Significance
On the basis of the amino acid sequence, a polypeptide folds into a unique structure. In contrast, aggregation-prone proteins often misfold into distinct aggregate conformations. Interestingly, each distinct aggregate conformation can dictate different phenotypic consequences, such as cellular toxicity. However, the underlying mechanism of the origin of such structural diversity of protein aggregates has been a critical open question. Using the intrinsically disordered domain of yeast prion protein Sup35, we revealed the structural diversification of prion aggregate is triggered by thermodynamic fluctuations within the monomeric protein prior to aggregation. Our findings not only provide a clue to explain previously puzzling observations in prion biology but also have important implications for therapeutic approaches for human diseases in which protein aggregation is involved.
Keywords: yeast prion, amyloid, protein misfolding, protein dynamics, aggregate
Abstract
Self-propagating β-sheet–rich fibrillar protein aggregates, amyloid fibers, are often associated with cellular dysfunction and disease. Distinct amyloid conformations dictate different physiological consequences, such as cellular toxicity. However, the origin of the diversity of amyloid conformation remains unknown. Here, we suggest that altered conformational equilibrium in natively disordered monomeric proteins leads to the adaptation of alternate amyloid conformations that have different phenotypic effects. We performed a comprehensive high-resolution structural analysis of Sup35NM, an N-terminal fragment of the Sup35 yeast prion protein, and found that monomeric Sup35NM harbored latent local compact structures despite its overall disordered conformation. When the hidden local microstructures were relaxed by genetic mutations or solvent conditions, Sup35NM adopted a strikingly different amyloid conformation, which redirected chaperone-mediated fiber fragmentation and modulated prion strain phenotypes. Thus, dynamic conformational fluctuations in natively disordered monomeric proteins represent a posttranslational mechanism for diversification of aggregate structures and cellular phenotypes.
Formation of protein aggregates including amyloid fibers is widely observed in a variety of organisms, including microorganisms and vertebrates, and is often associated with human diseases (1, 2). In contrast to the fact that polypeptides fold into unique native structures, amyloidogenic proteins frequently misfold into distinct aggregate conformations which can dictate different physiological consequences, such as cellular toxicity or tissue specificity (3–5). A classic example of this phenomenon is prion strains where prion protein particles apparently composed of the same protein lead to phenotypically distinct infectious and transmissible states (6–8). Previous studies show that distinct prion strains are caused by structural polymorphism in infectious prion amyloid (9–11). For example, amyloid fibers of Sup35NM, the intrinsically disordered N-terminal and middle (M) domain of yeast prion protein Sup35, the protein determinant of [PSI+] prion states (7, 8), adopt distinct conformations at 4 °C (WT-4) and 37 °C (WT-37), and introduction of these conformational variants into yeast cells leads to mitotically stable strong and weak [PSI+] strains, respectively (11, 12). Moreover, single point mutations in the amino acid sequence may alter the cellular phenotypes associated with the amyloidogenic protein. A single point mutation of Sup35 where serine 17 is replaced with arginine (S17R) induces characteristic sectoring (mitotically unstable), weak [PSI+] strains (13, 14), and a codon polymorphism at residue 129 in human prion protein causes clinical diversity in Creutzfeldt–Jakob disease (6, 15). While distinct cellular phenotypes are likely caused by conformational differences in protein aggregates, it has been a critical open question how distinct structures of protein aggregates such as prions are created from the same monomeric protein. One leading hypothesis is that the difference in aggregate conformations may originate from structural diversity and fluctuation within the monomeric protein. In particular, Sup35NM, which is intrinsically disordered, might show monomer conformational heterogeneity (16, 17). Nonetheless, it remains unknown whether or how genetic mutations or environmental factors alter the dynamics or conformational equilibrium of intrinsically disordered Sup35NM monomers to induce the formation of distinct amyloid conformations with different phenotypic effects.
Results
Sup35NM Can Misfold into Two Strikingly Distinct Amyloid Conformations.
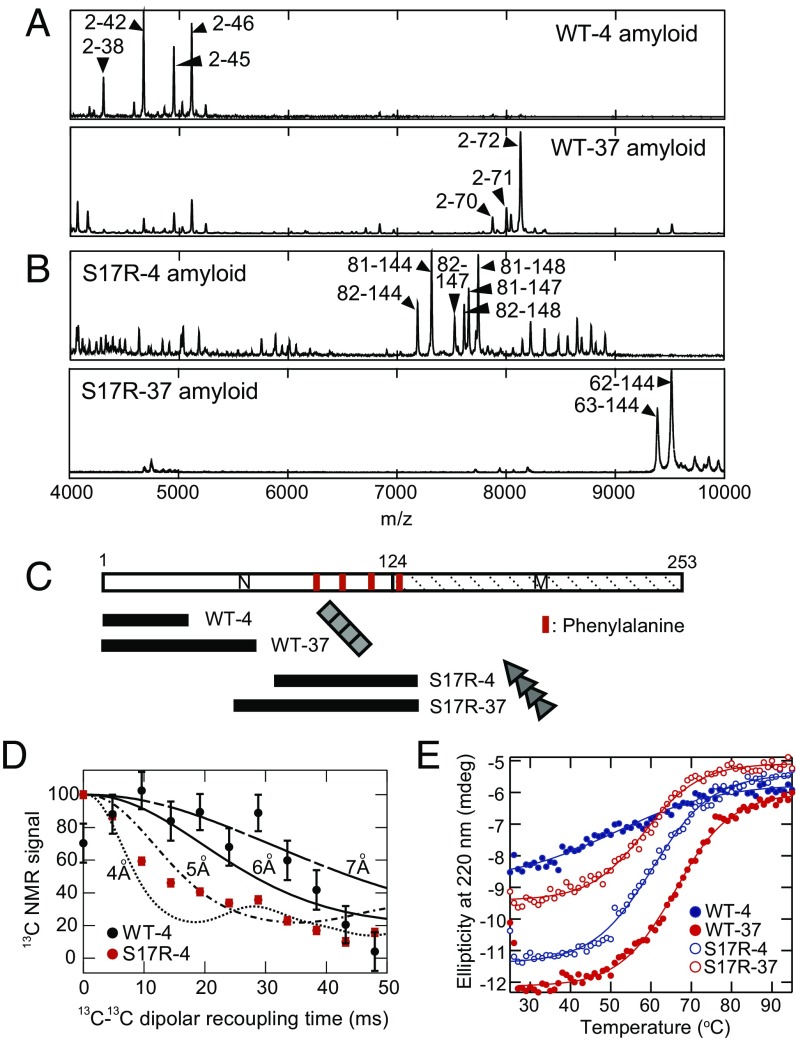
To address this question, we first established methods to identify amyloid core regions, which are critical determinants of prion strain phenotypes (18), by limited proteolysis with proteinase K followed by mass spectral analysis for high and low molecular weight regions. When we analyzed WT-4 and WT-37 amyloid core regions of WT Sup35NM, the main peptides in the high molecular weight region were assigned to the amino acid regions 2–38, 2–42, 2–45, and 2–46 for WT-4 amyloid and 2–70, 2–71, and 2–72 for WT-37 amyloid. (Fig. 1 A and C and Table S1). This result mirrored previous results by hydrogen/deuterium exchange NMR experiments (18). With mass spectrometry of proteinase K-treated fibrils, we remarkably found that the S17R amyloid formed at 4 °C (S17R-4) and 37 °C (S17R-37) showed core regions of amino acids 81, 82–144, 147, and 148 and amino acids 62–144 and 63–144, respectively (Fig. 1 B and C and Table S1).
Fig. 1.
Sup35NM can misfold into two strikingly distinct amyloid conformations. (A) High-m/z-range MALDI-TOF-MS spectra (m/z 4,000–10,000) of core peptides derived from WT-4 and WT-37 amyloids. Arrowheads indicate distinctive signals (amino acids 2–42/45/46 for WT-4 and amino acids 2–70/71/72 for WT-37). (B) High-m/z-range MALDI-TOF-MS spectra (m/z 4,000–10,000) of core peptides derived from S17R-4 and S17R-37 amyloids. Arrowheads indicate distinctive signals (amino acids 81/82–144/147/148 for S17R-4 and amino acids 62/63–144 for S17R-37). (C) Alignment of main core regions of WT and S17R amyloids detected by high-m/z-range MALDI-TOF-MS analysis. The red lines in the upper panel show the location of phenylalanine residues. (D) Solid-state NMR 13C–13C dipolar dephasing curves of Phe-1-13C–labeled amyloids of Sup35NM WT (black) and S17R (red) formed at 4 °C. Simulated finite-pulse RFDR-CT (fpRFDR-CT) curves are shown for ideal linear chains of 13C nuclei with distances of 4, 5, 6, and 7 Å. (E) Thermal disassembly of WT-4, WT-37, S17R-4, and S17R-37 amyloid fibers was monitored by the ellipticity at 220 nm with a CD spectrophotometer. Resulting plots were fitted by a sigmoidal curve for determination of Tm.
The unexpected existence of a C-terminal prion domain (PrD-C) amyloid core was further supported by solid-state NMR spectroscopy and proline-scanning analyses. Taking advantage of the fact that all four phenylalanines at positions 92, 104, 117, 129 are located in the PrD-C (Fig. 1C), we labeled Sup35NM WT and S17R proteins with Phe-1-13C, formed amyloid fibers, and measured 13C–13C dipolar dephasing by constant-time RF-driven dipolar recoupling (CT-RFDR) of the amyloid by solid-state magic angle spinning (MAS) NMR spectroscopy (Fig. 1D) (19, 20). The S17R fibrils labeled with Phe-1-13C showed a rate of signal decay characteristic for a distance of 4.5−5.0 Å to the nearest neighbor for all labeled Phe residues. This result indicated that all four Phe residues must be located in parallel, in-register β-sheets and thus confirmed the nature of the PrD-C core of S17R amyloid. In-register parallel β-sheet structures had been observed for WT fibrils with the N-terminal core before (19, 20). Furthermore, we performed proline scanning with thioflavin T (ThT) fluorescence, in which various single-proline Sup35NM mutants were polymerized in the presence of WT-4, WT-37, S17R-4, or S17R-37 seeds, and examined whether the replacement with proline, which cannot adopt β-sheet conformations, in Sup35NM monomers impairs seeding activity (Fig. S1A). We observed that mutational positions which showed seeding defects matched the amyloid core region identified by mass spectral analysis (Fig. S1 B and C). These results demonstrated that our method of limited proteolysis and mass spectrometry readily identified amyloid core regions without assignment of NMR signals of dimethyl sulfoxide-denatured protein (18).
The conformational differences between WT and S17R amyloids were further investigated by the analyses of thermal stability using circular dichroism (CD) spectroscopy. The CD spectrophotometer monitored changes of the ellipticity at 220 nm of amyloid solutions in the presence of SDS upon the increase of temperature from 25 °C to 95 °C. This experiment reports on amyloid disassembly as evidenced by the spectral changes from β-sheet/random coil-containing amyloid structures to random coil-rich monomer structures (Fig. S1D). Notably, the low secondary structure content estimated from the CD spectra of Sup35NM amyloid indicates that Sup35NM amyloid is constituted by both β-sheet–rich amyloid core and random coil regions. The Tm values calculated from the melting curves were different in WT-4 and WT-37 amyloids (WT-4: 55.7 ± 1.2, WT-37: 64.8 ± 0.3) but were similar in S17R-4 and S17R-37 amyloids (S17R-4: 57.9 ± 0.5, S17R-37: 56.2 ± 0.5) (Fig. 1E). Such different characteristics of Tm in WT and S17R amyloids are consistent with their distinct amyloid core regions (Fig. 1 A–C). Furthermore, our atomic force microscopy (AFM) analysis revealed that the S17R-4 and S17R-37 amyloid fibers have the propensity to exhibit more periodically twisted morphology than WT-4 and WT-37 fibrils, respectively (Fig. S2), implying that the WT and S17R amyloid structures are different. Finally, to examine the physiological consequences of the S17R amyloid, we performed a protein infection assay (11). The infectivities of in vitro-generated S17R-4 and S17R-37 amyloids to nonprion [psi−] yeast were 2.8% and <0.1%, respectively, which were much lower than those of WT-4 and WT-37 amyloids (77.7% and 20.5%, respectively) (Fig. S1E). In most cases (>80%), the resulting [PSI+(S17R)] strains showed sectoring/weak phenotypes (Fig. S1F), indicating unstable prion transmission, as previously reported (13, 14). Collectively, these results support the notion that the S17R mutation induces an amyloid structure that is markedly different from the amyloid conformation of WT Sup35NM.
WT Sup35NM Can Adopt the PrD-C Amyloid Core.
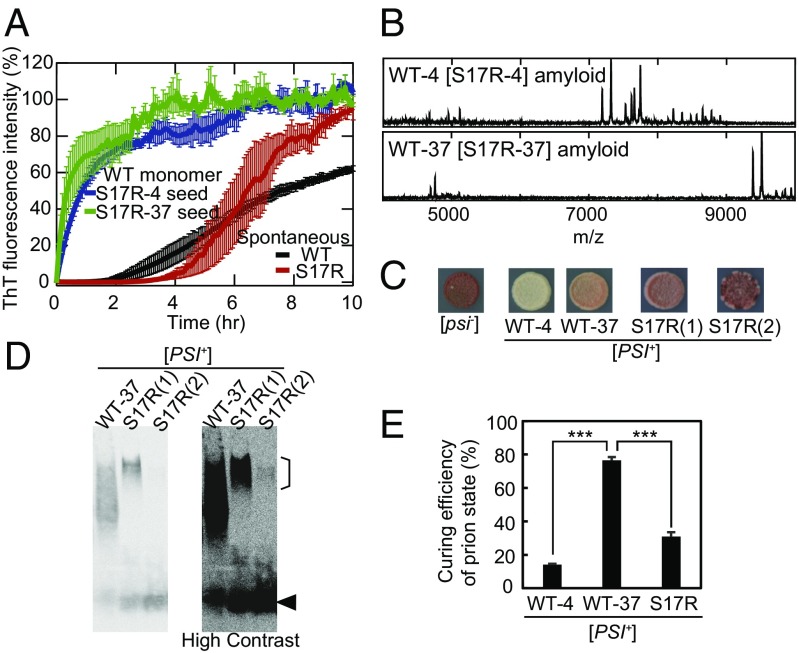
To examine whether WT Sup35NM is able to adopt the S17R-type PrD-C amyloid core, we polymerized WT Sup35NM monomer in the presence of S17R amyloid seeds [5% (mol/mol)] and examined the resulting amyloid structures. First, we found that the S17R amyloid efficiently seeded WT Sup35NM (Fig. 2A), suggesting that WT Sup35NM is able to form the S17R-type amyloid conformation. Importantly, using the WT fibrils that were formed with S17R seeds [5% (mol/mol)] (denoted as “WT[S17R]”), we confirmed this hypothesis by the peaks in the high-m/z region of mass spectra (Fig. 2B and Table S1) and the low infectivity accompanied by the appearance of sectoring/weak [PSI+] phenotypes in protein infection assays (Fig. S3 A and B), both of which are quite similar to the results obtained with S17R amyloid (Fig. S1 E and F). Furthermore, we found that the addition of 1 M sodium chloride to the amyloid formation buffer induced a S17R-type amyloid conformation even for WT Sup35NM (Fig. S3C and Table S1). In addition, the Sup35NM mutant lacking residues 2–34 (Δ2–34) could form an amyloid conformation with a PrD-C core (Fig. S3D and Table S1). These results established that WT Sup35NM has the intrinsic potential to form two strikingly different amyloid conformations, either of which is eventually selected depending on amino acid sequences and/or solvent conditions for amyloid formation.
Fig. 2.
Wild-type Sup35NM can adopt the PrD-C amyloid core. (A) Kinetics of spontaneous WT and S17R amyloid formation and seeding reaction with WT monomer and S17R-4 and S17R-37 seeds monitored by ThT fluorescence at 25 °C. Error bars denote SEM. (B) High-m/z-range MALDI-TOF-MS spectra (m/z 4,000–10,000) of core peptides derived from cross-seeded WT[S17R] amyloid formed at 4 °C (WT-4[S17R-4]) and 37 °C (WT-37[S17R-37]). (C) Weak/sectoring phenotypes of two examples of [PSI+(S17R)] strains obtained by overexpression of the Sup35NM S17R mutant in [psi−] yeast. Phenotypes of [PSI+(WT-4)] and [PSI+(WT-37)] strains and [psi−] yeast are also shown as controls. (D) SDD-AGE analysis with yeast lysates of [PSI+(WT-37)] and [PSI+(S17R)] strains probed by an anti-Sup35NM antibody. A high-contrast image is shown on the right. An arrowhead and a line in the high molecular region indicate the positions of monomeric Sup35NM and S17R-type prions, respectively. Note that [PSI+(S17R (2))] strains also contain a large size of prion aggregates. (E) Efficiency of [PSI+]-state curing of [PSI+(WT-4)], [PSI+(WT-37)], and [PSI+(S17R)] strains by Hsp104 overexpression. [PSI+(S17R)] strains were generated by in vitro infection of Sup35NM S17R-4 or S17R-37 amyloid fibers in [psi−] yeast. The curing efficiency of [PSI+(S17R)] is an average of the data from the two [PSI+(S17R)] strains. Error bars denote SEM. ***P < 0.001 by a Student’s t test; n ≥ 3.
How does the PrD-C core of S17R amyloid induce the characteristic sectoring phenotypes in [PSI+(S17R)] strains which are obtained when the GFP-fused S17R mutant is overexpressed (13) or when in vitro-generated S17R amyloid is introduced in [psi−] yeast (Fig. 2C and Fig. S1F)? The mitotically unstable/sectoring phenotypes of [PSI+(S17R)] strains suggest reduced chaperone-mediated fiber fragmentation. If this is the case, we would expect to observe an increased size of prion aggregates in [PSI+(S17R)] strains. Therefore, we examined the size of prion aggregates in two representative weak [PSI+(S17R)] strains as well as weak [PSI+(WT-37)] strains (Fig. 2 C and D). We employed a semidenaturing detergent agarose gel electrophoresis (SDD-AGE) (21) to address it, since SDD-AGE previously revealed a difference in the prion size between [PSI+(WT-4)] and [PSI+(WT-37)] strains (12). As we expected, the average size of [PSI+(S17R)] prions was larger than that of WT-37 prions (Fig. 2D). Importantly, the increased prion size did not result from enhanced seeding extension reactions of [PSI+(S17R)] prions, as the seeding activity of S17R amyloid to WT monomer was similar to that of WT amyloid to WT monomer at 30 °C in vitro (Fig. S3E). These results suggested impairment of chaperone-mediated fragmentation of [PSI+(S17R)] prion aggregates, although the fragmentation defect was not so severe as to eliminate the [PSI+(S17R)] prion state. Since Hsp70/Hsp40 chaperones first bind to prion aggregates and then recruit Hsp104 (22), an AAA+ ATPase chaperone essential to prion propagation (23), our observations imply that the PrD-C region, which is buried in [PSI+(S17R)] prion aggregates, partly includes a binding site of Hsp70/Hsp40.
A recent study showed that residues 129–148 in Sup35 are involved in binding to overexpressed Hsp104 (24). Interestingly, the PrD-C core region of S17R amyloid overlaps with this binding site, suggesting that the curing of [PSI+(S17R)] strains by Hsp104 overexpression may be compromised due to residues 80–148 being buried in the amyloid core. Indeed, we found that [PSI+(S17R)] strains were more resistant to [PSI+] curing by Hsp104 overexpression than [PSI+(WT-37)] strains (Fig. 2E), although the [PSI+(S17R)] strains had been expected to be cured more easily due to their larger size and smaller amounts of prions than the [PSI+(WT-37)] strains (Fig. 2D). Overexpressed Hsp104 is suggested to bind to Sup35 prion aggregates in the initial step of [PSI+] curing, which prevents Hsp70/Hsp40 from having access to prion aggregates, resulting in a loss of [PSI+] states (24). The acquired resistance of [PSI+(S17R)] yeast to the curing would be caused by impaired binding of overexpressed Hsp104 to the [PSI+(S17R)] prions, as observed for Δ129–148 Sup35 prions (24).
Sup35NM Monomers Form Compact Local Structures with Long-Range Interactions.
We asked how such a dramatic difference in amyloid conformation might be caused by the single S17R mutation although both WT and S17R Sup35NM monomers have similar disordered structures (Fig. S4A). To address this question at amino acid resolution, we performed a comprehensive assignment of protein backbone 1H-15N NMR chemical shifts of Sup35NM. This had been a challenge, because Sup35NM is a relatively large protein (253 residues) for signal assignment, the aggregation-prone property limits the measurement time of NMR, and both the intrinsically disordered structure and the presence of 5.5 degenerate oligopeptide repeats cause severely impaired signal separation. Nonetheless, we completed the amino acid assignment of more than 90% of 1H-15N HSQC signals for the Sup35NM monomer.
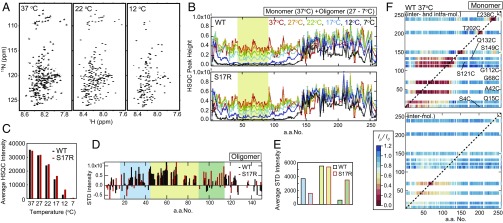
First, we investigated the amino acid residues required for oligomer formation by NMR, since oligomers play an important role in determining amyloid conformation (25). We acquired 1H-15N HSQC spectra at several temperatures from 37 °C to 7 °C and quantified temperature-dependent changes in NMR signal intensities (Fig. 3 A and B). While analytical ultracentrifugation showed that Sup35NM under the NMR measurement condition is solely monomeric at 37 °C (Fig. S4B), the temperature-dependent reduction of NMR signal intensities from 37 °C to 7 °C implies protein assembly due to decreased tumbling rates of protons (Fig. 3 B and C). This result is consistent with our previous observation that Sup35NM is a monomeric form at 37 °C but forms oligomers at low temperatures (25). In particular, residues 45−90 showed a gradual reduction in signal intensity as the temperature decreased (yellow area in Fig. 3B), indicating that these residues are involved in oligomer formation. We observed a slight decrease in the amount of oligomers in the S17R mutant, but no major difference between WT and the S17R mutant was detected in the amino acid region required for oligomer formation (Fig. 3 B and C).
Fig. 3.
Sup35NM monomers form latent compact local structures with long-range interactions. (A) Temperature-dependent changes of 1H-15N HSQC spectra of WT Sup35NM. (B) Signal intensities for 1H-15N cross-peaks in an HSQC spectrum of WT and S17R Sup35NM at 37 °C (red), 27 °C (orange), 22 °C (green), 17 °C (cyan), 12 °C (blue), and 7 °C (black). The yellow area indicates the amino acid region with significantly reduced signal intensities at low temperature, indicating the oligomer core region. (C) Temperature-dependent change of average 1H-15N HSQC signals in the yellow area in B (amino acids 45–90). (D) STD signal intensity of the N domain of WT (black) and S17R Sup35NM (red) at 22 °C. The blue and green areas indicate the amino acid regions with increased STD signals specific to WT and S17R Sup35NM, respectively. (E) Averaged STD signal intensities in the blue (amino acids 15–44), yellow (amino acids 45–90), and green (amino acids 91–115) areas in D are indicated by blue, yellow, and green bars, respectively. (F) Paramagnetic relaxation enhancement of WT Sup35NM monitored by 1H-15N HSQC signal intensity. A MTSL spin probe was introduced into each selected single-cysteine Sup35NM mutant (S4C, Q15C, A42C, G68C, G112C, S121C, Q132C, S149C, T202C, and L238C). Inter/intramolecular (Upper) and intermolecular (Lower) PRE values at 37 °C are shown as a color scale showing the ratio of the HSQC intensity of PRE measurement to the HSQC intensity of control measurement (Ip/I0 ratio). The red color of high PRE values indicates the presence of interactions between a MTSL spin probe and an amino acid residue of interest.
Next, we examined oligomer structures by saturation transfer difference (STD) NMR spectroscopy at 22 °C, the temperature at which both Sup35NM monomer and oligomer are populated (Fig. 3C). By mixing unlabeled (1H14N) Sup35NM with 2H15N-labeled Sup35NM proteins and saturating the 1H signals in unlabeled Sup35NM, STD signals can be observed only if the two molecules directly interact with each other due to the transfer of magnetization from excited unlabeled Sup35NM to 2H15N-labeled Sup35NM. Therefore, the STD measurements provide information on monomer–monomer contacts in oligomers. Fig. 3D shows the STD signal intensities in the N-terminal prion domain and a part of the M domain, which are involved in prion propagation (26). As expected from the results shown in Fig. 3B, residues 45–90, which are located in the oligomer core region, showed STD signals for both the WT and the S17R mutant (yellow area in Fig. 3 D and E and Fig. S5A). Interestingly, we found that amino acid regions 15−44 and 91−115 specifically showed STD signals in the WT and S17R mutant, respectively (blue and green areas in Fig. 3 D and E and Fig. S5A). This result indicated that the different regions are involved in intermolecular interactions and thus may represent initiation sites for amyloid formation in WT and S17R mutant Sup35NM, respectively. In addition, the STD analysis showing that the amino acid residues required for intermolecular interactions are different in WT and S17R mutant Sup35NM implies that their nucleus structures are distinct from each other.
Furthermore, we measured 1H-15N heteronuclear Overhauser effects (NOE) at 37 °C (Fig. S5B), the temperature at which Sup35NM is populated only in a monomeric state (Fig. S4B), to examine structural fluctuation of Sup35NM monomer. Remarkably, the positive heteronuclear NOE values in the region of residues 30–110 suggested the presence of a locally compact structure (Fig. S5B) despite the overall disordered Sup35NM conformation (Fig. S4A). In contrast, negative heteronuclear NOE values throughout the M domain showed that the M domain is mostly unfolded (Fig. S5B). We performed further NMR experiments that report on two different time scale dynamics, T1 and T2 relaxation time measurements (nanoseconds and picoseonds) (Fig. S5 C and D) and paramagnetic relaxation enhancement (PRE) (microseconds) experiments with a paramagnetic spin probe, 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate (MTSL), which was specifically introduced into a variety of single-cysteine Sup35NM mutants (Fig. 3F and Fig. S5E). The shorter relaxation times and the large PRE effects in the N-terminal prion domain revealed the presence of local compact structures by long-range interactions, while the longer relaxation times and the absence of PRE effects in the M domain showed that the M domain is largely unfolded. Taken together, these results show that the prion domain in Sup35NM adopts local compact structures by long-range interactions despite its overall intrinsically disordered structure (Fig. S4A); this was hypothesized in a previous single-molecule FRET study (17), but its biological significance had not been fully addressed.
Exposed Asparagine Residues in Sup35NM Monomers Are Scaffolds for Amyloid Formation.
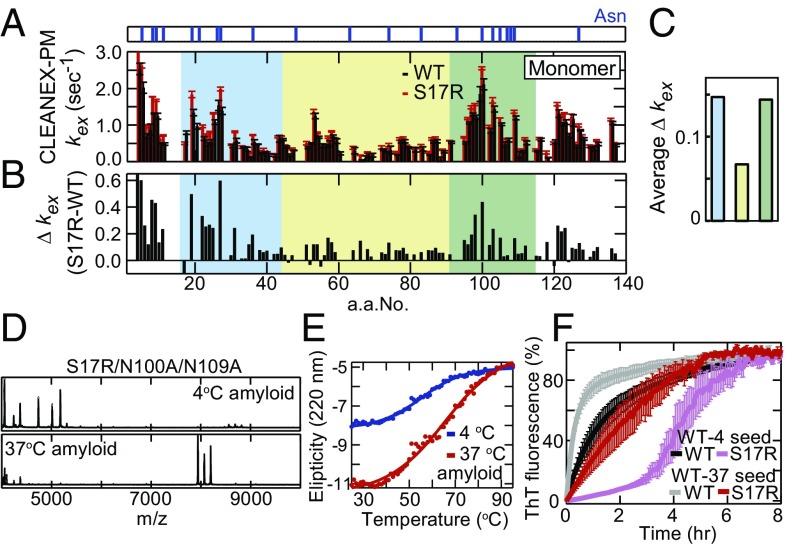
The fast time-scale protein dynamics was not significantly different between Sup35NM WT and the S17R mutant by our NMR analyses with PRE, T1, and T2 measurements (Fig. 3F and Fig. S5 C–E). Therefore, to gain insights into slow time-scale dynamics (in milliseconds) of the Sup35NM monomer at 37 °C, we applied phase-modulated CLEAN chemical exchange (CLEANEX-PM) NMR spectroscopy which allows us to examine backbone amide 1H exchange rates with the solvent protons and thereby identify which residues are solvent-exposed or buried/hydrogen-bonded (27). We revealed that most of the amino acids in the prion domain of the S17R mutant monomer showed higher exchange rates than those in the WT monomer (Fig. 4 A–C and Fig. S5F), indicating that the local compact structure of the S17R mutant monomer is more unfolded than that of the WT monomer. Therefore, the S17R mutation altered the conformational space of the Sup35NM monomer, resulting in an increased population of the monomer conformation containing a less compact prion domain.
Fig. 4.
Exposed asparagine residues outside the compact region in disordered Sup35NM monomers serve as a scaffold for amyloid formation. (A) Hydrogen exchange rates of the N domain of WT (black) and S17R Sup35NM (red) at 37 °C measured by CLEANEX-PM NMR. The positions of asparagine residues are shown as blue lines in the upper panel. (B) Difference in the exchange rates (kex S17R – kex WT) between WT and S17R Sup35NM. Color coding is as in Fig. 3D. (C) Averaged differences in the exchange rates in amino acids 15–44, 45–90, and 91–115 are indicated by blue, yellow, and green bars, respectively. (D) High-m/z-range MALDI-TOF-MS spectra (m/z 4,000–10,000) of core peptides derived from S17R/N100A/N109A amyloid formed at 4 °C and 37 °C. (E) Thermal disassembly of S17R/N100A/N109A amyloid fibers formed at 4 °C (blue) and 37 °C (red) was monitored by the ellipticity at 220 nm with a CD spectrophotometer. (F) Kinetics of amyloid formation of WT (black) and S17R (pink) monomers in the presence of WT-4 seeds and WT (gray) and S17R (red) monomers in the presence of WT-37 seeds at 25 °C. Error bars denote SEM.
Interestingly, the asparagine residues located at both ends (residues 15–44 and 91–115) of the local compact region in the Sup35NM monomer showed significantly higher exchange rates in the S17R mutant (blue and green areas in Fig. 4 A–C), indicating that specific asparagine residues are more solvent-exposed in the S17R mutant than in the WT monomer. To examine effects of the exposed asparagine residues (Asn8, 19, 26, and 27 in the N-terminal core region and Asn100, 103, 105, and 109 in the C-terminal core region) on the formation of the WT and S17R-type amyloid cores, we prepared a range of Sup35NM mutants in which single or a few exposed asparagine residues were replaced with alanine together with or without the S17R mutation. For asparagine mutants in the N-terminal core region, we found that the N8A/N19A/N26A mutant partially acquired the PrD-C amyloid core at 4 °C (Fig. S6 A and B). Remarkably, we further found that the triple mutant S17R/N100A/N109A restored the WT-type N-terminal amyloid core both at 4 °C and 37 °C (Fig. 4D and Table S1), and the mutant amyloids which formed at 4 °C and 37 °C showed melting curves similar to WT-4 and WT-37 amyloids, respectively (Fig. 4E). Importantly, the S17R/N100A/N109A mutant still has the capability to form a PrD-C amyloid core when seeded by S17R amyloid (Fig. S6C and Table S1), excluding the possibility that the two N100A/N109A mutations introduced into the S17R mutant exert their effects on the amyloid state simply by preventing the mutant from having access to the PrD-C core. These results show that the exposed Asn100 and Asn109 residues are required for the S17R mutant to form the PrD-C amyloid core.
If the S17R mutant is more unfolded and both potential amyloid core regions are exposed, why does the S17R mutant selectively adopt the PrD-C amyloid core but not the N-terminal core? The STD data revealed that intermolecular interactions of S17R monomers resulted from the contacts between amino acids 91–115, suggesting that Arg17 in the S17R mutant reduced intermolecular interactions involving amino acids 15–44 while increasing intermolecular contacts involving amino acids 91–115 (Fig. 3 D and E). Consistently, the seeding assay showed that S17R monomers are incorporated into the WT-4 or WT-37 amyloid seeds, albeit more slowly than WT monomers (Fig. 4F), indicating that the S17R mutant still has the capability to adopt (Fig. S6D and Table S1) but does not prefer to form an N-terminal amyloid core. These factors would allow the S17R mutant to eventually select a PrD-C amyloid core upon aggregation.
Discussion
In this study, we revealed that Sup35NM is able to form two strikingly different amyloid conformations that can be selectively propagated and result in alternate strain phenotypes. These conformational subtypes of amyloid derive from the disinhibition of local compact structures of monomeric Sup35NM. Our comprehensive NMR analysis of Sup35NM at amino acid resolution indicates that exposed asparagine residues in the Sup35NM monomer serve as initiation sites for amyloid elongation and constitute an amyloid core. In WT Sup35NM, Asn100 and Asn109 are buried in the local compact structure, and thereby the N-terminal amyloid core region is selected (Fig. S7). In contrast, the S17R mutation alters the conformational equilibrium of monomer and increases the population of an unfolded form with exposed asparagine residues in the C-terminal region of the prion domain. The exposure of Asn100/Asn109 and the preference of the intermolecular contacts of the PrD-C region allow the S17R mutant to select Asn100 and Asn109 as an initiation site for amyloid formation (Fig. S7). The altered intermolecular contacts due to the unfolded nature of the S17R Sup35NM monomer would result in the formation of different nucleus structures, which could also contribute to the selection of the PrD-C amyloid core. Taken together, our results highlight that exposed asparagine residues at the edge of the local compact region in Sup35NM monomer play pivotal roles in determining Sup35 amyloid conformation and suggest that the unfolding of the local compact structure by the S17R mutation triggers selective formation of the PrD-C amyloid core.
A previous report showed that Ser17 is involved in a tight-turn structure in Sup35NM fibrils and thus is located at a strategic position (28). Importantly, the selection of the C-terminal amyloid core of the S17R mutant is not caused simply by its inability to accommodate the bulky and charged Arg17 residue into an N-terminal amyloid core. We revealed that both the triple S17R/N100A/N109A mutant and the single S17R mutant have the capability to adopt an N-terminal amyloid core (Fig. 4D and Fig. S6D and Table S1). This result shows that such a charged and/or bulky residue at the strategic position 17 can be accommodated into an N-terminal amyloid core. Furthermore, we found that the mutation of Ser17 to either bulky tryptophan or tyrosine, which might destabilize the tight-packing structures within the amyloid core, maintained the formation of an N-terminal amyloid core (Fig. S6 E and F and Table S1). Together, these results show that potential destabilization of the N-terminal core by Arg17 would not be a major factor in the formation of the PrD-C amyloid core in the S17R mutant. Rather, our data suggest that disruption of the latent compact structure in the S17R monomer and resulting exposure of Asn100 and Asn109 in the monomeric state are involved in the selection of the PrD-C amyloid core in the S17R mutant. The PrD-C amyloid core would decrease the efficiency of chaperone-mediated fiber fragmentation, resulting in the large prion size and weak/sectoring (mitotically unstable) phenotypes of [PSI+(S17R)] strains. This result agrees well with our previous finding that fiber fragmentation rates have a greater impact on prion strain phenotypes (11). The present study indicates that dynamic structural fluctuation in natively disordered monomeric proteins is involved in the diversification of amyloid conformations dictating distinct physiological consequences.
Notably, the prion strain conformation harboring a PrD-C core that we showed in this study provides a molecular explanation for many previous findings whose underlying mechanisms had remained elusive. First, it is suggested that the first 137 amino acids of Sup35 are required for faithful maintenance of weak [PSI+] strains (26). This observation agrees well with the PrD-C core region of the S17R-type weak prion strain conformation. Furthermore, it should be noted that protein fluctuation of Sup35NM monomer is dramatically increased from residue 140 (Fig. S5 B–D), which may be associated with the C-terminal end position (residues 144–148) (Table S1) of the PrD-C amyloid core. Next, the Sup35 mutant lacking a region between positions 22 and 69 forms highly unstable prions in vivo (29). The deletion of residues 22–69 is likely to disrupt the latent local compact structure, potentially generating a PrD-C amyloid core upon aggregation of the deletion mutant. In addition, a wild-type [PSI+] variant that is insensitive to excess Hsp104 for its curing has been reported (30), and chaotropic anions including Cl− favor the formation of weaker Sup35NM prion variants (31). These observations could result from a prion strain conformation harboring a PrD-C core.
Furthermore, our results resolve the long-standing, puzzling observation that Sup35NM forms amyloid in a head-to-head and tail-to-tail manner (32) although this amyloid conformation may not reconcile with the unique N-terminal amyloid core demonstrated by hydrogen/deuterium exchange NMR experiments (18) or the in-register parallel structure demonstrated by solid-state NMR spectroscopy (19, 20). Rather, our findings suggest that Sup35NM may be able to adopt both N-terminal and PrD-C amyloid core regions simultaneously, since solvent conditions or protein modification by chemicals such as pyrene could modulate the conformational equilibrium of Sup35NM monomers. The present study underscores the critical role of conformational space in natively disordered proteins for determination of amyloid conformation. Therefore, the regulation of conformational fluctuation can be a target for therapeutic intervention of pathologic protein aggregates. More broadly, our findings suggest that structural fluctuation in intrinsically disordered monomeric proteins, and its controlling factors, can account for individual variation in cellular and organism-wide behaviors, adding to posttranslational mechanisms for phenotypic diversification in biological systems.
Methods
Amyloid Formation.
Sup35NM amyloid formed spontaneously in 5 mM potassium phosphate buffer including 150 mM NaCl at pH 7.4 (buffer C) with mild agitation [by eight end-over-end rotations per minute (Labquake, Theromo Fisher Scientific)] within 24 h. The second generation (G2) of amyloid is formed by polymerization of Sup35NM in the presence of 5% (mol/mol) sonicated amyloid that was spontaneously formed without agitation within several hours, as previously described (11–13). Spontaneously formed amyloids were used in kinetics experiments for amyloid formation, and G2 or third-generation (G3) amyloids were used in the other amyloid experiments.
NMR Measurement of HSQC Spectra.
Lyophilized Sup35NM was dissolved in 50 mM Mes buffer including 10% D2O (pH 5.0). After filtration by a 100-kDa-cutoff spin filter, Sup35NM concentration and pH were adjusted to 100 μM and pH 5.2. HSQC spectra were acquired with an Avance III 600 spectrometer equipped with a cryogenic probe. The NMR data were processed by TopSpin (Bruker BioSpin) or XWINNMR (Bruker BioSpin) and analyzed by SPARKY (https://www.cgl.ucsf.edu/home/sparky).
Supplementary Material
Acknowledgments
We thank Naomi Takahashi for preparation of reagents; Drs. Herry Martadinata, Santhosh Ayalur Karunakaran, Manuel Etzkorn, Vadim Zorin, and the members of the M.T. laboratory for discussions; and Charles Yokoyama for critical reading and editing of the manuscript. DNA sequencing and mass spectrometry were performed by the RIKEN Brain Science Institute Research Resources Center. Funding was provided by the Next Program Grant LS129 from the Japan Society of the Promotion of Science (to M.T.); Grant-in-Aid for Scientific Research on Innovative Area “Molecular Science of Fluctuations toward Biological Functions” 23107734 (to Y.O.); Grant-in-Aid for Young Scientists (B) 23770193 (to Y.O.); Grant-in-Aid for Scientific Research (B) 15H04345 (to M.T.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and Grants-in-Aid from the Ministry of Health, Labour, and Welfare, Japan (the Research Committee of Prion Disease and Slow Virus Infection) (Y.O., K.K., and M.T.), RIKEN Pioneering Projects, Cellular Evolution (M.T.), and the Mochida Foundation (M.T.). This work was also funded in part by the Heinrich Heine International Graduate School of Protein Science and Technology of the Heinrich Heine University Dusseldorf and a Forschungszentrum Julich Germany Fellowship (T.P.). B.U. was supported by the International Helmholtz Research School of Biophysics and Soft Matter.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu/ (accession no. 12016).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715483115/-/DCSupplemental.
References
- 1.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Komi Y. Layers of structure and function in protein aggregation. Nat Chem Biol. 2015;11:373–377. doi: 10.1038/nchembio.1818. [DOI] [PubMed] [Google Scholar]
- 5.Tycko R. Amyloid polymorphism: Structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 7.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuite MF, Serio TR. The prion hypothesis: From biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 10.Legname G, et al. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 13.Chien P, DePace AH, Collins SR, Weissman JS. Generation of prion transmission barriers by mutational control of amyloid conformations. Nature. 2003;424:948–951. doi: 10.1038/nature01894. [DOI] [PubMed] [Google Scholar]
- 14.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 15.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 16.Marchante R, Rowe M, Zenthon J, Howard MJ, Tuite MF. Structural definition is important for the propagation of the yeast [PSI+] prion. Mol Cell. 2013;50:675–685. doi: 10.1016/j.molcel.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci USA. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 19.Shewmaker F, Kryndushkin D, Chen B, Tycko R, Wickner RB. Two prion variants of Sup35p have in-register parallel beta-sheet structures, independent of hydration. Biochemistry. 2009;48:5074–5082. doi: 10.1021/bi900345q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- 22.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 24.Helsen CW, Glover JR. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104) J Biol Chem. 2012;287:542–556. doi: 10.1074/jbc.M111.302869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6:225–230. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradley ME, Liebman SW. The Sup35 domains required for maintenance of weak, strong or undifferentiated yeast [PSI+] prions. Mol Microbiol. 2004;51:1649–1659. doi: 10.1111/j.1365-2958.2003.03955.x. [DOI] [PubMed] [Google Scholar]
- 27.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 28.Luckgei N, et al. The conformation of the prion domain of Sup35p in isolation and in the full-length protein. Angew Chem Int Ed Engl. 2013;52:12741–12744. doi: 10.1002/anie.201304699. [DOI] [PubMed] [Google Scholar]
- 29.Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borchsenius AS, Müller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet. 2006;49:21–29. doi: 10.1007/s00294-005-0035-0. [DOI] [PubMed] [Google Scholar]
- 31.Rubin J, et al. Ion-specific effects on prion nucleation and strain formation. J Biol Chem. 2013;288:30300–30308. doi: 10.1074/jbc.M113.467829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.