hsp82 as the wild-type strain at 30 and 37 °C (Fig. S2B). Together, these experiments and previous work (26, 34, 35) show that four of the five yeast Hsp90 residues that were hypothesized to be important for interaction with yeast Hsp70 are important or essential for growth at high temperature.
Previously, it had been shown that yeast sti1Δ strains expressing an Hsp90 mutant in one of our residues of interest, Hsp82-G313S or Hsc82-G309S, as the sole source of Hsp90 were unable to grow at the permissive temperature (25, 26). Therefore, we tested our mutants, one in Hsc82 and three in Hsp82, for the ability to grow in the absence of Sti1. We observed that sti1Δ cells expressing Hsc82-K394E, like cells expressing Hsc82-G309S, were unable to grow at 30 °C (Fig. 3C and Table S1) (26). Similarly, sti1Δ cells expressing Hsp82-K399C were nonviable at 30 °C (Fig. 3D). sti1Δ cells expressing Hsp82-K394C grew at 30 °C; however, the growth rate was significantly reduced compared with cells expressing wild-type Hsp82 (Fig. 3 D and E). sti1Δ cells expressing Hsp82-P281C grew similarly to cells expressing the wild type (Fig. 3 D and E). Taken together, four of the five mutants containing substitutions in Hsp90 residues predicted to be important for interaction with Ssa1 exhibited a growth defect in the absence of Sti1.
Interestingly, the three yeast mutants with substitutions in residues conserved in E. coli (Hsc82-G309S/Hsp82-G313S, Hsc82-K394E, and Hsp82-K399C) were very defective in vivo (Fig. 2A and Table S1), while the two yeast mutants in residues not conserved in E. coli were only partially defective (Hsp82-K394C) or were like the wild type (Hsp82-P281C).
Next, the ability of the Hsp82 mutants to perform specific protein-remodeling activity in vivo was assessed by monitoring mammalian glucocorticoid receptor (GR) maturation (36). Strains expressing the Hsp82 mutant proteins were transformed with a plasmid that constitutively expresses GR and carries a glucocorticoid-regulated lacZ reporter gene. Using this assay, prior work showed that cells expressing Hsp82-G313S exhibited ∼25% the level of GR activity compared with cells expressing wild-type Hsp82 (35). We observed that cells expressing Hsp82-K399C also exhibited ∼25% the level of GR activity as cells expressing wild-type Hsp82, and cells expressing Hsp82-K394C displayed ∼80% of the GR activity of the wild-type Hsp82-expressing cells (Fig. 3F and Table S1). The activity of Hsp82-P281C–expressing cells was indistinguishable from cells expressing wild-type Hsp82 in this assay (Fig. 3F). These results suggest that Hsp82 mutations in K394 and K399 cause defects in chaperone function in vivo.
Yeast Hsp90 Mutants Are Defective in Interaction with Hsp70 in Cell Lysates.
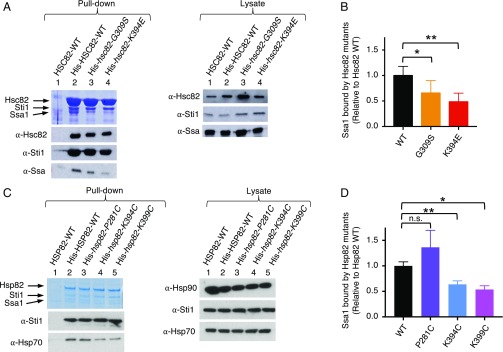
We next tested if the Hsp90 mutants were defective in their ability to interact with Ssa1 in cell lysates. One mutant, Hsc82-G309S, had been previously shown to interact less well with Ssa1 than wild-type Hsc82 by a pull-down assay using cell lysates (26). To test if our other Hsc82 and Hsp82 mutant proteins were also defective, we constructed His-tagged fusions of the mutant proteins. The wild-type or mutant proteins were expressed as the only Hsp90 source in the yeast cells, and pull-down assays were performed. Complexes containing His–Hsc82 or His–Hsp82 wild-type or mutant proteins were isolated from crude lysates using metal affinity resin and were separated by SDS/PAGE followed by Coomassie staining and immunoblot analysis. Hsc82-K394E bound ∼50% less Ssa1 than wild-type Hsc82, and Hsc82-G309S bound ∼35% less (Fig. 4 A and B and Table S1) (26). Hsp82-K399C bound ∼50% less, and Hsp82-K394C bound ∼40% less Ssa1 than wild-type Hsp82 (Fig. 4 C and D and Table S1). Hsp82-P281C interacted with Ssa1 similarly to wild-type Hsp82 (Fig. 4 C and D and Table S1). Thus the four mutants that were defective for Hsp90 function in vivo were defective in Ssa1 interaction in cell lysates.
Fig. 4.
Hsp82 and Hsc82 mutants exhibit defective Ssa1 interaction in vivo. Pull-down assays from yeast cell lysates were carried out as described in Materials and Methods using lysates from cells expressing wild-type or mutant His–Hsc82 (A and B) or wild-type or mutant His–Hsp82 (C and D). (A and C) The proteins from the pull down and the cell lysate were analyzed by SDS/PAGE and visualized by Coomassie staining and immunoblot analysis; shown are representatives of at least three independent experiments. (B and D) Quantification of Ssa1 bound to wild-type or mutant His–Hsc82 (B) or to wild-type or mutant His–Hsp82 (D). The ratio of Ssa1 bound to each of the His–Hsc82 or His–Hsp82 mutants relative to wild type is presented as the mean ± SEM. *P < 0.05, **P < 0.01, n.s., not significant (ANOVA; see Materials and Methods).
Hsp90 Mutant Proteins Are Defective in Interaction with Yeast Hsp70 in Vitro.
To determine if the Hsp90 mutant proteins that were defective in interaction with Ssa1 in vivo were defective in vitro, substitution mutations of Hsp82, including P281C, G313S, K394C, K398E, and K399C, were introduced into pET–His6–Hsp82 and were expressed in E. coli (shown in the model in Fig. 2C). Hsp82-K398 and Hsp82-G313 are homologous to Hsc82-K394 and -G309, respectively (Fig. 2A). The mutant proteins were purified and shown to have physical properties similar to those of the wild-type protein, including size-exclusion elution profiles and partial protease digestion patterns (Fig. S3 A and B). In addition, all the mutant proteins were able to hydrolyze ATP (Fig. S3C and Table S2); however, Hsp82-P281C and Hsp82-K399C hydrolyzed ATP at ∼65% and ∼80% the rate of wild-type protein, respectively. The significance of these differences is not known, since a direct relationship has not been observed between the rate of ATP hydrolysis and activities of Hsp90 (37).
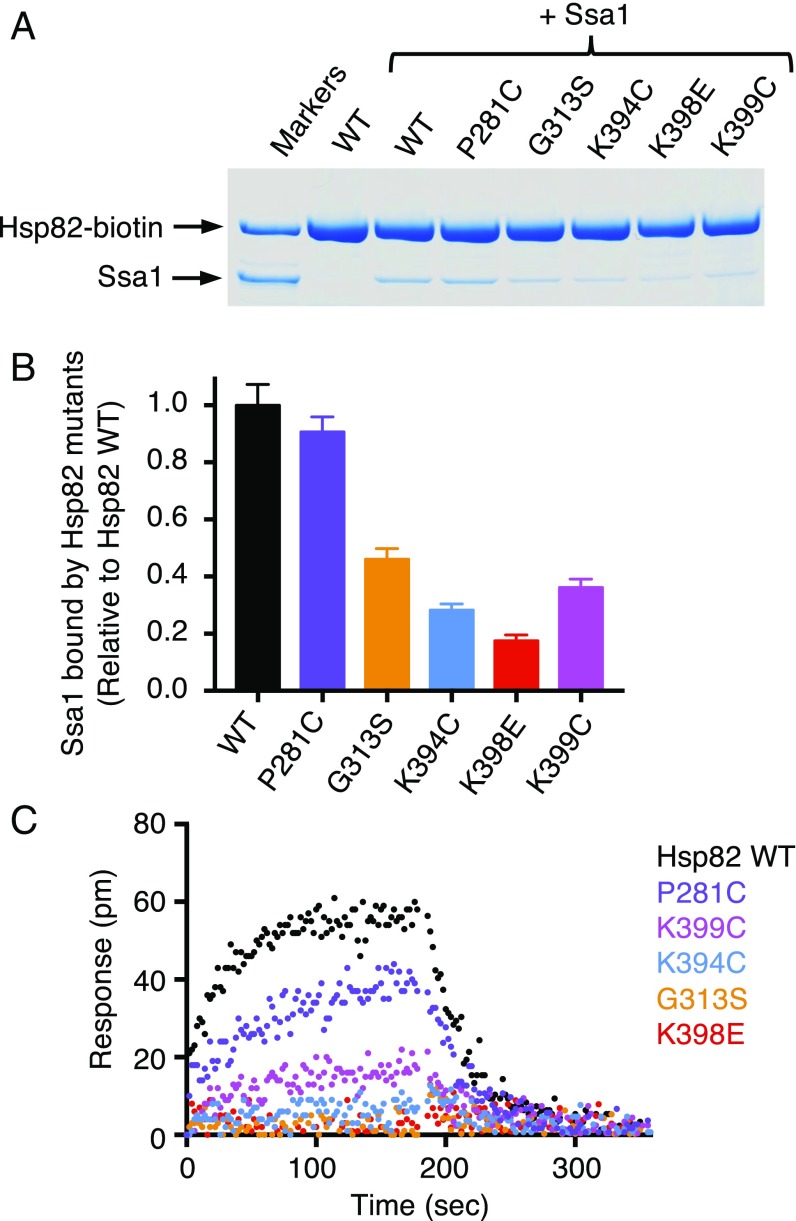
We biotinylated each of the five Hsp82 mutant proteins and monitored the ability of the mutant proteins to interact directly with Ssa1 in the pull-down assay that was used to show interaction between wild-type Hsp82 and Ssa1 in vitro (see Materials and Methods and Fig. 1A). Four of the Hsp82 mutants, G313S, K394C, K398E, and K399C, bound less Ssa1 than the wild-type protein (Fig. 5A and Table S2), retaining 20–45% the amount of Ssa1 as wild-type Hsp82 (Fig. 5B). Hsp82-P281C and wild-type Hsp82 bind similar amounts of Ssa1 (Fig. 5 A and B). Thus, the same four yeast Hsp90 mutants that were defective in Ssa1 interaction in cell lysates were defective in binary interaction with Ssa1 using purified proteins.
Fig. 5.
Hsp82 mutant proteins exhibit defective interaction with Ssa1 in vitro. (A) The interaction between biotin-labeled wild-type or mutant Hsp82 (1 μM) and Ssa1 (4 μM) was measured in a pull-down assay as described in Materials and Methods. The proteins were analyzed by SDS/PAGE and visualized by Coomassie staining; the gel shown is a representative of at least three independent experiments. (B) Quantification of Ssa1 bound to wild-type or mutant Hsp82-biotin from A. The ratio of Ssa1 bound to each of the biotin-labeled Hsp82 mutants relative to wild-type Hsp82 is presented as mean ± SEM. (C) The kinetics of association and dissociation for the interaction between 60 μM Ssa1 and biotinylated wild-type (black) or mutant (colored) Hsp82 was monitored by BLI as described in Materials and Methods. Representative curves are shown for three or more independent experiments.
We also monitored the direct interaction between Hsp82 and Ssa1 using BLI. For these experiments, the biotinylated Hsp82 mutant proteins were loaded onto biosensors to the same response level (Fig. S4 A and B), and Ssa1 binding was measured using a concentration of Ssa1 (60 μM) that was saturating for wild-type Hsp82 (Fig. 5C and Table S2). We observed that the Hsp82 mutants that were the most defective for interaction with Ssa1 in the pull-down assay, including G313S, K394C, K398E, and K399C, were the most defective in interaction with Ssa1 as monitored by BLI (Fig. 5). Hsp82-P281C appeared slightly defective compared with the wild-type protein in this assay (Fig. 5C). In control experiments, all five mutants bound Sti1 similarly to wild-type Hsp82 in this assay (Fig. S4C). Taken together, these results show that this region of the Hsp82 M-domain is important for direct interaction with Ssa1. These residues may participate in the direct interaction between the two chaperones or may be important for a conformational change in Hsp82 necessary for the interaction with Ssa1 elsewhere on Hsp82.
Hsp82 Mutants That Are Defective in Interaction with Ssa1 Are Defective in Protein Reactivation in Vitro.
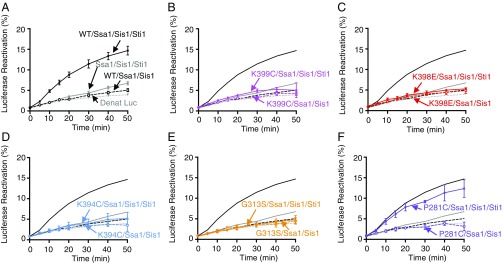
We next wanted to assess if the Hsp82 mutants collaborated functionally with Ssa1 in vitro. For these experiments a model system, reactivation of heat-inactivated luciferase, was used. The reactivation assay was optimized to show the maximal stimulation by Hsp82 in mixtures containing Ssa1, Sis1, and Sti1. Hsp82 stimulated luciferase reactivation approximately threefold over the rate of luciferase refolding by Ssa1, Sis1, and Sti1 (Fig. 6A and Table S2). In control experiments Hsp82, Ssa1, and Sis1 were unable to stimulate reactivation in the absence of Sti1 (Fig. 6A). Additional controls showed that each of the four components was required for maximal luciferase reactivation (Fig. S5). Also, reactivation by Hsp82, Ssa1, Sis1, and Sti1 was inhibited by geldanamycin, a specific Hsp90 inhibitor (Fig. S5).
Fig. 6.
Hsp82 mutants defective in Ssa1 interaction are defective in functional collaboration with Ssa1 in luciferase reactivation in vitro. Heat-inactivated luciferase was reactivated as described in Materials and Methods using 2 μM Ssa1, 0.4 μM Sis1, 0.2 μM Sti1, and 0.5 μM wild-type (A) or mutant (B–F) Hsp82 as indicated. Luciferase reactivation by Ssa1, Sis1, Sti1 with wild-type Hsp82 (black closed symbols and solid lines) or Hsp82 mutants (colored closed symbols and solid lines), Ssa1 and Sis1 in the presence of Sti1 (gray closed symbols and solid lines), Ssa1 and Sis1 with wild-type Hsp82 (black open symbols and dashed lines) or mutants (colored open symbols and dashed lines). Gray dotted lines indicate luciferase-alone control. In A–F, data from three or more replicates are presented as mean ± SEM. For some points, the symbols obscure the error bars.
When we tested the Hsp82 mutants for the ability to collaborate with Ssa1, Sis1, and Sti1 in luciferase reactivation, we found that Hsp82-K399C, -K398E, -K394C, and -G313S were unable to stimulate luciferase reactivation (Fig. 6 B–E). Hsp82-P281C reactivated luciferase with Ssa1, Sis1, and Sti1 at a rate similar to wild-type Hsp82 (Fig. 6 A and F). Thus, the mutants that were defective in direct binding to Ssa1 were also defective in functional collaboration with Ssa1.
Hsp82 Mutant Proteins Are Defective in Complex Formation with Ssa1 and Sti1.
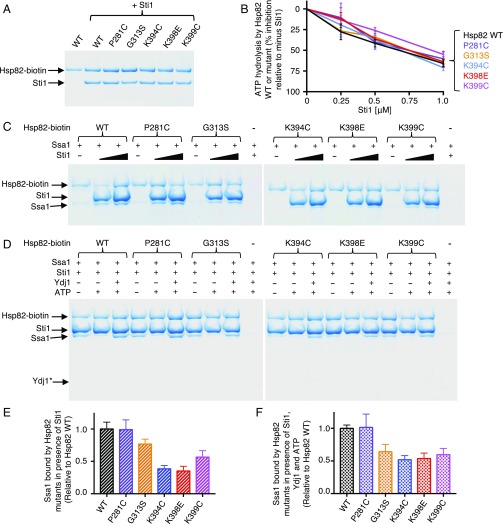
It is well established that Hop/Sti1 interacts with Hsp90 and Hsp70, thereby facilitating a stable Hsp90-Hop/Sti1-Hsp70 complex (3, 4, 38, 39). In addition, the observation that Sti1 was essential for growth of three of the Hsc82 and Hsp82 mutants at 30 °C (Fig. 3 C and D) suggested that Sti1 might compensate for the defective interaction between Ssa1 and the Hsp82 mutant proteins. To explore this possibility, we examined the effect of Sti1 on the interaction between the Hsp82 mutant proteins and Ssa1 in vitro.
In control experiments, the biotinylated Hsp82 mutants and the wild-type protein bound Sti1 similarly in pull-down assays, as was observed by BLI (Fig. 7A, Fig. S4C, and Table S2). In addition, the ATPase activity of the Hsp82 mutant proteins was inhibited by Sti1 (Fig. 7B), as has been shown for the wild-type protein (40, 41). These results suggest that the Hsp82 mutant proteins are not defective in physical or functional interaction with Sti1 and are consistent with the pull-down experiments in cell lysates (Fig. 4). Using in vitro pull-down assays, we observed that more Ssa1 was bound by wild-type Hsp82 in the presence of Sti1 than in the absence of Sti1 (Fig. 7C). However, less Ssa1 was associated with Hsp82-G313S, -K394C, -K398E, and -K399C compared with the wild-type protein in the presence of Sti1 (Fig. 7 C and E and Table S2). Hsp82-P281C and the wild-type protein retained similar amounts of Ssa1 in the presence of Sti1 (Fig. 7 C and E). The results show that the Hsp82 residues that are important for the direct interaction with Ssa1 are also important for the ternary complex involving Hsp82, Sti1, and Ssa1. Together with the in vivo data showing that Sti1 is essential in cells expressing Hsc82-G309S, Hsc82-K394E, or Hsp82-K399C, these data suggest that Sti1 stabilizes the interaction between Ssa1 and Hsc82-G309S, Hsc82-K394E, and Hsp82-K399C sufficiently to allow the mutant proteins to promote viability.
Fig. 7.
Effect of cochaperones on Hsp82–Ssa1 interaction. (A) The interaction between wild-type or mutant biotin-labeled Hsp82 (1 μM) and Sti1 (0.5 μM) was monitored using a pull-down assay and analyzed by Coomassie blue staining following SDS/PAGE as described in Materials and Methods. (B) ATP hydrolysis by wild-type or mutant Hsp82 was measured in the absence or presence of increasing concentrations of Sti1, and the percent of inhibition by Sti1 is shown. In the absence of Sti1 (0% inhibition) the rate of ATP hydrolysis is 0.11 ± 0.007 nmol/min for wild-type Hsp82, 0.086 ± 0.008 nmol/min for Hsp82-P281C, 0.12 ± 0.002 nmol/min for Hsp82-G313S, 0.12 ± 0.01 nmol/min for Hsp82-K394C, 0.11 ± 0.007 nmol/min for Hsp82-K398E, and 0.09 ± 0.01 nmol/min for Hsp82-K399C. The results of three or more replicates are presented as mean ± SEM. (C) Effect of increasing Sti1 concentration on interaction between Hsp82 mutants and Ssa1. Pulldowns were carried out as in A with 1 μM of biotin-labeled wild-type or mutant Hsp82, 6 μM Ssa1, and 2 μM or 4 μM Sti1 as indicated. (D) Effect of Ydj1 on the interaction between Hsp82 mutants and Ssa1 in the presence of ATP and Sti1. Pull-down experiments were performed with 1 μM Hsp82-biotin, 6 μM Ssa1, 2 μM Sti1, 0.35 μM Ydj1, and 2 mM ATP where indicated. Ydj1* indicates the expected position for Ydj1. (E) Quantification of Ssa1 bound to wild-type or mutant Hsp82-biotin in the presence of 2 μM Sti1 from C and replicates. The ratio of Ssa1 bound to each of the Hsp82 biotin-labeled mutants relative to wild-type Hsp82 is presented as mean ± SEM. (F) Quantification of Ssa1 bound to wild-type or mutant Hsp82-biotin in the presence of Sti1, Ydj1, and ATP from D and replicates. The ratio of Ssa1 bound to each of the biotin-labeled Hsp82 mutants relative to wild-type Hsp82 is presented as mean ± SEM. In A, C, and D, a representative of three replicate experiments is shown.
Involvement of Hsp40 in Ssa1–Sti1–Hsp82 Wild-Type or Mutant Complex Formation.
In previous experiments, it was shown that Hsp40 cochaperones, in substoichiometric concentrations, facilitate complex formation among Hsp90, Hsp70, and Hop/Sti1 (42–45). Therefore, we wanted to test if the mutant Hsp82 proteins were defective in forming complexes with Ssa1 in the presence of Hsp40 in addition to Sti1. Using a pull-down assay, we observed that significantly less Ssa1 associated with wild-type Hsp82 and Sti1 in the presence of ATP than in the absence of ATP (Fig. 7D and Fig. S6A). Moreover, in the presence of ATP and Sti1, Hsp40 proteins, including Ydj1, Sis1, or DnaJ, stimulated the association of Ssa1 with Hsp82 (Fig. 7D and Fig. S6A). In the absence of ATP, Hsp40 did not significantly affect the association of Ssa1 with wild-type Hsp82 in the presence of Sti1 (Fig. S6B).
When we tested the Hsp82 mutants for ability to interact with Ssa1 in the presence of ATP, Ydj1, and Sti1, we observed that Hsp82-G313S, -K394C, -K398E, and -K399C were partially defective in complex formation compared with the wild-type protein, while P281C was like the wild type (Fig. 7 D and F and Table S2). These results suggest that the same residues that are important for Hsp82–Sti1–Ssa1 complex formation in the absence of ATP are also important for complex formation facilitated by Ydj1 in the presence of ATP.
Discussion
In this work we showed that yeast Hsp90 and Hsp70 interact directly and that yeast mutants with substitutions in a surface-exposed region of the M-domain of Hsp82 are defective in functional and physical interaction with Ssa1 in vivo and in vitro. The simplest interpretation of these results is that yeast Hsp90 and Hsp70 interact directly via this surface on the M-domain. These Hsp82 residues align with residues on Hsp90Ec that were previously shown to be important for binding DnaK (30). Another interpretation of the results is that the Hsp90 region identified as important for interaction with Hsp70 might be involved in conformational changes that alter the binding to Hsp70 at a different site.
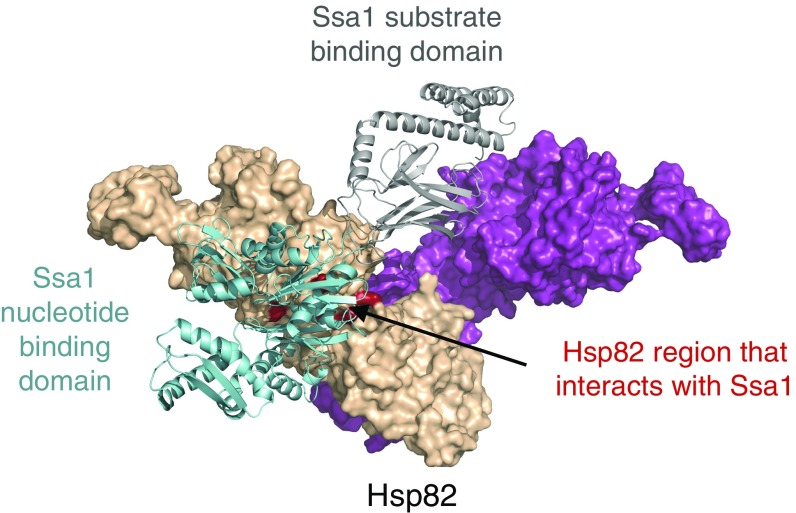
To visualize the potential direct interaction between the M-domain of Hsp82 and Ssa1, we made a docked model of the two chaperones (Fig. 8). First, individual models of apo Hsp82 and ADP-bound Ssa1 were constructed (Materials and Methods) based on the homology between Hsp82 and Hsp90Ec and between Ssa1 and DnaK. Next, the two models were docked, and the top model that included the region identified here by mutation was selected (Materials and Methods and Fig. 8). The model shows the M-domain of Hsp82 interacting with the nucleotide-binding domain of Ssa1. Due to the homology between Ssa1 and DnaK, the region on Ssa1 that interacts with Hsp82 may also be involved in interactions with Hsp40 and with the Ssa1 substrate-binding domain, as has been observed for the homologous region of DnaK (46–50).
Fig. 8.
Modeled interaction between Hsp82 and Ssa1. Computational docking of homology models of apo-Hsp82 and ADP-bound Ssa1 was carried out using ClusPro as described in Materials and Methods. Hsp82 is shown as a surface rendering with one protomer in magenta and the other in wheat. The residues tested in this study are shown in red. Ssa1 is shown as a ribbon model with the nucleotide-binding and substrate-binding domains colored blue and gray, respectively.
The region of the Hsp90 M-domain we identified to be important for interacting with Ssa1 has also been shown to be involved in binding other cochaperones and client proteins. Based on structural studies, Hsp82-K394, -K398, and -K399 are implicated in binding both Aha1 and Sba1 cochaperones (3, 6, 17, 51). Furthermore, residues of Hsp90β homologous to Hsp82-K394 and -K399 were shown to interact with the cochaperone Cdc37 during the formation of the Hsp90–Cdc37–Cdk4 ternary complex (52). Just as Hsp70 recruits client proteins to Hsp90, the cochaperone Cdc37 is hypothesized to bind the Cdk4 kinase and then interact with Hsp90 to promote substrate transfer (52). Therefore, this region on the M-domain of Hsp90 may be involved in the transfer of client proteins from cochaperones to the client-binding site of Hsp90. Additionally, client proteins such as Tau, p53, and Cdk4 bind in regions of the Hsp90 M-domain that include or are located near the region important for binding Ssa1 (3). These results suggest that there is a multiprotein binding site on the M-domain of Hsp90 and that collaboration between Hsp70 and Hsp90 in protein remodeling may be modulated through competition between Hsp70 and additional proteins for the interaction surface.
Previous studies have characterized multiprotein assemblies that involve various combinations of Hsp90, Hsp70, Hop/Sti1, and client proteins (3, 6, 42–45, 52). In recent reports, cryo-electron microscopy, electron microscopy, or chemical crosslinking followed by mass spectrometry were used to resolve complexes involving Hsp90, Hsp70, Hop, and a fragment of the GR (42, 44, 45, 52). However, the conformational states of the chaperones and the sites of interaction between the various components differ in the various reports. One interpretation of the apparent discrepancies in the arrangements of the proteins in the multiprotein complexes is that the various methods have captured different steps in the pathway of protein remodeling.
In summary, much progress has been made in recent years toward understanding the transient and dynamic physical interactions between Hsp90 and its interacting proteins, including the Hsp70 chaperone, Hsp90 cochaperones, and client proteins, during the process of protein remodeling. The information gained from these studies will be invaluable in designing inhibitors that target Hsp90 and its interacting proteins for the treatment of diseases, including cancer and diseases associated with protein misfolding.
Materials and Methods
Yeast Strains, Plasmids, and Growth Conditions.
In studies with HSP82 and hsp82 mutants, yeast strains were derived from MR318 (53). Strain 1670 is identical to MR318 but is also sti1::HIS3. It was created through standard yeast genetic techniques (54). Strains used in this study were made via plasmid shuffling as described (12). In studies with HSC82 and hsc82 mutants, the strains were derived from W303, JJ816 (MATa hsc82:: LEU2 hsp82::LEU2/YeP24–HSP82), and JJ832 (MATa hsc82::LEU2 hsp82::LEU2 sti1::MET2/YeP24–HSP82) (55). Except where indicated, standard growth medium and conditions were used (54).
Generation of hsc82 Mutants.
The hsc82-K394E mutation was identified in a genetic screen. We constructed a library of hsc82 alleles using error-prone PCR. Plasmids expressing mutant hsc82 were screened for the inability to support growth of an hsc82hsp82/YeP24-HSP82 strain at 37 °C on medium containing 5-fluoroorotic acid (5-FOA) using the plasmid-shuffle method. One of the mutants obtained was hsc82-K394E. This mutation was verified by DNA sequencing.
In Vivo His6–Hsc82 and His6–Hsp82 Protein–Protein Interaction Assay.
His6–Hsc82 (56) and His6–Hsp82 (53) complexes were isolated from yeast cells grown at 30 °C and lysated as described. Samples were separated by SDS/PAGE and were either stained with Coomassie blue or transferred to PVDF membranes and blotted with antibodies specific to Hsp82 (Enzo ADI-SPA-840) (57), Hsc82 (generated in the J.L.J. laboratory), Hsp70 (Enzo ADI-SPA-757), Ssa1 (a gift from E. Craig, University of Wisconsin, Madison, WI), or Sti1 (generated in the J.L.J. laboratory or the D.C.M. laboratory) (58). Prism 7.0a for Mac (GraphPad Software, https://www.graphpad.com/) was used for statistical analyses. One-way ANOVA followed by Dunnett’s multiple comparisons test was used to establish significant differences (P < 0.05).
In Vivo Glucocorticoid Maturation Assay.
The assay was performed as previously described (53). Briefly, 5-FOA–resistant STI1 cells expressing the indicated hsp82 allele were transformed with plasmids encoding the rat GR and β-galactosidase driven by a glucocorticoid response element. Log-phase liquid cultures were incubated at 30 °C for 5 h with 10 μM deoxycorticosterone, and the β-galactosidase activity was measured. Prism 7.0a for Mac (GraphPad Software) was used for statistical analyses. One-way ANOVA followed by Dunnett’s multiple comparisons test was used to establish significant differences (P < 0.05).
Proteins.
Substitution mutations of Hsp82 were introduced into pET24b–Hsp82 with the QuikChange Lightning Mutagenesis Kit (Agilent). All mutations were verified by DNA sequencing. Hsp82 wild-type and mutant proteins were purified as previously described (12). Sis1 was purified as described (24) with minor modifications. Briefly, following chromatography using an SP-Sepharose Fast Flow column (GE Healthcare), Sis1 was chromatographed on a Sephacryl S-200 column (GE Healthcare) in 20 mM Mes (pH 6.0), 0.1 mM EDTA, and 500 mM NaCl. Ydj1 and Ssa1 were purified as described (59, 60). All proteins were >95% pure as determined by SDS/PAGE. Concentrations given are for Hsp82, Ydj1, and Sis1 dimers and for Ssa1, Sti1, and luciferase monomers. Hsp82 was labeled using a 1.5-fold excess of NHS-PEG4-Biotin (Thermo Scientific, Life Technologies). Excess biotin reagent was removed using 7K molecular weight cut off Zeba Spin Desalting Columns (Thermo Scientific).
Luciferase Reactivation.
Luciferase reactivation was performed as previously described (12). Heat-denatured luciferase (20 nM), prepared as described (27), was incubated at 24 °C in reaction mixtures (75 μL) containing 25 mM Hepes (pH 7.5), 50 mM KCl, 0.1 mM EDTA, 2 mM DTT, 10 mM MgCl2, 50 μg/mL BSA, 1 mM ATP, an ATP-regenerating system (25 mM creatine phosphate, 6 μg creatine kinase), 2 µM Ssa1, 0.4 µM Sis1, 0.2 µM Sti1, and 0.5 µM wild-type or mutant Hsp82, as indicated. Aliquots were removed at the indicated times, and light output was measured using a Tecan Infinite M200Pro in luminescence mode with an integration time of 1,000 ms following the injection of luciferin (50 µg/mL). Reactivation was determined compared with a non-denatured luciferase control. Luciferase and luciferin were from Promega.
ATPase Activity.
Steady-state ATP hydrolysis was measured at 37 °C in 25 mM Hepes (pH 7.5), 20 mM KCl, 5 mM DTT, 5 mM MgCl2, and 2 mM ATP using a pyruvate kinase/lactate dehydrogenase enzyme-coupled assay as described (14) and using 1 μM wild-type or mutant Hsp82 and cochaperone concentrations as indicated.
BLI Assay.
BLI was used to monitor the interaction between Hsp82 and Ssa1 using a ForteBio Octet RED96 instrument and streptavidin biosensors at 30 °C. Each assay step was 200 μL containing BLI buffer [20 mM Tris (pH 7.5), 25 mM NaCl, 0.01% Triton X-100 (vol/vol), 0.02% Tween-20 (vol/vol), 1 mg/mL BSA, 1 mM ATP, and 10 mM MgCl2]. Wild-type and mutant Hsp82-biotin was loaded on the biosensors to a BLI response signal of 1.1 nm. The biosensors were blocked in BLI buffer containing 10 μg/mL biocytin (Sigma) for 1 min, and a baseline was established in BLI buffer alone. Association of Ssa1 (2.5–70 μM) with the Hsp82-biotin–bound sensor tip in BLI buffer was monitored over time. Last, dissociation was monitored in BLI buffer alone. For each experiment, nonspecific binding was monitored using a reference biosensor subjected to each of the above steps in the absence of biotinylated Hsp82, and the nonspecific binding signal was subtracted from the corresponding experiment. For steady-state analysis of kinetic association data, the association curve at each Ssa1 concentration was fit using a single exponential equation without constraints in Prism 7.0a for Mac (GraphPad Software), and the plateau value determined from the fit was plotted versus the concentration of Ssa1. The resulting binding curve was analyzed using a one-site specific binding model in Prism to determine the Kd (equilibrium binding constant) and Bmax (maximum specific binding) values.
In Vitro Protein–Protein Interaction Assay.
Interaction of Hsp82 with Ssa1 with or without Sti1 was measured using a pull-down assay. Wild-type or mutant Hsp82-biotin (1 µM) was incubated for 5 min at 23 °C in reaction mixtures (50 μL) containing PD buffer [20 mM Tris⋅HCl (pH 7.5), 50 mM KCl, 5% glycerol (vol/vol), 0.01% Triton X-100 (vol/vol), 2 mM DTT] with Ssa1 and/or Sti1 as indicated. NeutrAvidin agarose (40 μL; 1:1 slurry) (Thermo Scientific, Pierce) was then added and incubated for 5 min at 23 °C with mixing. The reactions were diluted with 0.4 mL PD buffer and centrifuged for 1 min at 1,000 × g, and the recovered agarose beads were washed once with 0.4 mL PD buffer. Bound proteins were eluted with buffer containing 2 M NaCl and analyzed by Coomassie blue staining following SDS/PAGE. For assays in the presence of Ydj1 and ATP, reactions mixtures contained 10 mM MgCl2 and 2 mM ATP where indicated and were incubated in the presence of NeutrAvidin agarose for 10 min at 30 °C with mixing. Reactions mixtures were centrifuged for 1 min at 1,000 × g, and the recovered agarose beads were washed twice with 0.5 mL PD buffer and processed as above. Where indicated, protein band intensities from replicate gels were quantified using ImageJ (https://imagej.nih.gov/ij/). The ratio of Ssa1 bound to each of the Hsp82-biotin mutants relative to Ssa1 bound to wild-type Hsp82-biotin was calculated and plotted.
Modeling Ssa1–Hsp82 Interaction.
Starting with the ADP-bound conformation of DnaK [Protein Data Bank (PDB ID code 2KHO] (61) or, separately, the apo conformation of Hsp90Ec (PDB ID code 2IOQ) (16), we constructed homology models of the yeast proteins Ssa1, Hsp82, and Hsc82 using the I-TASSER structure-prediction software (62). To generate dimer structures of Hsp82 and Hsc82, two copies of each resulting full-length monomer were then oriented as to minimize the rmsd with each monomer of the Hsp90Ec dimer. The predicted structures of Ssa1, Hsp82, and Hsc82 were very similar to the E. coli homologs, with rmsd values of ∼1.5 Å. The resulting structures of Hsp82 and Hsc82 predicted that the MEEVD-containing regions were disordered and protruded from the organized portion of the C-terminal domains. Similarly, the VEEVD-containing region of Ssa1 was also disordered and protruded from the C terminus. Since these disordered termini clashed with domains within the protein (either the substrate binding domain of Ssa1 or the dimerization domain of Hsp82 or Hsc82), the disordered regions were truncated beginning at residue 611 of Ssa1, residue 676 of Hsp82, or residue 670 of Hsc82.
Hsp82–Ssa1 complexes were generated using ClusPro (63) without restraining the interaction between Ssa1 and Hsp82, as described previously (30). The resulting models were then filtered based on the Hsp82 residues tested and found defective in this study.
Supplementary Material
Acknowledgments
We thank Matthew Stack for purifying and characterizing some Hsp82 proteins and Grzegorz Piszczek (National Heart, Lung, and Blood Institute Biophysical Core Facility) for helpful comments and suggestions about BLI experiments and analysis. This research was supported by the Intramural Research Program of the NIH, the National Cancer Institute Center for Cancer Research, the National Institute of Diabetes and Digestive and Kidney Diseases, and NIH Grant P30 GM103324 (to J.L.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719969115/-/DCSupplemental.
References
- 1.Mayer MP, Le Breton L. Hsp90: Breaking the symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Röhl A, Rohrberg J, Buchner J. The chaperone Hsp90: Changing partners for demanding clients. Trends Biochem Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim Biophys Acta. 2012;1823:607–613. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearl LH. Review: The HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers. 2016;105:594–607. doi: 10.1002/bip.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, Buchner J. The plasticity of the Hsp90 co-chaperone system. Mol Cell. 2017;67:947–961. doi: 10.1016/j.molcel.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Taipale M, et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 10.Hainzl O, Lapina MC, Buchner J, Richter K. The charged linker region is an important regulator of Hsp90 function. J Biol Chem. 2009;284:22559–22567. doi: 10.1074/jbc.M109.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsutsumi S, et al. Hsp90 charged-linker truncation reverses the functional consequences of weakened hydrophobic contacts in the N domain. Nat Struct Mol Biol. 2009;16:1141–1147. doi: 10.1038/nsmb.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genest O, et al. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol Cell. 2013;49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Street TO, et al. Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J Mol Biol. 2012;415:3–15. doi: 10.1016/j.jmb.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krukenberg KA, Förster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: Insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 17.Ali MMU, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- 20.Johnson BD, et al. Hsp90 chaperone activity requires the full-length protein and interaction among its multiple domains. J Biol Chem. 2000;275:32499–32507. doi: 10.1074/jbc.M005195200. [DOI] [PubMed] [Google Scholar]
- 21.Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperones protein folding in vitro. Nature. 1992;358:169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- 22.Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol Biol Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morishima Y, Murphy PJM, Li DP, Sanchez ER, Pratt WB. Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J Biol Chem. 2000;275:18054–18060. doi: 10.1074/jbc.M000434200. [DOI] [PubMed] [Google Scholar]
- 24.Reidy M, et al. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet. 2014;10:e1004720. doi: 10.1371/journal.pgen.1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang HC, Nathan DF, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genest O, Hoskins JR, Camberg JL, Doyle SM, Wickner S. Heat shock protein 90 from Escherichia coli collaborates with the DnaK chaperone system in client protein remodeling. Proc Natl Acad Sci USA. 2011;108:8206–8211. doi: 10.1073/pnas.1104703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genest O, Hoskins JR, Kravats AN, Doyle SM, Wickner S. Hsp70 and Hsp90 of E. coli directly interact for collaboration in protein remodeling. J Mol Biol. 2015;427:3877–3889. doi: 10.1016/j.jmb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamoto H, et al. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. J Biol Chem. 2014;289:6110–6119. doi: 10.1074/jbc.M113.524801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kravats AN, et al. Interaction of E. coli Hsp90 with DnaK involves the DnaJ binding region of DnaK. J Mol Biol. 2017;429:858–872. doi: 10.1016/j.jmb.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Röhl A, et al. Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules. Nat Commun. 2015;6:6655. doi: 10.1038/ncomms7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 33.Sung N, et al. 2.4 Å resolution crystal structure of human TRAP1NM, the Hsp90 paralog in the mitochondrial matrix. Acta Crystallogr D Struct Biol. 2016;72:904–911. doi: 10.1107/S2059798316009906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohen SP, Yamamoto KR. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Zierer BK, et al. Importance of cycle timing for the function of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2016;23:1020–1028. doi: 10.1038/nsmb.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baindur-Hudson S, Edkins AL, Blatch GL. Hsp70/Hsp90 organising protein (hop): Beyond interactions with chaperones and prion proteins. Subcell Biochem. 2015;78:69–90. doi: 10.1007/978-3-319-11731-7_3. [DOI] [PubMed] [Google Scholar]
- 39.Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 40.Prodromou C, et al. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J Biol Chem. 2003;278:10328–10333. doi: 10.1074/jbc.M213094200. [DOI] [PubMed] [Google Scholar]
- 42.Alvira S, et al. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat Commun. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 43.Hernández MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J Biol Chem. 2002;277:38294–38304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- 44.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgner N, et al. Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 2015;11:759–769. doi: 10.1016/j.celrep.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmad A, et al. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci USA. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gassler CS, et al. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 49.Qi R, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20:900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suh W-C, et al. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer P, et al. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23:1402–1410. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verba KA, et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 2016;352:1542–1547. doi: 10.1126/science.aaf5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuehlke AD, et al. An Hsp90 co-chaperone protein in yeast is functionally replaced by site-specific posttranslational modification in humans. Nat Commun. 2017;8:15328. doi: 10.1038/ncomms15328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 55.Flom G, Weekes J, Williams JJ, Johnson JL. Effect of mutation of the tetratricopeptide repeat and asparatate-proline 2 domains of Sti1 on Hsp90 signaling and interaction in Saccharomyces cerevisiae. Genetics. 2006;172:41–51. doi: 10.1534/genetics.105.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson JL, Halas A, Flom G. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol Cell Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reidy M, Masison DC. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol Cell Biol. 2010;30:3542–3552. doi: 10.1128/MCB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1) J Biol Chem. 2005;280:34178–34185. doi: 10.1074/jbc.M505420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miot M, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci USA. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reidy M, Sharma R, Roberts BL, Masison DC. Human J-protein DnaJB6b cures a subset of Saccharomyces cerevisiae prions and selectively blocks assembly of structurally related amyloids. J Biol Chem. 2016;291:4035–4047. doi: 10.1074/jbc.M115.700393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozakov D, et al. The ClusPro web server for protein-protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure. 2004;12:1087–1097. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.