Significance
Three-dimensional histology of soft-tissue samples has proven to provide crucial benefits for the understanding of tissue structure. Nevertheless, the low attenuation of soft tissue has impaired the use of computed tomography (CT) as a tool for the nondestructive and 3D visualization of external and internal structural details. We present a cytoplasm-specific staining method tailored for X-ray CT that enables a routine and efficient 3D volume screening at high resolutions. Our technique is fully compatible with conventional histology and allows further histological investigations, as demonstrated for a mouse kidney. The comparative analysis of our CT data with conventional 2D histology highlights the future applicability of the cytoplasm-specific X-ray staining method for modern histological and histopathological investigations using laboratory X-ray CT devices.
Keywords: 3D virtual histology, microCT, nanoCT, cytoplasm-specific X-ray stain, 3D imaging
Abstract
Many histological methods require staining of the cytoplasm, which provides instrumental details for diagnosis. One major limitation is the production of 2D images obtained by destructive preparation of 3D tissue samples. X-ray absorption micro- and nanocomputed tomography (microCT and nanoCT) allows for a nondestructive investigation of a 3D tissue sample, and thus aids to determine regions of interest for further histological examinations. However, application of microCT and nanoCT to biological samples (e.g., biopsies) is limited by the missing contrast within soft tissue, which is important to visualize morphological details. We describe an eosin-based preparation overcoming the challenges of contrast enhancement and selectivity for certain tissues. The eosin-based staining protocol is suitable for whole-organ staining, which then enables high-resolution microCT imaging of whole organs and nanoCT imaging of smaller tissue pieces retrieved from the original sample. Our results demonstrate suitability of the eosin-based staining method for diagnostic screening of 3D tissue samples without impeding further diagnostics through histological methods.
Currently, histology is an essential technique when biological tissue is studied at microscopic scales to provide instrumental and essential information for medical diagnostics and research (1, 2). However, conventional histology involves very often time-consuming, complicated, and moreover, destructive sample preparation protocols. The individual 2D slices of one studied object are observed under the microscope, images are digitalized, evaluated, and stored. This procedure allows for the assembling of a volumetric data set. There are different histological section-based 3D imaging methods around and, despite the developments made with regards to the semiautomated and automated section realignment, histological section-based 3D reconstruction techniques hold limitations and require specialists (3−6).
On the other hand, X-ray computed tomography (CT) has become an invaluable tool for the nondestructive investigation of objects in many fields such as material science, industrial testing, or medical diagnostics (7). With demand for higher resolutions in laboratory-based X-ray CT imaging in the submicrometer range, the fields of microcomputed tomography (microCT) (8−10) and nanocomputed tomography (nanoCT) (11) imaging have been established (within this work microCT refers to voxel sizes >1 µm and nanoCT refers to voxel sizes from >100 nm to <1 µm). To become almost comparable to conventional histology with regard to resulting contrast, nanoCT imaging makes use of X-ray optics (12) or X-ray transmission tubes with very small focal spot sizes (13, 14). The application of microCT and nanoCT for biological sample screening, however, remains limited due to very low intrinsic contrast of soft tissue. The utilization of contrast agents is necessary to visualize microscopic structures (15−23). At present, the most widely used staining method is based on the inorganic iodine staining method IKI (iodine potassium iodide). Here, the triiodide and iodide anion allow for an ionic interaction with (poly-)cationic structures within nonmineralized tissues. This makes IKI an optimal overview stain being able to penetrate entire organisms such as animal embryos. However, the current available staining methods are often time-consuming, elaborate, toxic, and inhibit further investigation of the sample by other methods such as histology. The eosin-based staining method in combination with a recently developed laboratory-based X-ray nanoCT device (24) presented here resolves these shortcomings by offering a remarkably improved contrast of the stained soft tissue within a much faster staining procedure at resolutions ranging from a few micrometers down to 200 nm. Furthermore, the presented method still enables compatibility with conventional histological examinations, i.e., the possibility of counterstaining by the histologist. Overall, this work demonstrates the powerful potential of 3D histology as a tool for modern histological and histopathological applications.
Results
In histology, staining of the cell cytoplasm is of great importance as it provides an overview of cell shape, size, and changes within cells giving distinct indications for diagnosis or further diagnostic investigations. The current histological standard used as plasma or counterstain is eosin. It is applied as 0.1% (wt/vol) aqueous solution to 2D microscopic slices of soft tissue (generally cut with a thickness of 2−10 µm) (25). At present, such a tissue-specific and histological compatible staining agent or staining protocol is not available for X-ray CT imaging. However, application of the described standardized histological staining procedure to 3D tissue samples, e.g., whole-organ mouse kidney, resulted in similar image quality compared with the unstained tissue sample (Fig. S1). The very same mouse kidney was used for X-ray CT imaging before and after staining using the same imaging parameters. A contrast enhancement was not achieved. The result for the unstained mouse kidney was expected, highlighting the low intrinsic attenuation properties of soft tissue for typically used X-ray energies of laboratory-based microCT systems, which consists of mainly carbon, hydrogen, oxygen, and nitrogen (26). As for the stained mouse kidney, the low concentration of eosin used for staining was seen as the limiting factor. Here, sensitivity levels were not met for X-ray CT imaging with regard to the high atomic number element bromine (Z = 35) (26), of which one eosin molecule holds four bromide atoms being covalently bound to the fluorescein core.
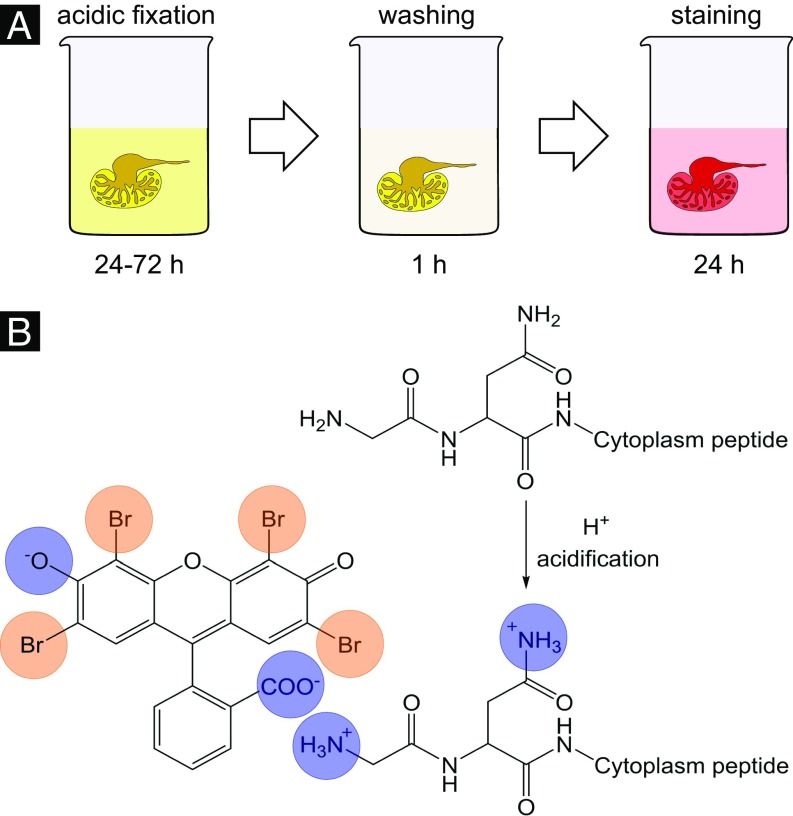
To improve the contrast enhancement within the soft tissue, several concentrations of eosin were tested (with maximum solubility of eosin in water as the end point). As expected, the best contrast enhancement within the soft tissue was observed with the highest eosin concentration. Therefore, the final staining protocol was carried out with the highest concentration. Further improvements to reach the final staining protocol, which consists of only three steps, were made (Fig. 1A). Here, the acidification of the soft-tissue sample during fixation or before staining was found to be crucial. This was also shown by Hong et al. (27). The soft tissue is optimally prepared on molecular level for the staining procedure with the eosin disodium salt. Amino acid side chains of proteins and peptides present within the cell cytoplasm are protonated by the acid, which allows improved ionic interaction with the applied stain leading to a higher accumulation of staining agent within the cell cytoplasm (Fig. 1B).
Fig. 1.
Protocol and chemistry of the proposed eosin-based staining protocol for X-ray microCT. (A) The eosin-based staining protocol shows the individual steps involved. Because of its efficiency, it is particularly suited for larger samples (e.g., whole organs). (B) The stain interacts noncovalently with soft tissue on the molecular level (highlighted through blue circles). Shown is the negatively charged eosin with cationic amino acid side chains of cytoplasm proteins/peptides. The optimized ionic interaction is achieved by acidification of the soft tissue during fixation or before staining, and allows for a higher accumulation of eosin within the cytoplasm of cells. The eosin molecule of the disodium salt contains four bromine atoms (highlighted through orange circles). The high atomic number (Z = 35) component generates a particularly high-contrast enhancement for X-rays.
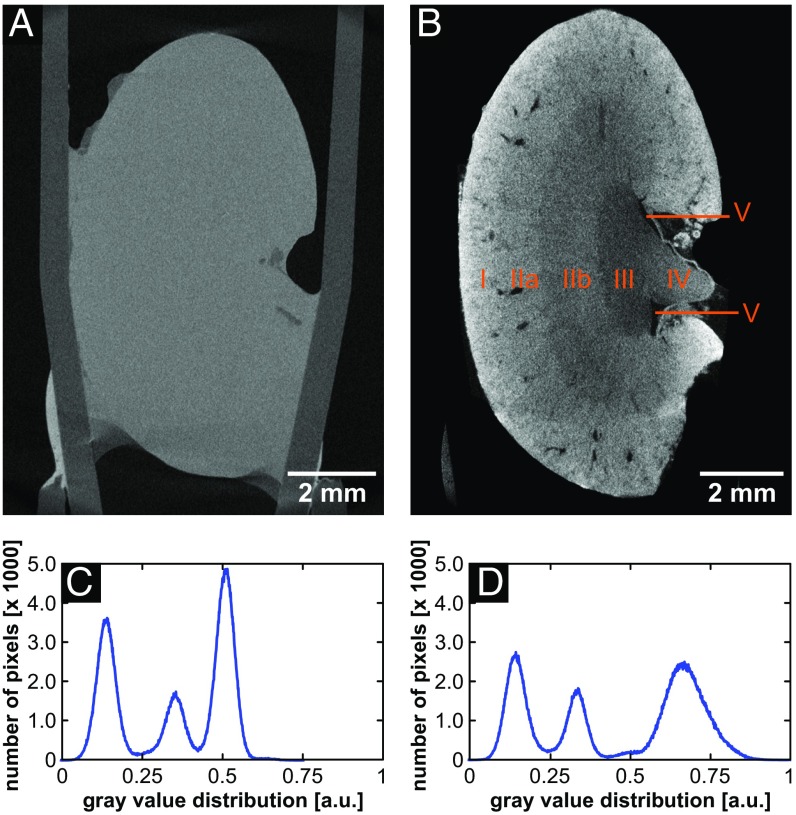
Before staining was applied using the final eosin-based staining protocol, a mouse kidney was imaged with microCT. As expected, anatomical structural information was not visible (Fig. 2A; for an explanation see above). The very same mouse kidney was subjected to the eosin-based staining protocol and again imaged with the microCT. The actual staining step is quite fast, resulting in a complete staining of a whole organ, such as a mouse kidney, within 24 h. The overview microCT scan provided a clear distinction of anatomical structural regions of a mouse kidney, such as cortex, inner and outer medulla, papilla, and renal pelvis (Fig. 2B). Furthermore, the staining was homogeneous within one anatomical structural region. The remarkable contrast enhancement compared with the unstained mouse kidney is further seen in the histograms of the microCT images (Fig. 2 C and D). The histograms display the present materials of air, sample holder, and soft tissue, which are represented by the peaks from the left to the right with gray values increasing proportional to the effective attenuation of the material. Same materials are expected to result in the same gray-value distribution, which is seen in the histograms for the materials' air and sample holder. The signal corresponding to the soft tissue is shifted to a higher gray value and the width of the peak is broadened for the stained soft-tissue sample (Fig. 2 C and D). This demonstrates the influence of the staining agent on the attenuation properties of soft tissue, which results in visualization of the different anatomical structural regions of a mouse kidney.
Fig. 2.
CT slices and histogram distributions of the same whole mouse kidney before and after staining highlighting the contrast enhancement obtained after application of the eosin-based staining protocol. Both data sets were acquired with the Xradia Versa 500 microCT using identical acquisition parameters. The voxel size in both data sets is 12 µm. (A) Overview image of the unstained mouse kidney. (B) Overview image of the same mouse kidney sample shown in A after staining. The following anatomic structural regions could be identified and were labeled: cortex (B, I), outer medulla (B, II) with further distinction in outer stripes of outer medulla (B, IIa) and inner stripes of outer medulla (B, IIb), inner medulla (B, III), papilla (B, IV), and renal pelvis (B, V). (C) Histogram of CT slice shown in A, and (D) Histogram of CT slice shown in B.
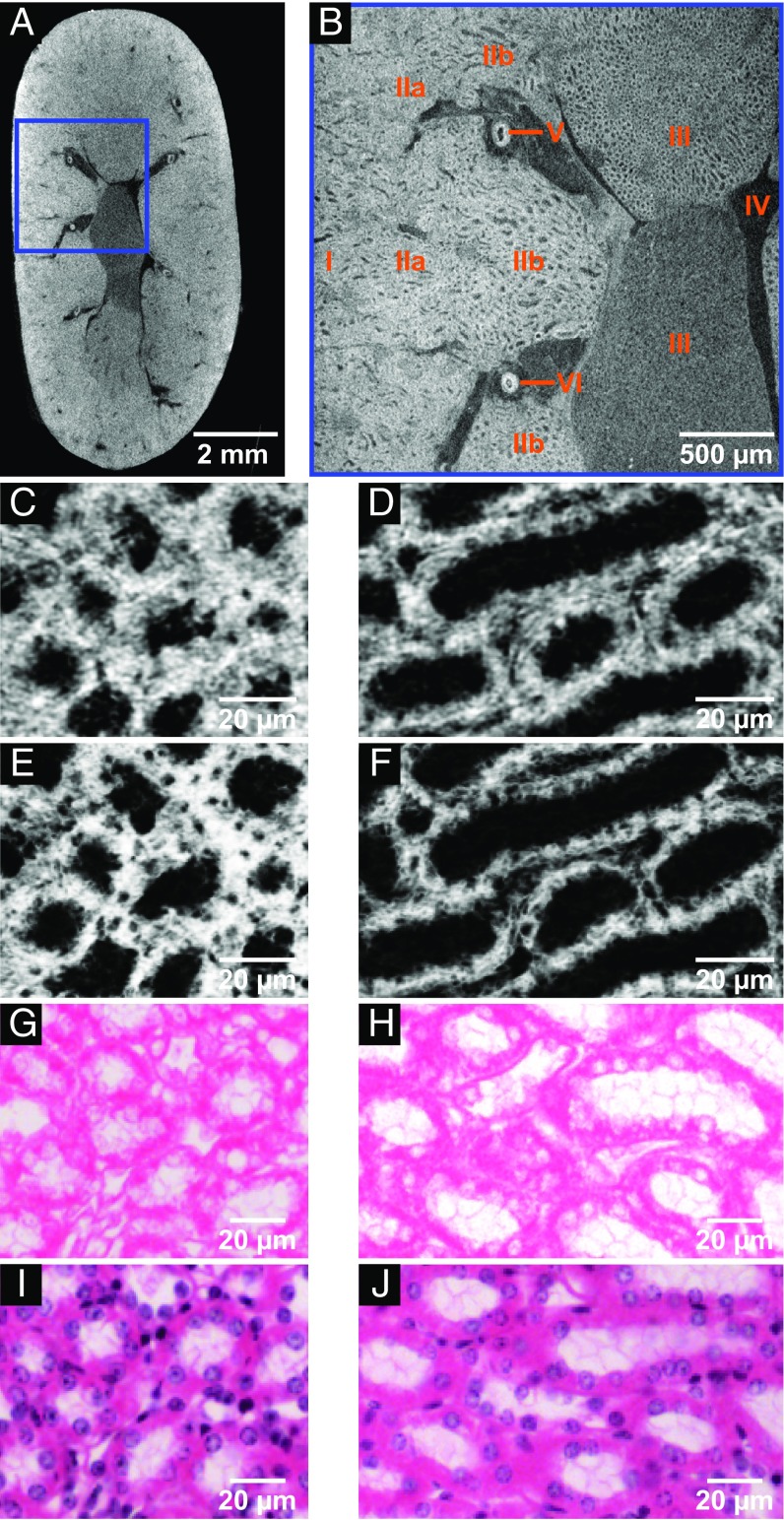
Using a high-resolution microCT scan [region of interest (ROI) highlighted in Fig. 3A] of the very same mouse kidney sample without destruction of the whole organ revealed an even more detailed view of the anatomical structures of the mouse kidney (Fig. 3B). Furthermore, the regions of cortex (Fig. 3, B, I) and inner stripes of outer medulla (Fig. 3, B, IIb) were chosen for nanoCT investigations, whereby favored anatomical regions were dissected and subjected to critical point drying (CPD). Both regions contain important anatomical structures such as the renal corpuscle (cortex) and the loop of Henle (inner stripes of outer medulla), respectively, which offer valuable parameters for diagnosis (28). The nanoCT images of the medulla region (Fig. 3, C, IIb and Fig. 3, D, IIb) seen in Fig. 3 C and D show thick ascending limbs of the loop of Henle. The nanoCT slices have a virtual thickness of ∼400 nm and compare already very well with the histological microscopic slides (Fig. 3 G−J) derived from the very same mouse kidney sample. Since the histological microscopic slides have a thickness of ∼7−10 µm, minimum intensity projection slices of the nanoCT data (Fig. 3 E and F), which correspond to a virtual slice thickness of ∼7 µm, were generated for reasons of better comparison with the histological microscopic slides (Fig. 3 G−J). The minimum intensity projection slices of the nanoCT data reveal now clearly the cell nuclei as nonattenuation area. This result is confirmed by the histological microscopic slides seen in Fig. 3 G and H, which have been directly prepared from the developed eosin-based stained mouse kidney sample without further staining procedures. The obtained contrast was very well suitable for the assignment of anatomical structures through a histologist. The cell nuclei here appeared white being recessed from eosin, which specifically stains proteins and peptides of the cell cytoplasm (25).
Fig. 3.
CT slices of microCT (A and B) and nanoCT (C−F) data in comparison with the histological microscopic slides (G−J) derived from the same mouse kidney after application of the developed eosin-based staining protocol. These demonstrate in particular the compatibility with conventional histological methods, i.e., that it is possible to counterstain with hematoxylin. (A) Overview microCT image highlighting the ROI (blue box) for the high-resolution image seen in B. (B) High-resolution microCT image showing the following anatomic structural regions: cortex (B, I), outer medulla (B, II) with further distinction in outer stripes of outer medulla (B, IIa) and inner stripes of outer medulla (B, IIb), inner medulla (B, III), minor calyx (B, IV), and vessels (B, V and B, VI). (C and D) The nanoCT images of the same mouse kidney sample, which were obtained after staining, dissecting, and CPD shows detailed structures of region (B, IIb) seen in B. These are known as thick ascending limbs of the loop of Henle. (E and F) Minimum intensity projection slices of the same nanoCT data set shown in C and D. The slices correspond to a virtual slice thickness of ∼7 µm, which allows for clear visualization of the cell nuclei. (G and H) Representative histological microscopic slides with an approximate thickness of 7 µm obtained from the same mouse kidney sample after the applied eosin-based staining and embedding in a paraffin block. Displayed are thick ascending limbs of the loop of Henle with clear visualization of cell nuclei and cilia (ciliated epithelium). (I and J) Representative histological microscopic slides prepared close to slides shown in G and H with approximate thickness of 7 µm. Counterstain hematoxylin was applied highlighting the cell nuclei in purple.
To allow for an even better comparison with the histological results and showcase the compatibility of the developed eosin-based staining protocol with standard histological methods, the cell-nuclei−specific staining with Mayer’s sour hematoxylin (in the following referred to as hematoxylin) was applied to the histological microscopic slides (Fig. 3 I and J). As expected, the cell nuclei appear purple in color. Furthermore, the eosin staining was not disturbed, resulting in an expected histological microscopic slide treated with standard H&E staining procedures.
A second sample, the region of the cortex (I), was investigated (Fig. S2) and the nanoCT data (Fig. S2 A and B) analyzed in comparison with a representative histological microscopic slide (Fig. S2C). Significant anatomical structures such as the renal corpuscle including the glomerulus, the Bowman’s capsule, and the renal cortex with convoluted tubules could be assigned from the nanoCT images (Fig. S2A). Minimum intensity projection images (Fig. S2B) allowed again for better comparison of the nanoCT data with a representative histological microscopic slide (Fig. S2C). Very good agreement between nanoCT and histological data was achieved, confirming again the suitability of the developed eosin-based staining protocol for further histological investigations (without further procedures by the histologist) as well as compatibility with the histological counterstain hematoxylin.
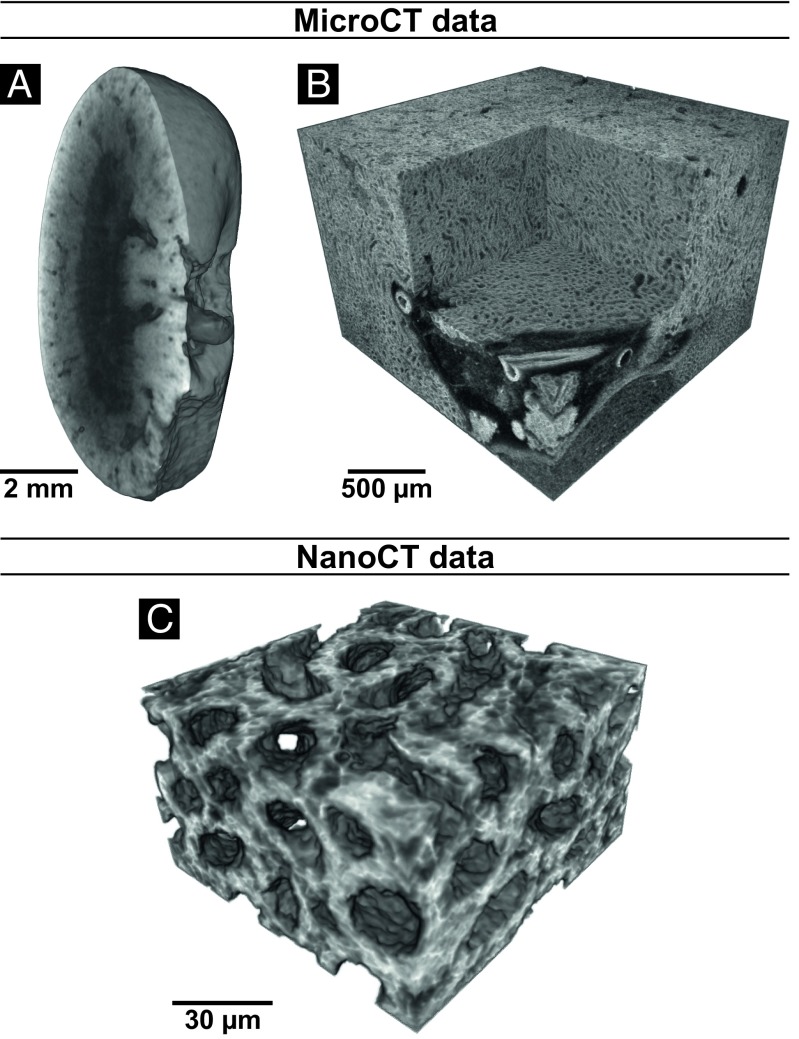
The enormous potential of CT technology lies in 3D visualization, which has been demonstrated in Fig. 4. Renderings of microCT (Fig. 4 A and B) and nanoCT (Fig. 4C) data are displayed. The overview of the whole mouse kidney (Fig. 4A) allowed the identification of an ROI (Fig. 4B), which was further investigated with nanoCT, revealing a very detailed view into the 3D structure of the ascending limbs of the loop of Henle (Fig. 4C).
Fig. 4.
Volume renderings of microCT and nanoCT data from the same mouse kidney after application of the developed eosin-based staining protocol. (A) Virtual sagittal section through the whole mouse kidney acquired with a voxel size of 12 µm. (B) Region of interest microCT data showing the medulla region and a virtual section through a vessel. A local tomography of the whole kidney was performed with a voxel size of 3.3 µm. (C) Region of interest nanoCT data visualizing the 3D structure of thick ascending limbs of the loops of Henle. The data were acquired from a small piece of the kidney with a voxel size of ∼400 nm.
Discussion
Soft-tissue samples prepared with the developed eosin-based protocol meet all of the requirements for a complete and homogeneous staining enabling CT contrast enhancement of a whole mouse organ such as a kidney (Fig. 2). Throughout the soft-tissue sample, the morphology is preserved well enough to identify important anatomical regions and structures, which can be assigned by a pathologist. Moreover, for microCT imaging prepared soft-tissue samples are suitable for further histological investigations, whereby the histological microscopic slides are derived from the very same stained soft-tissue samples (Fig. 3). The eosin-based staining method does not impede further histological treatment by the pathologists and enables the direct evaluation of the tissue by the pathologist with an optical microscope and the counterstaining with the H stain. Even though the concentration of the staining solution is much higher compared with the histological staining solution, the contrast enhancement of the histological microscopic slides is very well suited for histological studies (Fig. 3 G and H).
The staining properties of the eosin stain remain intact, which is seen by the visualization of the cell nuclei in the nanoCT data as nonattenuation area (Fig. 3 C−F) and confirmed by histology (Fig. 3 G and H). Here, the cell nuclei appear white, which reflects the localization of the eosin stain within the cell cytoplasm of the cell. The possibility of counterstaining of the histological microscopic slides by the pathologist is demonstrated by applying the cell-nuclei−specific hematoxylin stain, which resulted in an H&E-stained histological microscopic slide (Fig. 3 I and J) displaying the expected form of appearance. It should be pointed out that standard H&E-prepared histological microscopic slides are firstly stained with hematoxylin and secondly with eosin. Compatibility is still given and quality of the staining result remains very high, even though the staining order was reversed starting first with the developed eosin-based staining protocol for CT, followed by counterstaining of those eosin-based histological microscopic slides with hematoxylin.
The developed eosin-based staining protocol allows for high-resolution CT visualization of soft tissue down to the submicrometer range. In combination with the recently developed nanoCT devices [such as SkyScan 2211 (Bruker), Phoenix nanotom m (GE), and Xradia ZEISS 520 Versa and 810 Ultra (ZEISS)] (12, 13, 29), nondestructive generation of virtual histological slices that are comparable in contrast and resolution to conventional histological data is rendered possible. In the future, this will enable the pathologist to directly identify anatomical structures and regions of interest without the time-consuming preparation of individual slices required for standard histology.
Furthermore, this method provides real 3D information of a soft-tissue bulk sample that supports the pathologist with additional information regarding anatomical structures, which may facilitate improved diagnosis. It should be mentioned here that the developed nanoCT setup used for this work provides much faster scan times and offers a larger field of view (FOV) (allowing for studying of bigger samples) compared with current available laboratory-based nanoCT X-ray devices that can achieve comparable resolutions (12).
The required staining agents for the developed eosin-based staining procedure are easily accessible. The staining protocol is simple to apply, nontoxic, and fast for a whole-organ CT staining, which enables 3D visualization of soft-tissue samples (Fig. 4). Thus, by looking at pathological soft-tissue samples, overview scans will provide valuable insights into altered anatomical regions and structures, which allow for the determination of ROIs. Those can be studied in 3D by microCT or nanoCT and evaluated in 2D with histology. Here, the application of further counterstaining is possible. This work demonstrates the potential of 3D X-ray histology as a tool for future histological and histopathological applications.
Methods
Animals Used.
Animal housing was carried out at the Klinikum rechts der Isar, Technical University of Munich in accordance with the European Union guidelines 2010/63. Organ removal was approved from an internal animal protection committee of the Center for Preclinical Research of Klinikum rechts der Isar, Munich, Germany (internal reference number 4−005-09). All procedures were in accordance with relevant guidelines and regulations. All laboratories are inspected for accordance with the Organisation for Economic Co-operation and Development (OECD) principles of good laboratory practice. We prepared a whole mouse kidney using the final version of the eosin-staining procedure. The soft-tissue sample was then used to evaluate structural preservation and to assess stain quality, identify morphological structures, compare with conventional histological methods, and evaluate for further histological staining. The remaining organs (mainly mouse liver for sample screening during staining method development and optimization and mouse kidney for final optimization and final data acquisition) were used for the development of the staining protocol and the optimization of parameters (the other organs such as the heart, lung, spleen, and brain will be used for future studies).
Sample Screening.
We purchased all reagents from Sigma-Aldrich unless otherwise indicated. Whole mouse organs were fixated and preserved under conditions described below. Cuboidal soft-tissue samples from mouse liver (2−3-mm edge length) were used for stain development and optimization. The small cuboidal tissue samples were cut with a scalpel (Aesculap). Temperature was controlled by placing samples in a refrigerator (4 °C) or in ambient conditions of the laboratory. Incubations were done in sample holders with a flat bottom, which were replaced after each step but not after rinse or dehydration steps. For stain development and optimization several parameters such as fixative, concentration of fixative or staining agent, incubation time, or pH of fixative or staining agents were tested. The stained soft-tissue samples were investigated on the phoenix v|tome|x s 240 CT scanner with typical settings of 50-kV peak voltage, 6.0-W, and with 1,001 projections distributed over 360°. The low-resolution CT data were acquired with an exposure time of 1 s per projection with an effective pixel size of ∼30 µm. The microCT data were reconstructed with the integrated phoenix datos x CT software and analyzed for (i) completeness of staining, (ii) appearance of diffusion rings, (iii) contrast enhancement, (iv) appearance of CT artifacts as streaks, and (v) homogeneity of the staining.
Eosin-Staining Protocol.
The mouse organ was surgically removed and immediately placed in a 50-mL Falcon Centrifuge Tube (neoLab), which was filled with a fixative solution containing 9.5 mL of 4% (vol/vol) formaldehyde solution (FA, derived from a 37% acid-free FA solution stabilized with ∼10% methanol from Carl Roth; further dilution with Dulbecco’s phosphate-buffered saline (DPBS; Thermo Fisher Scientific) without calcium and magnesium) and 0.5 mL glacial acetic acid (Alfa Aesar). The sample was refrigerated for 24−72 h and then washed with phosphate saline buffer solution for 1 h (DPBS without calcium and magnesium). The mouse organ was placed in the staining solution of eosin y [30% (wt/vol) in distilled water; Sigma-Aldrich, product number E4382, stain certified by the Biological Stain Commission]. The soft-tissue sample was stained with 2 mL of staining solution for 24 h (the soft-tissue sample was moving freely within the sample container). During the incubation time the soft-tissue sample was kept on a horizontal shaking plate allowing for a smooth rocking of 60 rpm. After staining, the soft-tissue sample was carefully removed from the sample container and access of staining agent was softly patted with a cellulose tissue paper. The soft-tissue sample was stored in an Eppendorf tube above an ethanol vapor phase [the Eppendorf tube contained a few drops of 70% (vol/vol) ethanol at the bottom of the tube].
X-Ray MicroCT Imaging.
The stained mouse kidney was transferred to a sample holder, which allows the anchorage of the mouse organ above 70% (vol/vol) ethanol vapor. The X-ray microCT measurements were performed with the ZEISS Xradia Versa 500. All shown images were acquired at 50-kV peak voltage, 3.5 W, and with 1,601 projections distributed over 360°. The low-resolution CT data were acquired with the 0.39× objective and an exposure time of 2 s per projection with an effective pixel size of 12 µm. These overview CT data were used to identify the ROI for the high-resolution CT scan, which was selected using the integrated scout and scan software tool of the ZEISS Xradia Versa 500. The high-resolution CT data were acquired with the 4× objective and an exposure time of 15 s per projection with an effective pixel size of 3.3 µm. The CT data were reconstructed using the integrated software.
The volume renderings of the microCT data shown in Fig. 4 were generated with Avizo Fire 8.1 (FEI Visualization Sciences Group).
Sample Preparation for NanoCT.
The stained mouse kidney was dehydrated and CPD before nanoCT imaging. Before the first step, the stained mouse kidney was cut into two halves along the longest axis. One of the halves was further sectioned for nanoCT imaging analysis. The two anatomical regions of renal cortex and renal medulla were separated from each other and cut into very small tissue pieces (scalpel from Aesculap) of ∼0.5-mm edge length. The dehydration incubations were performed for 1 h each. Before the first dehydration step, the small pieces were transferred to a new Petri dish, where they remained for all subsequent steps. The used concentrations (all vol/vol) for the dehydration series were in %: 50, 60, 70, 80, 90, 96, and 100 ethanol balanced with distilled water. The dehydrated mouse tissue pieces were then CPD using a Bal-TEC CPD 030 with CO2 as drying agent. The mouse tissue pieces were kept in a silicon basket, which was placed in the vacuum chamber being half prefilled with 100% ethanol. After cooling to 6−8 °C the chamber was filled up with liquid CO2. While stirring, a waiting period of 3 min was given for mixing of the two components before the chamber got drained until the sample holder was still covered. This procedure was repeated 10 times to ensure complete replacement of ethanol with CO2 within the sample. After the final filling of the chamber with CO2, the machine was heated to the critical point of CO2 (31 °C and 73.8 bar) followed by very slow release of the gaseous CO2 over a time of 30 min. The CPD mouse tissue pieces were stored in a Petri dish kept in a desiccator before further use.
X-Ray NanoCT Imaging.
The X-ray nanoCT measurements were performed with an in-house−developed nanoCT system that consists of a nanofocus X-ray source (prototype Nano Tube; Excillum) (30) and a single-photon counting detector (PILATUS 300K-W 20 Hz; Dectris) (31, 32). The lens-free device is based on mere geometrical magnification and can generate 3D data with resolutions down to 100 nm (24). The presented data were acquired at a peak voltage of 60 kV with 1,599 projections distributed over 360° and a voxel size of ∼400 nm. For this voxel size, the FOV of a single CT measurement is given by ∼560 µm in the direction perpendicular to the rotation axis (horizontal) and 75 µm in the direction of the rotation axis (vertical). However, the FOV can be extended along the rotation axis by combining multiple scans at different vertical positions to a larger volume. Furthermore, the possibility of local tomography measurements allows for larger sample diameters perpendicular to the rotation axis than given by the FOV of a global CT measurement. The exposure time per image was 4 s and the total acquisition time per dataset was ∼3.5 h. After normalization of the acquired projections with flat-field images, the sharpness of the projections was further enhanced by using a Richardson−Lucy deconvolution algorithm (33, 34). The kernel for the deconvolution was a rotationally symmetric Gaussian function with an SD of 1 pixel. To increase the soft-tissue contrast Paganin’s phase-retrieval algorithm was applied to the sharpened images (35). The input energy was 20 keV, which corresponds to the mean energy of the nanoCT setup at 60 kVp. The ratio between the attenuation and the phase coefficient was optimized for the best image quality.
The preprocessed projections were reconstructed with a state-of-the-art filtered backprojection algorithm. The minimum intensity projection slices shown in Fig. 4 and Fig. S2 were generated by calculating the minimum value for each pixel in 18 adjacent slices, which corresponds to a virtual slice thickness of ∼7 µm.
The volume renderings of the nanoCT data shown in Fig. 4 were generated with Avizo Fire 8.1 (FEI Visualization Sciences Group).
Histological Analysis.
The stained mouse kidney was dehydrated and embedded in paraffin according to standard procedures before histological sectioning and further histochemical counterstaining. Before the first step the stained mouse kidney was cut into two halves along the sagittal axis. One of the halves was further histologically investigated. Before the first dehydration step, the mouse kidney was transferred to a new 50-mL Falcon centrifuge tube (neoLab), where it remained for all subsequent steps. For the dehydration, samples were dehydrated in 70% and 96% ethanol twice for 1 h and cleared in xylol twice for 1 h. Subsequent the sample was incubated overnight in paraffin wax at 50 °C to infiltrate the complete sample with wax, embedded in paraffin wax, and sections of 7-µm thickness were cut using a microtome (Leica). Sections were rehydrated and either directly embedded (Eukitt; Merck) or counterstained for 6 min with Mayer’s sour hematoxylin (Morphisto) according to the manufacturer’s protocol. Histological analysis was performed using an Axio Imager 2 microscope and AxioVision Software (ZEISS).
Supporting Information accompanies this article.
Supplementary Material
Acknowledgments
We thank Dr. Enken Drecoll for histological discussions and the extremely helpful team at Excillum AB, Sweden. We acknowledge financial support through the DFG Gottfried Wilhelm Leibniz Program. This research project has received funding from the European’s Union Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie Grant Agreement H2020-MSCA-IF-2015-703745-CONSALT.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720862115/-/DCSupplemental.
References
- 1.Kiernan JA. Histological and Histochemical Methods: Theory and Practice. Scion Publishing Limited; Banbury, UK: 2015. pp. 141–174. [Google Scholar]
- 2.Suvarna SK, Layton C, Bancroft JD. Theory and Practice of Histological Techniques. 7th Ed. Churchill Livingstone Elsevier; Oxford: 2013. pp. 173–186. [Google Scholar]
- 3.Gerneke DA, et al. Surface imaging microscopy using an ultramiller for large volume 3D reconstruction of wax- and resin-embedded tissues. Microsc Res Tech. 2007;70:886–894. doi: 10.1002/jemt.20491. [DOI] [PubMed] [Google Scholar]
- 4.Geyer SH, Mohun TJ, Weninger JW. Visualizing vertebrae embryos with episcopic 3D imaging techniques. Sci World J. 2009;16:1423–1437. doi: 10.1100/tsw.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohun TJ, Weninger WJ. Imaging heart development using high-resolution episcopic microscopy. Curr Opin Genet Dev. 2011;21:573–578. doi: 10.1016/j.gde.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenthal J, et al. Rapid high resolution three dimensional reconstruction of embryos with episcopic fluorescence image capture. Birth Defects Res C Embryo Today. 2004;72:213–223. doi: 10.1002/bdrc.20023. [DOI] [PubMed] [Google Scholar]
- 7.Slaney M, Kak AC. Principles of Computerized Tomographic Imaging. Society for Industrial and Applied Mathematics; Philadelphia: 2001. pp. 114–134. [Google Scholar]
- 8.Landis EN, Keane DT. X-ray microtomography. Mater Charact. 2010;61:1305–1316. [Google Scholar]
- 9.Senter-Zapata M, et al. The role of micro-CT in 3D histology imaging. Pathobiology. 2016;83:140–147. doi: 10.1159/000442387. [DOI] [PubMed] [Google Scholar]
- 10.Stock SR. Micro-Computed Tomography−Methodology and Applications. CRC Press Boca Raton; FL: 2009. pp. 39–84. [Google Scholar]
- 11.Withers PJ. X-ray nanotomography. Mater Today. 2007;10:26–34. [Google Scholar]
- 12.Tkachuk A, et al. X-ray computed tomography in Zernike phase contrast mode at 8 keV with 50-nm resolution using Cu rotating anode X-ray source. Z Kristallogr. 2007;222:650–655. [Google Scholar]
- 13.Dierick M, et al. Recent micro-CT scanner developments at UGCT. Nucl Instrum Methods Phys Res Sect B. 2014;324:35–40. [Google Scholar]
- 14.Salomon M, Hanke R, Krüger P, Uhlmann N, Voland V. Realization of a computed tomography setup to achieve resolutions below 1 µm. Nucl Instrum Methods Phys Res Sect A. 2008;591:50–53. [Google Scholar]
- 15.Jeffery NS, Stephenson RS, Gallagher JA, Jarvis JC, Cox PG. Micro-computed tomography with iodine staining resolves the arrangement of muscle fibres. J Biomech. 2011;44:189–192. doi: 10.1016/j.jbiomech.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JT, et al. Virtual histology of transgenic mouse embryos for high-throughput phenotyping. PLoS Genet. 2006;2:e61. doi: 10.1371/journal.pgen.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Anton N, Zuber G, Vandamme T. Contrast agents for preclinical targeted X-ray imaging. Adv Drug Deliv Rev. 2014;76:116–133. doi: 10.1016/j.addr.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 18.de S e Silva JM, et al. Three-dimensional non-destructive soft-tissue visualization with X-ray staining micro-tomography. Sci Rep. 2015;5:14088. doi: 10.1038/srep14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metscher BD. MicroCT for developmental biology: A versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn. 2009;238:632–640. doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- 20.Metscher BD. MicroCT for comparative morphology: Simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009;9:11. doi: 10.1186/1472-6793-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizutani R, Suzuki Y. X-ray microtomography in biology. Micron. 2012;43:104–115. doi: 10.1016/j.micron.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Pai VM, et al. Coronary artery wall imaging in mice using osmium tetroxide and micro-computed tomography (micro-CT) J Anat. 2012;220:514–524. doi: 10.1111/j.1469-7580.2012.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharir A, Ramniceanu G, Brumfeld V. High resolution 3D imaging of ex-vivo biological samples by micro CT. J Vis Exp. 2011;52:2688. doi: 10.3791/2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller M, et al. Myoanatomy of the velvet worm leg revealed by laboratory-based nanofocus X-ray source tomography. Proc Natl Acad Sci USA. 2017;114:12378–12383. doi: 10.1073/pnas.1710742114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedelsheimer B, Büchl-Zimmermann S. In: Mikroskopische Technik. 19th Ed. Mulisch M, Welsch U, editors. Spektrum; Heidelberg: 2015. pp. 193–194. [Google Scholar]
- 26.Hubbell JH, Seltzer SM. 1996 Tables of X-ray mass attenuation coefficients and mass energy-absorption coefficients from 1 keV to 20 MeV for elements Z = 1 to 92 and 48 additional substances of dosimetric interest, Table 3. Available at physics.nist.gov/PhysRefData/XrayMassCoef/tab3.html. Accessed January 1, 2018.
- 27.Hong HY, Yoo GS, Choi JK. An eosin Y method for protein determination in solution. Anal Lett. 1999;32:2427–2442. [Google Scholar]
- 28.Thomas C, et al. Histopathologie−Lehrbuch und Atlas zur Befunderhebung und Differenzialdiagnostik. 14th Ed. Schattauer; Stuttgart: 2006. pp. 188–212. [Google Scholar]
- 29.Kastner J, Plank B, Heinzl C. Proceedings of Digital Industrial Radiology and Computed Tomography. Ghent BC Administratieve informatie, Ghent; Belgium: 2015. Advanced X-ray computed tomography methods: High resolution CT, quantitative CT, 4DCT and phase contrast CT; pp. 120–132. [Google Scholar]
- 30.Nachtrab F, et al. Development of a Timepix based detector for the NanoXCT project. J Instrum. 2015;10:C11009. [Google Scholar]
- 31.Kraft P, et al. Performance of single-photon-counting PILATUS detector modules. J Synchrotron Radiat. 2009;16:368–375. doi: 10.1107/S0909049509009911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft P, et al. Characterization and calibration of PILATUS detectors. IEEE Trans Nucl Sci. 2009;56:758–764. [Google Scholar]
- 33.Lucy LB. An iterative technique for the rectification of observed distributions. Astron J. 1974;79:745–765. [Google Scholar]
- 34.Richardson WH. Bayesian-based iterative method of image restoration. J Opt Soc Am. 1972;62:55–59. [Google Scholar]
- 35.Paganin D, Mayo SC, Gureyev TE, Miller PR, Wilkins SW. Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J Microsc. 2002;206:33–40. doi: 10.1046/j.1365-2818.2002.01010.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.