Significance
The biological consequences of climate change are determined by the responses of individual species and interactions among species. Hybridization, or interbreeding between related species, is an interaction that affects how species evolve in response to environmental change. Here we provide evidence that climatic warming has caused a geographic shift of a butterfly hybrid zone and that strong selection and/or genetic incompatibilities maintain species boundaries during this movement. Through simulations, we show that as climate change progresses, the rate and geographic configuration of future hybrid zone movement will vary across space and time. This geographic variation in future hybrid zone movement may lead to divergent ecological and evolutionary outcomes, and thus has implications for local conservation and management.
Keywords: climate change, hybridization, diapause, genomic, cline
Abstract
Climate-mediated changes in hybridization will dramatically alter the genetic diversity, adaptive capacity, and evolutionary trajectory of interbreeding species. Our ability to predict the consequences of such changes will be key to future conservation and management decisions. Here we tested through simulations how recent warming (over the course of a 32-y period) is affecting the geographic extent of a climate-mediated developmental threshold implicated in maintaining a butterfly hybrid zone (Papilio glaucus and Papilio canadensis; Lepidoptera: Papilionidae). These simulations predict a 68-km shift of this hybrid zone. To empirically test this prediction, we assessed genetic and phenotypic changes using contemporary and museum collections and document a 40-km northward shift of this hybrid zone. Interactions between the two species appear relatively unchanged during hybrid zone movement. We found no change in the frequency of hybridization, and regions of the genome that experience little to no introgression moved largely in concert with the shifting hybrid zone. Model predictions based on climate scenarios predict this hybrid zone will continue to move northward, but with substantial spatial heterogeneity in the velocity (55–144 km/1 °C), shape, and contiguity of movement. Our findings suggest that the presence of nonclimatic barriers (e.g., genetic incompatibilities) and/or nonlinear responses to climatic gradients may preserve species boundaries as the species shift. Further, we show that variation in the geography of hybrid zone movement could result in evolutionary responses that differ for geographically distinct populations spanning hybrid zones, and thus have implications for the conservation and management of genetic diversity.
Evidence from the fossil record and contemporary studies demonstrate that changes in climate profoundly alter species’ geographic distributions (1). Further, biotic interactions such as competition and predation influence how species’ distributions track changes in climate (1, 2). One species interaction that has received a great deal of attention in evolutionary biology, but less in the context of species responses to climate change, is hybridization. Although hybrid zones are ubiquitous in nature (3), we know very little about how these contact zones influence the way organisms respond to climate change. In fact, it has only recently been appreciated (and documented) that hybrid zones are mobile and that observed movement of these zones could act as biological indicators of climate change (4).
Here we leverage an ecologically well studied and historically well sampled hybrid zone between eastern (Papilio glaucus) and Canadian (Papilio canadensis) tiger swallowtail butterflies to explore whether recent climatic warming has resulted in hybrid zone movement. Divergence between these two species is estimated to have occurred 0.5–0.6 million years ago (5–7). Although there are a number of divergent physiological (8–10), behavioral (11–13), and morphological (14, 15) traits between these two species, the traits most strongly implicated in maintaining the location of this hybrid zone are those associated with seasonal adaptations, including diapause and growth (16). A climate-mediated developmental threshold (length of the growing season required for P. glaucus to complete two generations) is correlated with the northern boundary of P. glaucus and the southern range boundary of P. canadensis. Along this boundary, a narrow (<50 km) east–west hybrid zone forms, extending from Minnesota to New England (Fig. 1) (17). Geographic variation in climate (length of the growing season) appears to be a primary factor in maintaining the location of this hybrid zone by limiting the ability of genotypes with facultative diapause, a trait characteristic of the southerly species, P. glaucus, to complete a second generation. In multiple, independent portions of the hybrid zone (i.e., latitudinal transects from longitudinally distant regions of the hybrid zone), variation in genetic markers, physiological traits, and allozymes exhibit steep clines that coincide with this geographic gradient in climate (18–20), and not the distribution of other ecological factors, such as the distribution of differentially detoxified host plants or the Batesian model Battus philenor for which female P. glaucus are a mimic (16).
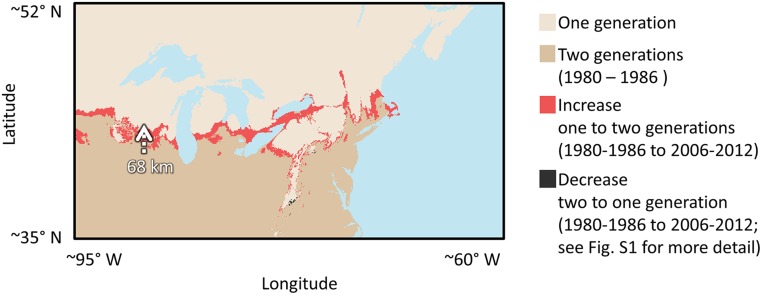
Fig. 1.
Simulated changes in the location of a climate-mediated developmental threshold and the P. glaucus–P. canadensis hybrid zone between historic (1980–1986) and contemporary (2006–2012) periods. Estimate of the spatial distribution of the ability of P. glaucus to complete two generations [implicated in maintaining the location of the hybrid zone (16, 21)], based on simulation modeling parameterized from growth chamber experiments. The highlighted arrow indicates the predicted 68-km northward expansion of this climate-mediated developmental threshold in Wisconsin between historic (1980–1986) and contemporary (2006–2012) periods.
Recently observed changes (increase) in the frequency of a physiological trait (Liriodendron tulipifera detoxification ability) and an allozyme (Pgd-100) north of the hybrid zone (21) suggest this hybrid zone may be shifting. To evaluate the potential for climate-mediated hybrid zone movement, we set out to first test whether recent warming (22) resulted in the expansion of the climate-mediated developmental threshold implicated in maintaining the location of this hybrid zone. To evaluate spatiotemporal changes in the geography of this climate-mediated developmental threshold during a 32-y period of documented warming, we constructed a simulation model parameterized with developmental data (from P. glaucus) collected from a series of growth chamber experiments. Then, we empirically measured hybrid zone movement during this same 32-y period, using a combination of morphological and molecular markers.
Empirical examination of dynamically shifting hybrid zones provides a unique opportunity to explore the interplay between reproductive isolation and climatic variation (23). Unfortunately, such evaluations are lacking, largely because of inadequate historical sampling (23, 24). Museum and contemporary specimens sampled across a latitudinal gradient spanning the hybrid zone in Wisconsin provided us with a rare opportunity to test predictions of hybrid zone movement, using male butterflies collected over the course of a 32-y period (1980–2012), both for two species diagnostic molecular markers (e.g., a Z-linked SNP in the gene Ldh and a restriction fragment-length polymorphism within the mtDNA gene COI) and one morphological marker that has been previously shown to exhibit steep clinal variation across the hybrid zone, and for a genome-wide random subsampling of single-nucleotide polymorphisms (SNPs) derived from double-digest restriction-associated DNA sequencing (ddRADseq) (25). The last allows for a comprehensive assessment of how the genomes in these two species are changing through time and an evaluation of the potential for the breakdown or buildup of reproductive barriers as a result of hybrid zone movement. All marker types were used in a geographic cline analysis to measure temporal changes in the location of the hybrid zone.
Whether species boundaries break down or build up as a consequence of hybrid zone movement will have implications for conservation efforts and the management of natural resources, given that changes in gene flow can dramatically alter the genetic composition of interbreeding species (26–28). The presence of exogenous barriers (ecologically mediated selection) or endogenous barriers (nonecologically mediated selection; e.g., genetic incompatibilities) should act to maintain species boundaries during hybrid zone movement. We evaluated the prediction that previously identified barriers, distributed across the genomes of P. glaucus and P. canadensis (16), should act to maintain the integrity of this hybrid zone during movement by comparing estimates of admixture between periods.
Finally, the geography (spatiotemporal variation) of climatic change is predicted to be a critical factor in species’ responses to climate change (29). For example, because of topographic effects (e.g., mountainous vs. flat landscapes), the velocity of temperature change may vary markedly across geography, with species’ survival dependent on their ability to keep pace; regions with higher velocities may be more vulnerable to climate change. To gain insight as to how this concept may be important to hybrid zones specifically, we simulated future hybrid zone movement by modeling the spatial distribution of the climate-mediated developmental threshold (as a surrogate for the location of the hybrid zone) under a range of plausible future warming conditions (30). Using this 30,000-ft view of (simulated) hybrid zone movement, we explore how spatiotemporal variation in climate alters the geography (velocity and shape) of hybrid zone movement and discuss how the geography of hybrid zone movement itself is dynamic and underappreciated.
Results and Discussion
Simulating Historical Hybrid Zone Movement.
We reared and phenotyped 1,153 individuals, derived from 40 families, to fit seven models to the developmental data of each of life stage (egg, larva, and pupa). We compared model performance of each fitted model to predict the development of each life stage, using an independent dataset of growth data from 844 individuals derived from 22 families. From these data, we determined that the Briere (31), Taylor (32)/Beta (33), and Logan10 (34)/Taylor models performed best (lowest root mean squared error; RMSE) for egg (RMSE, 1.19), larva (first/second generation, respectively; RMSE, 3.47/4.67), and pupal (direct-developing/diapaused, respectively; RMSE, 6.10/4.33) development, respectively (SI Appendix, Table S1). Using these best performing models to simulate insect development of P. glaucus, we predicted that a climate-mediated developmental threshold-limiting bivoltinism (i.e., the ability to complete two generations) expanded geographically from 1980 to 2012 (Fig. 1). Specifically, this climate-mediated developmental threshold is expanding northward by an average of 10.6 ± 13.1 km/y. This trend toward a lengthening growing season allows P. glaucus to complete an additional generation in localities where it was previously not possible. This result is in line with other predictions that climatic warming will increase voltinism in many insect species (35–37) and supports studies already documenting a similar response in other Lepidoptera (38). The predicted geographic movement of this climate-mediated developmental threshold varied substantially across latitude and elevational gradients (SI Appendix, Table S2). In some regions, voltinism was not predicted to have changed during the last 32 y, and was even predicted to decrease in a few locations (e.g., in portions of the Appalachian Mountains; Fig. 1 and SI Appendix, Fig. S1). The slower rate of movement of this climate-mediated developmental threshold in areas of higher elevation fits with predictions that mountainous biomes will experience the slowest climate velocities (29), potentially providing a refuge for some species from climate change (39).
Empirically Evaluating Historic Hybrid Zone Movement.
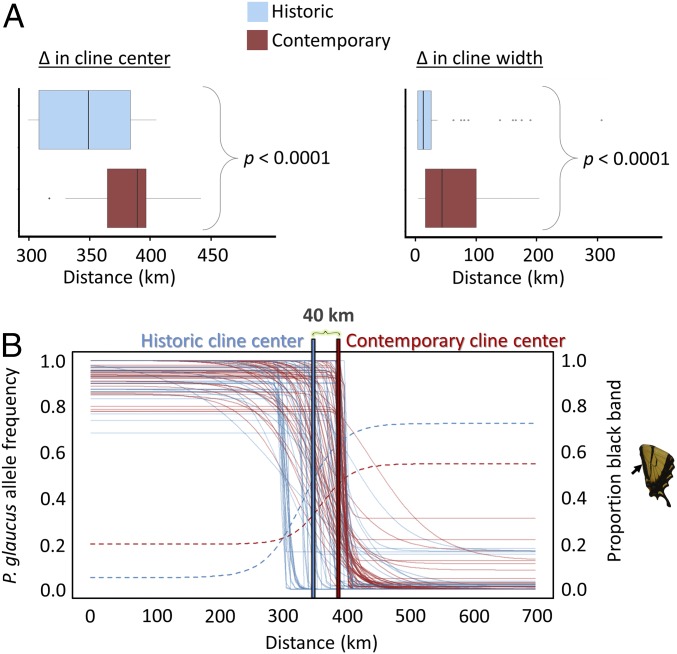
Although climate-mediated changes in voltinism could facilitate range expansion and contraction in many insects, this outcome has only been documented in a handful of species (40, 41). In Wisconsin, our simulation model predicts that the climate-mediated developmental threshold for a bivoltine life history has shifted ∼68 km northward from 1980 to 2012 (Fig. 1). If climate is a strong selective agent maintaining the location of this hybrid zone, as has been proposed (16), we would predict that the hybrid zone should be tracking the northward movement of this climate-mediated developmental threshold. From both museum and contemporary specimens collected from across a latitudinal transect in Wisconsin, we were able to develop 1,196 SNPs to test this hypothesis (1,194 ddRADseq SNPs and a SNP in Ldh and COI). Of these 56 SNPs distributed across 15 autosomes, the mitochondrial genome and the Z-chromosome appear to be playing a disproportionate role in maintaining divergence between these species (7, 16) and exhibited steep clinal variation (Methods). Comparing the median cline center and width of these markers between periods, we document a significant northward shift of the hybrid zone by 40 ± 4 km (Wilcoxon signed-rank test, P < 0.0001; Fig. 2). This northward shift also holds when looking at a subset of species diagnostic genetic markers (Z-linked Ldh and mtDNA COI) and a morphological trait (width of the anal band on the hindwing), for which we have larger sample sizes (n = 385, 564, and 1,110 respectively). We found these markers shifted significantly northward by 50, 60, and 30 km, respectively (Fig. 2 and SI Appendix, Table S3).
Fig. 2.
Empirical evaluation of hybrid zone movement. (A) Estimate of median cline center and width (for 56 clinal genetic markers; ddRADseq, COI and Ldh markers) between historic and contemporary periods (Wilcoxon signed-rank test; α = 0.05; n = 56). The scaling of the x-axis reflects a latitudinal transect spanning from ∼40°N (0 km) to 46°N (600 km). (B) Maximum likelihood cline for 56 clinal genetic markers (historic: solid blue and contemporary: solid red) and a morphological wing trait (historic: dashed blue and contemporary: dashed red).
Using the median cline estimates for all 56 clinal genetic markers, and assuming a constant rate of movement, the center of the P. glaucus–P. canadensis hybrid zone is advancing northward at a rate of ∼1.25 km/y. This rate of hybrid zone movement is slower than our estimated rate of expansion in the climate-mediated developmental threshold of 2.11 km/y, but falls within the range of documented movement of other hybrids zones spanning a broad range of taxa (∼0.2–5.8 km/y). For the majority of these hybrid zones, however, the reason for movement was either unknown or a result of assortative mating, not climate (4). The only two other butterfly hybrid zones with documented movement, Heliconius erato hydara–Heliconius erato petiverana and Anartia fatima–Anartia amathea, had rates estimated at 2.76 and 2.5 km/y, respectively (42, 43), which are nearly twice the rate we observed. In these cases, the movement was best explained by competitive interactions and dispersal, not climate.
Given that the hybrid zone is formed at the range boundaries of these two species, the movement of the zone could be considered a parallel range shift of the two species. From this perspective, the observed range expansion of P. glaucus of ∼1.25 km/y would be moving slower than the average range shift documented in other butterflies of ∼1.4–1.8 km/y (44).
Whether this apparent lag in the shift of the hybrid zone relative to the expanding climate-mediated developmental threshold is a result of hybridization or other ecological factors is difficult to determine. It could be that as P. glaucus expands northward, the overlap between P. glaucus and P. canadensis increases, resulting in a higher incidence of hybridization. Given the apparent lower fitness of hybrids (16, 45), a rise in the rate of hybridization could result in a short-term population sink that diminishes as P. canadensis is displaced. Alternatively, given the large amount of interannual variation in the geographic position of this climate-mediated developmental threshold during the last 32 y, it is possible that movement slowly advances northward in warmer years, but shifts back in cold years, such as waves advancing further inland during a rising tide. Similar to this scenario would be a ratcheting of movement northward (46), with northward expansion in warm years and persistence achieved in cooler years through diapause (i.e., individuals do not attempt a second generation). Evidence of oscillating hybrid zones has only been documented in a single case (47). Unfortunately, as is true with most historical datasets, we lack the fine-scale temporal resolution to test these alternative hypotheses.
Understanding whether and how isolating barriers within the genomes of hybridizing species are altered as a consequence of changes in climate is critical to predicting how hybridization will affect genetic diversity in the context of climate change (26). In the case of P. glaucus and P. canadensis, we find little evidence of any substantial breakdown in species boundaries. As the hybrid zone shifted, broadly distributed genomic regions exhibiting steep clines maintained strong overlap in both their center (coincidence: 84% in historic, 74% in contemporary) and width (concordance: 84% in historic and contemporary) between periods (Fig. 2A). In other words, we found little discordance in the movement among divergent regions within these genomes. These genomic regions are shifting largely in concert with one another and with a morphological trait.
The hybrid zone has become slightly wider, by 31 ± 8 km, during this period of hybrid zone movement (Wilcox signed-rank test, P < 0.0001; Fig. 2A). A widening hybrid zone could result from an increase in hybridization and/or increased overlap between the two species. Although our dataset is not ideal for estimating hybridization rates, we find no evidence for substantial changes in hybridization between periods based on Q-values in individual populations (two-tailed Fisher’s exact test, P > 0.05) or across the entire transect (two-tailed Fisher’s exact test, P = 0.367). Examining the frequency of heterozygous males for the species diagnostic SNP in the gene Ldh (indicative of an early generation hybrid), we find a significantly greater proportion of heterozygous males in location number 3 than was found historically (historic, 0/35; contemporary, 6/41; two-tailed Fisher’s exact test, P = 0.028) (SI Appendix, Table S4). However, this increase corresponds to a decrease in heterozygous males in the population south of this location (location number 2), decreasing from 4/36 to 0/33 of individuals being heterozygous for this locus in the historic and contemporary period, respectively. The implication of this observation is that the overall number of early-generation hybrids is not increasing over time in the hybrid zone (historic, 7/193; contemporary, 6/192; two-tailed Fisher’s exact test, P = 1.0). Instead, the contact zone has shifted further north. Taken together, these patterns further suggest that hybrid zone movement is the result of a parallel range shift, with P. glaucus undergoing a northern range expansion and P. canadensis a southern range contraction, and not the result of an increase in hybridization.
Our observation that this hybrid zone remained narrow as it shifted in response to climatic change is similar to that observed in a black-capped chickadee hybrid zone believed to be maintained by a combination of endogenous and exogenous barriers that has also shifted in response to changes in climate (16). Our observation also contrasts with a rapid spread in hybridization between a nonnative rainbow trout and threatened westslope trout in response to changes in temperature and precipitation (48). It is possible that climate-mediated developmental thresholds, such as voltinism, that result in a steep (binomial response) rather than a gradual selective landscape may act to maintain steep clines as the climate changes. However, the presence of endogenous barriers, when isolation between species is maintained by fitness consequences of hybridization independent of the environment, should also act to reduce introgression (49) and could help maintain species boundaries during hybrid zone movement. It has recently been proposed that endogenous barriers may exist between P. glaucus and P. canadensis (16). Thus, a combination of exogenous and endogenous barriers may be acting to maintain the integrity of the P. glaucus–P. canadensis hybrid zone as it shifts. Additional studies of climate-mediated hybrid zone movement in systems that range in their mixture of endogenous and exogenous barriers (as well as vary in the genetic architecture of these barriers) and steepness in climate selection gradients will aid in making broader generalizations about the genomic consequences of such movement.
Although there are a number of exogenous and endogenous factors other than climate that could be responsible for the hybrid zone movement we observe, climate remains the most likely explanation. It is possible that the geographic range boundaries of other ecological factors associated with divergent traits between these species (i.e., distribution of host plants and/or a Batesian model species) have changed, causing movement in this hybrid zone. We are unaware of any documented evidence implicating these factors. Further, given that the distribution of the ranges of these other species are >100 km north or south of the hybrid zone, Ryan et al. (16) argued that this geographic discord is evidence that these other ecological factors do not play a major role in maintaining the location of this hybrid zone.
In a number of other systems, bias in mating preference has been implicated in hybrid zone movement (50–52). Mating preferences favoring P. glaucus could result in the pattern we observe. There is some evidence that males of both species slightly prefer P. glaucus females (for unknown reasons) (12). However, these studies rely on the assumption that sexual selection is driven by male choice in the wild, an assumption that has not been formerly evaluated. Moreover, there is some evidence that females are driving sexual selection through female choice (53). Perhaps the most compelling argument that changes in climate are responsible for the movement we observe, and not mating preferences or changes in other ecological factors, is that this same climate gradient coincides with clinal variation in genetic markers and behavioral and physiological traits in other independent portions of the hybrid zone across the eastern United States (reviewed in ref. 32).
Simulating Future Hybrid Zone Movement.
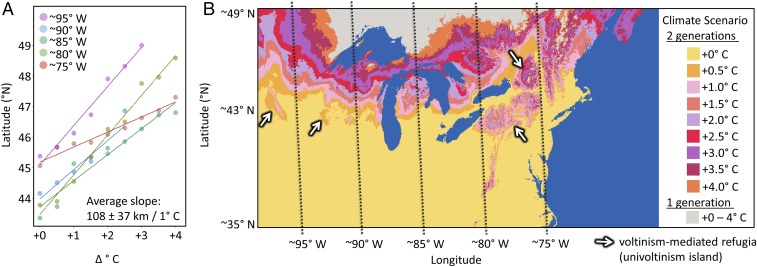
Hybrid zone movement is not unidimensional, even though that is how it is often viewed. Many hybrid zones traverse broad geographic distances spanning a great deal of abiotic (e.g., bodies of water, mountains) and biotic (e.g., biome) variation. The biological impacts of the velocity (km⋅y−1) and spatial direction of climatic changes (29) will depend critically on this abiotic and biotic variation, which has been referred to as the geography of climate change. We explored the geography of hybrid zone movement in P. glaucus and P. canadensis by modeling the spatial distribution of a climate-mediated developmental threshold (as a proxy for the location of the hybrid zone) under a range of warming conditions (future climate scenarios). These forecasts provide a two-dimensional view of how the hybrid zone might shift under continued warming and suggest there will be substantial variation in the rate and direction of continued hybrid zone movement (varying from 55–144 km/1 °C across the range), with the velocity of this change ranging from 0.4 to 9 km⋅y−1, depending on the rate of climatic warming (with warming ranging from +0.5 to 5 °C during an 80-y period; Fig. 3A). Similar to climate envelope modeling, these predictions assume that this climatic feature (length of the growing season) is the primary factor determining the location of this hybrid zone. Although our documenting of historical hybrid zone movement provides some evidence that this is a reasonable assumption in Wisconsin, it is less clear whether this assumption holds true across the entire hybrid zone.
Fig. 3.
Simulated spatial distribution of a climate-mediated developmental threshold to complete two generations (by P. glaucus) under climate warming scenarios. (A) Relationship between change in temperature and estimated most northern boundary of a climate-mediated developmental threshold. (B) Estimated spatial extent of a climate-mediated developmental threshold (proxy for location of the hybrid zone) within a given climate scenario (+0, 0.5, 1, 1.5, 2, and 2.5 °C from average 2010–2015).
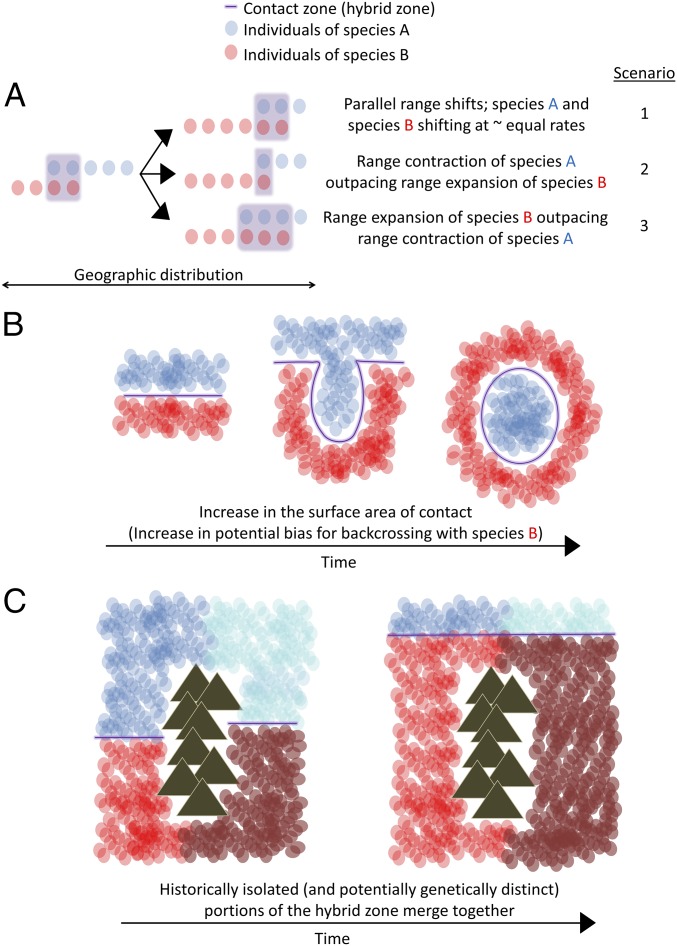
Spatial variation in the velocity of climate change has the potential to influence hybrid zone dynamics that feed back to influence further hybrid zone movement. Although at this point it is largely conjectural, we find it useful to highlight four possible nuances to climate-mediated range shifts when hybridization occurs. First, variation in the velocity of climate change could influence the spatial extent to which interbreeding species overlap. The concordance of parallel range shifts is of particular consequence in hybrid zones, because an increase or decrease in range overlap can result in altered rates of hybridization (conceptual example Fig. 4A). In the Wisconsin portion of the P. glaucus–P. canadensis hybrid zone, we find no change in overlap (Fig. 4A, scenario 2); however, given the predicted much faster rate of movement in the westernmost portion of the hybrid zone (Minnesota), it is reasonable to think one or both species may have difficulty keeping pace, which could result in an increase or decrease in overlap. In this system, increased overlap would likely result in a population sink, given that selection against hybrids is very strong (16). However, in other systems (or possibly other portions of this hybrid zone), where selection is weak or hybrids more fit, high velocities of hybrid zone movement could result in expanding hybrid zones (i.e., create conditions for a hybrid swarm) (48). Second, variation in the geographic movement of hybrid zones that increases the surface area or contact zone of interaction may increase biases in introgression, favoring the species for which the opportunity for dispersal into the contact zone is greater (Fig. 4B). Such biases could alter the evolution of the interbreeding populations as well as influence further movement of the hybrid zone, as asymmetry in introgression alone can lead to hybrid zone movement (49). Third, as hybrid zones shift, portions that were once isolated by physical barriers can come into contact, bringing together potentially locally adapted populations of both species (conceptual example in Fig. 4C). For example, with >3 °C of warming, portions of this hybrid zone (near latitudinal bands at 90 and 85° W) that have been physically isolated for potentially thousands of years will merge and form a contiguous boundary; this may already be occurring near latitudinal band 80° W (Fig. 3B). Fourth, our model predicts that local variation in the rate of warming will create “univoltinism islands” that allow isolated populations of P. canadensis to persist within the expanding range of P. glaucus (Figs. 3B and 4B). Although the effects of such changes in the spatial continuity of hybrid zones are not yet clear, they are likely to have both short- and long-term evolutionary consequences. These varying outcomes illustrate the importance of modeling the relationship between climate and the geography of hybrid zone movement in other systems to identify regions along a hybrid zone predicted to vary substantially in their movement. These populations can then be the primary focus for long-term monitoring and sampling efforts.
Fig. 4.
Conceptual illustrations of how variation in the geography of hybrid zone movement results in different hybrid zone dynamics. (A) Example of how differences in the rate of range expansion/shift and contraction between the species forming the hybrid zone can result in variation in the amount of overlap (and potential hybridization) between said species. (B) Example of how changes in the shape of the hybrid zone, viewed from left to right, can result in greater “surface area of contact” for one species that results in asymmetries in the contribution of parental species to the hybrid zone (favoring greater backcrossing of hybrids to species B in this example). (C) Example of how historically isolated (and potentially genetically distinct; indicated by different shades of blue and red) portions of a hybrid zone may merge, forming a more contiguous hybrid zone as a result of range shifts of one or both species.
This study, along with a few other documented cases of climate-mediated hybrid zone movement, provides support to the idea that hybrid zone movement may be a common response to climate change (23). Hybrid zone movement was associated with a shift in a voltinism transition zone, which is a climate-mediated phenomenon ubiquitous among insects, suggesting that movement in insect hybrid zones may be a common response to climate change. Our findings also suggest that climate-mediated hybrid zone movement may result in displacement, rather than blending of species boundaries. In the case of this hybrid zone, a steep exogenous selection gradient (length of growing season required to complete two generations), endogenous selection (previously identified potential genetic incompatibilities throughout the genome), or a combination of these two forces appears to be preserving the integrity of this hybrid zone as it shifts. However, our prediction that hybrid zone movement could vary substantially geographically underscores the importance of considering different regions of a hybrid zone. Given that many hybrid zones span large geographic areas that contain a diverse mix of ecological and topological gradients, the geography of hybrid zone movement in these other hybrid zones is also likely to be quite variable. From a conservation and resource/pest management perspective, it will be necessary to consider how features in the geographic landscape, as well as selection within the genomic landscape (exogenous and endogenous barriers), drive disparate evolutionary outcomes for different hybrid zones, and even different populations across a hybrid zone.
Methods
Model Fitting.
To predict the spatial extent of the climate-mediated developmental threshold to complete two generations by P. glaucus (Fig. 1 and SI Appendix, Fig. S1), we parameterized and evaluated a number of developmental models for each stage of development. Specifically, we measured growth rates from 1,153 offspring produced from 40 families created from wild-caught P. glaucus females collected from central Illinois and Indiana in 2012–2015 (SI Appendix, Fig. S3); for validation (see Model Evaluation/Selection), 22 wild-caught P. glaucus females were also collected from Illinois and Pennsylvania. Males were provided with a nutrient rich honey solution before mating to increase fertility. Females were placed in ∼15 × 15 cm cylindrical mesh cages containing tulip tree (Liriodendron tulipifera) leaves and kept in a temperature-controlled chamber cycling between 18 and 30 °C and 16:8 light:dark cycle. Females were fed a 10% honey water solution twice daily, and eggs were collected each morning and evening.
Freezers were modified into growth chambers by adjustments to the thermostat and the addition of heating blankets, and lights were controlled using a custom Matlab script that made regular adjustments to light (day length) and temperature (to stay within half a degree Celsius of the set point). Lighting was set to long-day conditions (16:8 light:dark cycle) to keep individuals from diapausing. Eggs from each female were evenly and randomly split among temperature treatments: 8, 12, 18, 24, 30, 36, and 40 °C. The 18, 24, and 30 °C treatments were kept constant. For the 8, 12, 36, and 40 °C treatments, we used a temperature transfer approach, as done in ref. 54, where individuals were moved to these temperatures for only a subset of their development (SI Appendix, Fig. S4). This approach reduces treatment-induced mortality (particularly near the upper threshold), and the potential for excessively long trials (particularly near the lower threshold) that can induce stress and, in the case of larvae, substantial changes in host plant quality over the course of the treatment. All individuals that died while in the “transfer treatments” (8, 12, 36, and 40 °C) were given a growth rate of 0, as these individuals were likely to have growth rates near 0 (54); to account for nontreatment mortality, the proportion of individuals that died in the 24 °C treatment (assumed rate of background mortality) were removed from each transfer treatment.
Eggs were kept in small ventilated plastic cups until hatching. Larvae were transferred to 15 cm diameter vented Petri dishes (1–2 per dish) and fed leaves of black cherry (Prunus serotina) in water piques that were changed every other day. Once larvae entered the “wandering” stage just before pupation (turned purple), they were placed in 24 °C and given 4 d to pupate before placing them in individual mesh cages for eclosion and moved to their pupal treatment. The treatment for each new developmental stage was chosen at random.
Developmental data from the growth chamber experiments (above) were fit to seven different commonly used models for insect development: Régnière (54), Taylor (32), Beta (33), Lactin (55), Logan-10 (34), Brière-1 (31), and Schoolfield (56) (SI Appendix, Figs. S5 and S6). Model fitting was performed using the general purpose optimization function optim from the stats package in the R environment (R 3.0.2). Each fit was checked for convergence and inspected visually by plotting each fit. This was done for each stage of development. For pupal development (time to eclosion), models were fit for only females (females develop slower than males and would be the limiting factor for completion of two generations). To account for the effect that change in host plant quality has on larval growth rate throughout the season, second-generation larvae were fit separately from first-generation larvae.
Model Evaluation/Selection.
The model for each stage was selected on the basis of its ability to predict developmental milestones from the independent growth chamber experiment. This evaluation step determines the degree to which our model accurately reflects development in more natural temperature conditions. In 2011, wild-caught males were collected from central Illinois (P. glaucus) and mated to field reared virgin females from southeastern Pennsylvania (P. glaucus; SI Appendix, Fig. S3). Matings from these pairings, along with eggs collected from wild-caught females in central Illinois, resulted in 844 individuals from 22 families. Eggs were collected from ovipositing females laying on tulip tree leaves (see Model Fitting) and were placed into one of four treatments that simulated temperature conditions representing northern (Vilas County), central (Marathon County), and southern (Rock County) Wisconsin and central Illinois (Peoria County), reflecting a broad natural range of climatic variation in the hybrid zone (SI Appendix, Fig. S3). Temperature profiles were created by averaging Tmin and Tmax for the years 1998–2008 (National Oceanic and Atmospheric Administration daily climate summaries) for each of the four sites. Tmin and Tmax were converted to hourly temperature profiles using the method of (57), where Th = Tavg + [(Tmax − Tmin)/2] × (cos[0.2618 × (h − d)]), with Th being the temperature at a given hour of the day, Tmax and Tmin the daily minimum and maximum temperature, and d the hour of day when the temperature is highest (Tmax); d was set to 15 for all locations. Therefore, these profiles included natural daily and seasonal variation. The temperatures were changed every other day (using the average temperature between the 2 d). Photoperiod was set as Coloma Wisconsin (putative hybrid zone) and was changed every other day (using the average photoperiod between the 2 d). Other rearing conditions were similar to those described in Model Fitting.
Parameter estimates for each of the seven fitted models were used to predict developmental milestones (completion of a developmental stage), using this independent dataset. Temperature data from each treatment were used as input in the model. The predicted days to completion of each developmental stage were then compared with the observed (known) number of days it took to complete a given stage of development. We calculated the RMSE for each developmental stage, and the model with the lowest RMSE was selected (SI Appendix, Fig. S7). Direct-developing and diapaused pupae were also evaluated separately because of differences in development between the two pupal conditions. All model validation was done in the R environment (R 3.0.2).
Simulations: Applying the Model to Climate Data.
Predicting historical changes 1980–2006.
For each period (historic, 1980–1986; contemporary, 2006–2012), using the selected models, we predicted whether P. glaucus was able to complete two generations [i.e., successfully develop from a diapausing pupae to eclosion, to egg, to larva, to pupae, to eclosion as a direct-developing pupae, to egg, to larvae, to pupae for each location (binary, yes/no); Fig. 1 and SI Appendix, Fig. S1]. This was done using daily minimum and maximum temperatures acquired from the Daymet 1 × 1 km gridded climatological dataset (58), resampled to 4 × 4 km spatial resolution, and averaged for each period as input. The latitudinal extent of this developmental threshold was then determined for each year as the highest latitude at which P. glaucus was able to complete two generations from within 1° wide (longitudinally; centered on 75, 80, 85, and 90° W) transects running north–south.
Predicting future changes (climate scenarios).
Using the same model fits and climate data used to predict changes in the climate-mediated development threshold (as described in Model Fitting and Model Evaluation/Selection), we calculated the highest latitude at which P. glaucus could complete two generations for a series of latitudinal transects (centered at 75, 80, 85, 90, 95° W; 1° wide) under a series of climate warming scenarios (by adding 0, 0.5, 1, 1.5, 2, and 2.5 °C to the average temperature of 2010–2015; Fig. 4). A linear regression was fit to these data (highest latitude to complete two generations x climate scenario for each latitudinal transect) to get an estimate of the rate of movement for every 1 °C warming (slope of each regression).
Assessing Historic Hybrid Zone Movement.
Specimens.
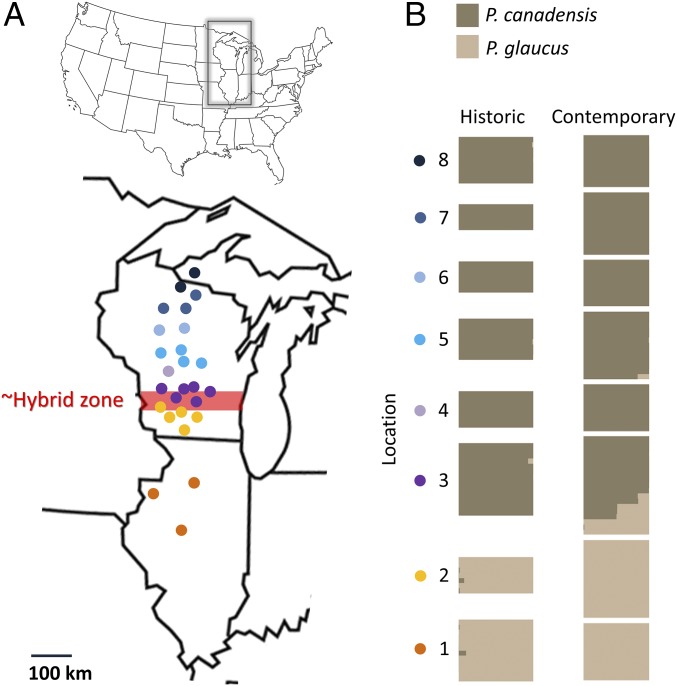
Specimens of wild-caught males used for genetic and morphological analyses were collected between historic (1979–1987) and contemporary (2007–2013) periods from a latitudinal transect that spans the P. glaucus–P. canadensis hybrid zone in Wisconsin (Figs. 2 and 5 and SI Appendix, Table S5); historic specimens were pinned, dried, and kept in a museum collection before use (these specimens are now maintained in the Scriber collection at the McGuire Center at the University of Florida, Gainesville, FL).
Fig. 5.
Assessing genetic structure and frequency of hybridization in the P. glaucus–P. canadensis hybrid zone for historic (1980–1984) and contemporary (2007–2012) periods. (A) Sampling locations for each population used for analyses of genetic structure; points with the same color were pooled for analysis. (B) Bayesian clustering of ancestry proportion to K = 2 populations (see SI Appendix, Fig. S2 for K = 1–5) based on 1,192 SNPs. Comparison of the proportion of hybrids (Q-value <0.95 for either species’ cluster) based on Fisher’s exact test (two-tailed, α = 0.05).
Genetic and morphological markers.
We measured 1,111 specimens for the species diagnostic morphological marker (59) (measured twice and the average value used), the width of the anal band in proportion to the width of the anal cell, and scaled to values between 0 and 1 [(value − minimum)/(maximum − minimum)]. We then genotyped 564 individuals for species diagnostic mtDNA (COI) haplotypes (60) and 385 individuals for a species diagnostic SNP in the lactase dehydrogenase (Ldh) gene. Additional, geographically clinal loci (allele frequencies changing from >0.8 to <0.2 across the hybrid zone) were identified from 200 (148 after filtering; see below) individuals from a reduced representation (ddRADseq) genome-wide set of SNPs; all contemporary samples were previously genotyped/sequenced (16).
Genomic DNA was isolated from leg tissue using a phenol:chloroform protocol (16); extractions of historic specimens were performed in a PCR-free room separately from contemporary samples. Reduced-complexity libraries were generated using a variation of the restriction site-associated DNA fragment (RAD) procedure, following that used in ref. 16, involving a double digest (ddRADseq) (25). Briefly, ∼400–600 ng genomic DNA was digested using the restriction enzymes EcoRI and MseI. Individuals were barcoded using an EcoRI adapter containing a unique 8- to 10-base pair (bp) index. PCR product was pooled, purified with QIAquick Purification Kit (Qiagen), and size-selected (400–600 bp range) on a BluePippin System (Sage Science), and six individuals and a negative control (sample processed through all steps, but with no tissue added during DNA extraction step) were sequenced on an Illumina MiSeq and 96 individuals on a HiSEq 2000 (BGI, University of California, Davis), using 150- and 100-bp paired-end sequencing, respectively.
Raw reads were demultiplexed using a custom Perl script. Reads were trimmed and cleaned with the program TRIMMOMATIC (v.0.32, ref. 61), using default settings and clipping the first 20 bp from all reads (to remove barcodes and cutsite) and keeping only reads >30 bp in length. Forward unpaired reads were of lower quality and were further trimmed to 80 and 60 bp for MiSeq and HiSeq reads, respectively. Cleaned reads were then aligned to the P. glaucus genome (glaucus_assembly_V1.1.fa; prodata.swmed.edu/LepDB/), using BWA aln (with default parameters; version: 0.7.8) (62). Aligned reads were then cleaned with Picard (ver 1.33) and realigned with GATK’s realigner (v3.4) (63, 64). SNPs were called using GATK’s HaplotypeCaller v3.4, and indels and multiallelic sites were removed. The following filters were then applied to historic and contemporary biallelic SNP datasets separately. A minimum of 0.05 and a maximum of 0.95 was applied as the allele frequency filter, using VCFtools v0.1.12b (65), followed by removal of individuals with a large amount of missing data (>95% and >90% for historic and contemporary samples, respectively). Sites not passing GATK’s hard filtering (quality by depth < 2, Fisher strand > 60, rms mapping quality < 40, mapping quality rank sum test < −12.5, and read position rank sum test < −8) and genotypes with a genotype quality <20 were removed. Only SNPs with minimum of 6× coverage in 75% of individuals and present in both historic and contemporary libraries were retained, resulting in 1,192 SNPs. The chromosomal location of each ddRADseq SNP was based on the putative assignment previously determined using the mapping of P. glaucus peptide sequences to the Bombyx mori genome (16). We also tested for the presence of DNA damage in historic samples, using the program mapDamage ver 2.0.5–1 and by using a nonparametric Kruskal-Wallis Rank Sum Test to compare the frequency of all C > T and G > T substitutions between periods. There was no evidence of significant DNA damage in historic samples; the significant differences observed were biologically irrelevant (<1%) and in the opposite direction predicted: there were slightly fewer C > T and G > T substitutions in DNA from historic specimens (SI Appendix, Fig. S8).
Cline analysis.
Clines were fit to 56 SNPs: 54 clinal (allele frequencies going from >0.8 to <0.2 across the transect in either period) RADseq SNPs found to be in Hardy-Weinberg equilibrium and the Z-linked (Ldh) and mtDNA (COI) markers. Cline fitting was done using a Metropolis-Hasting algorithm implemented in the HZAR package v2.5 in R (66) that fits a combination of equations (15 models) that describe the sigmoidal shape at the center of each cline and two exponential decay curves. The model with the lowest Akaike Information Criterion (67) was chosen for each SNP. We assessed whether the hybrid zone had significantly shifted or was widening/narrowing, using a Wilcox signed-rank test to compare the historic and contemporary distribution of cline center estimates for all genetic (ddRADseq, Ldh, COI) markers (Fig. 2). Loci were considered coincident or concordant if their high and low bound estimates for cline center or width (based on a 2-log-likelihood unit; comparable to 95% confidence intervals) overlapped with the average estimate (based on all 56 clinal genetic markers) for cline center or width, respectively.
Structure/admixture analysis.
Temporal changes in admixture/genetic structure between the two periods (Fig. 5 and SI Appendix, Fig. S2) was assessed with a Bayesian clustering method implemented in STRUCTURE v2.3.4 (68, 69), using 1,192 SNPs (6× in 75% of individuals and pruned 1 SNP per 1 kb) with default settings: a 100,000 burn-in, 500,000 iterations, and K values from one through eight. These analyses were permuted 10 times.
To assess whether hybridization has changed over time, a Fisher’s exact test (two-tailed) was used to compare the frequency of putative hybrids, using three different datasets: using Q-values (from STRUCTURE) to infer hybrids (Q-value <95% assignment to either cluster), putative F1s (individuals heterozygous for Ldh), and putative backcrosses (individuals P. glaucus-like for COI and P. canadensis-like for Ldh, or the inverse).
Supplementary Material
Acknowledgments
We thank Jason Dzurisin and Sarah Richman for their help in the collection of contemporary butterfly specimens and Mark Evans, Bob Hagen, Bob Lederhouse, Greg Lintereur, Heidi Luebke, Rick Lindroth, Maret Pajutee, Sally Rockey, and Jeff Thorne for their involvement in historical collections. Many thanks to Jacqueline Lopez and Melissa Stephens in the Notre Dame Genomics & Bioinformatics Core Facility for their advice in various aspects of experimental design of next-generation sequencing data collection. Historical collections were made possible through support from University of Wisconsin–Madison Agricultural Experiment Station and National Science Foundation (Grants DEB 79-21749 and DEB 8306063) and contemporary collections and molecular work supported by National Science Foundation Grants DEB-0918879 (to J.M.S.) and DEB-0919147 (to J.J.H.), and a grant from the Environmental Change Initiative at the University of Notre Dame (to S.F.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Demultiplexed RADseq reads generated in this study are available through NCBI’s Sequence Read Archive associated with Bioproject (PRJNA431486, SRA: SRP131962). All metadata and scripts associated with analyses in this study have been deposited on DRYAD (https://doi.org/10.5061/dryad.5vn76).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714950115/-/DCSupplemental.
References
- 1.Araújo MB, Luoto M. The importance of biotic interactions for modelling species distributions under climate change. Glob Ecol Biogeogr. 2007;16:743–753. [Google Scholar]
- 2.Van der Putten WH, Macel M, Visser ME. Predicting species distribution and abundance responses to climate change: Why it is essential to include biotic interactions across trophic levels. Philos Trans R Soc Lond B Biol Sci. 2010;365:2025–2034. doi: 10.1098/rstb.2010.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallet J. Hybridization, ecological races and the nature of species: Empirical evidence for the ease of speciation. Philos Trans R Soc Lond B Biol Sci. 2008;363:2971–2986. doi: 10.1098/rstb.2008.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buggs RJA. Empirical study of hybrid zone movement. Heredity (Edinb) 2007;99:301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- 5.Putnam AS, Scriber JM, Andolfatto P. Discordant divergence times among Z-chromosome regions between two ecologically distinct swallowtail butterfly species. Evolution. 2007;61:912–927. doi: 10.1111/j.1558-5646.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- 6.Kunte K, et al. Sex chromosome mosaicism and hybrid speciation among tiger swallowtail butterflies. PLoS Genet. 2011;7:e1002274. doi: 10.1371/journal.pgen.1002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong Q, Borek D, Otwinowski Z, Grishin NV. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 2015;10:910–919. doi: 10.1016/j.celrep.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scriber JM, Lindroth RL, Nitao J. Differential toxicity of a phenolic glycoside from quaking aspen to Papilio glaucus butterfly subspecies, hybrids and backcrosses. Oecologia. 1989;81:186–191. doi: 10.1007/BF00379804. [DOI] [PubMed] [Google Scholar]
- 9.Kukal O, Ayres MP, Scriber JM. Cold tolerance of the pupae in relation to the distribution of swallowtail butterflies. Can J Zool. 1991;69:3028–3037. [Google Scholar]
- 10.Mercader RJ, Scriber JM. Asymmetrical thermal constraints on the parapatric species boundaries of two widespread generalist butterflies. Ecol Entomol. 2008;33:537–545. [Google Scholar]
- 11.Scriber JM, Giebink BL, Snider D. Reciprocal latitudinal clines in oviposition behavior of Papilio glaucus and P. canadensis across the great lakes hybrid zone: Possible sex-linkage of oviposition preferences. Oecologia. 1991;87:360–368. doi: 10.1007/BF00634592. [DOI] [PubMed] [Google Scholar]
- 12.Deering MD, Scriber JM. Field bioassays show heterospecific mating preference asymmetry between hybridizing North American Papilio butterfly species (Lepidoptera: Papilionidae) J Ethol. 2002;20:25–33. [Google Scholar]
- 13.Mercader RJ, Aardema ML, Scriber JM. Hybridization leads to host-use divergence in a polyphagous butterfly sibling species pair. Oecologia. 2009;158:651–662. doi: 10.1007/s00442-008-1177-9. [DOI] [PubMed] [Google Scholar]
- 14.Luebke HJ, Scriber JM, Yandell BS. Use of multivariant discriminant analysis of male wing morphometrics to delineate a hybrid zone for Papilio glaucus glaucus and P. g. canadensis in Wisconsin. Am Midl Nat. 1988;119:366–379. [Google Scholar]
- 15.Lehnert MS, Scriber JM, Gerard PD, Emmel TC. The “converse to Bergmann’s rule” in tiger swallowtail butterflies: Boundaries of species and subspecies wing traits are independent of thermal and host-plant induction. Am Entomol (Lanham Md) 2012;58:156–165. [Google Scholar]
- 16.Ryan SF, et al. Patterns of divergence across the geographic and genomic landscape of a butterfly hybrid zone associated with a climatic gradient. Mol Ecol. 2017;26:4725–4742. doi: 10.1111/mec.14236. [DOI] [PubMed] [Google Scholar]
- 17.Scriber JM. Interaction of introgression from Papilio glaucus canadensis and diapause in producing “spring form” Eastern tiger swallowtail butterflies, P. glaucus. Gt Lakes Entomol. 1990;23:127–138. [Google Scholar]
- 18.Hagen RH, Scriber JM. Sex-linked diapause, color, and allozyme loci in Papilio glaucus: Linkage analysis and significance in a hybrid zone. J Hered. 1989;80:179–185. [Google Scholar]
- 19.Scriber JM. Evolution of insect-plant relationships: Chemical constraints, coadaptation, and concordance of insect/plant traits. Entomol Exp Appl. 2002;104:217–235. [Google Scholar]
- 20.Scriber JM, Stump A, Deering M. Ecology and Evolution Taking Flight: Butterflies as Model Study Systems. University of Chicago Press; Chicago: 2003. [Google Scholar]
- 21.Scriber JM. Impacts of climate warming on hybrid zone movement: Geographically diffuse and biologically porous “species borders.”. Insect Sci. 2011;18:121–159. [Google Scholar]
- 22.Kucharik CJ, Serbin SP, Vavrus S, Hopkins EJ, Motew MM. Patterns of climate change across Wisconsin from 1950 to 2006. Phys Geogr. 2010;31:1–28. [Google Scholar]
- 23.Taylor SA, Larson EL, Harrison RG. Hybrid zones: Windows on climate change. Trends Ecol Evol. 2015;30:398–406. doi: 10.1016/j.tree.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merilä J. Evolution in response to climate change: In pursuit of the missing evidence. BioEssays. 2012;34:811–818. doi: 10.1002/bies.201200054. [DOI] [PubMed] [Google Scholar]
- 25.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker M, et al. Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nat Clim Change. 2013;3:1039–1043. [Google Scholar]
- 27.Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- 28.Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: Setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- 29.Loarie SR, et al. The velocity of climate change. Nature. 2009;462:1052–1055. doi: 10.1038/nature08649. [DOI] [PubMed] [Google Scholar]
- 30.IPCC 2014. Climate Change 2013: The Physical Science Basis, Stocker T, et al. (Cambridge Univ Press, Cambridge, UK)
- 31.Brière J-F, Pracros P, Le Roux A-Y, Pierre J-S. Novel rate model of temperature-dependent development for arthropods. Environ Entomol. 1999;28:22–29. [Google Scholar]
- 32.Taylor F. Ecology and evolution of physiological time in insects. Am Nat. 1981;117:1–23. [Google Scholar]
- 33.Damos PT, Savopoulou-Soultani M. Temperature-dependent bionomics and modeling of Anarsia lineatella (Lepidoptera: Gelechiidae) in the laboratory. J Econ Entomol. 2008;101:1557–1567. doi: 10.1603/0022-0493(2008)101[1557:tbamoa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 34.Logan JA, Wollkind DJ, Hoyt SC, Tanigoshi LK. An analytic model for description of temperature dependent rate phenomena in arthropods. Environ Entomol. 1976;5:1133–1140. [Google Scholar]
- 35.Ziter C, Robinson EA, Newman JA. Climate change and voltinism in Californian insect pest species: Sensitivity to location, scenario and climate model choice. Glob Change Biol. 2012;18:2771–2780. doi: 10.1111/j.1365-2486.2012.02748.x. [DOI] [PubMed] [Google Scholar]
- 36.Stoeckli S, et al. Impact of climate change on voltinism and prospective diapause induction of a global pest insect–Cydia pomonella (L.) PLoS One. 2012;7:e35723. doi: 10.1371/journal.pone.0035723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jönsson AM, Pulatov B, Linderson M-L, Hall K. Modelling as a tool for analysing the temperature-dependent future of the Colorado potato beetle in Europe. Glob Change Biol. 2013;19:1043–1055. doi: 10.1111/gcb.12119. [DOI] [PubMed] [Google Scholar]
- 38.Altermatt F. Climatic warming increases voltinism in European butterflies and moths. Proc Biol Sci. 2010;277:1281–1287. doi: 10.1098/rspb.2009.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaak DJ, et al. Slow climate velocities of mountain streams portend their role as refugia for cold-water biodiversity. Proc Natl Acad Sci USA. 2016;113:4374–4379. doi: 10.1073/pnas.1522429113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomi T, Nagasaka M, Fukuda T, Hagihara H. Shifting of the life cycle and life-history traits of the fall webworm in relation to climate change. Entomol Exp Appl. 2007;125:179–184. [Google Scholar]
- 41.Aguirre-Gutiérrez J, et al. Functional traits help to explain half-century long shifts in pollinator distributions. Sci Rep. 2016;6:24451. doi: 10.1038/srep24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum MJ. Rapid movement of a Heliconius hybrid zone: Evidence for phase III of Wright’s shifting balance theory? Evolution. 2002;56:1992–1998. doi: 10.1111/j.0014-3820.2002.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 43.Dasmahapatra KK, et al. Inferences from a rapidly moving hybrid zone. Evolution. 2002;56:741–753. doi: 10.1111/j.0014-3820.2002.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 44.Hickling R, Roy DB, Hill JK, Fox R, Thomas CD. The distributions of a wide range of taxonomic groups are expanding polewards. Glob Change Biol. 2006;12:450–455. [Google Scholar]
- 45.Ryan SF, Valella P, Thivierge G, Aardema ML, Scriber JM. The role of latitudinal, genetic and temperature variation in the induction of diapause of Papilio glaucus (Lepidoptera: Papilionidae) Insect Sci. November 29, 2016 doi: 10.1111/1744-7917.12423. [DOI] [PubMed] [Google Scholar]
- 46.Jackson ST, Betancourt JL, Booth RK, Gray ST. Ecology and the ratchet of events: Climate variability, niche dimensions, and species distributions. Proc Natl Acad Sci USA. 2009;106:19685–19692. doi: 10.1073/pnas.0901644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy J-S, O’Connor D, Green DM. Oscillation of an anuran hybrid zone: Morphological evidence spanning 50 years. PLoS One. 2012;7:e52819. doi: 10.1371/journal.pone.0052819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muhlfeld CC, et al. Invasive hybridization in a threatened species is accelerated by climate change. Nat Clim Change. 2014;4:620–624. [Google Scholar]
- 49.Barton NH, Hewitt GM. Analysis of hybrid zones. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- 50.Konishi M, Takata K. Impact of asymmetrical hybridization followed by sterile F1 hybrids on species replacement in Pseudorasbora. Conserv Genet. 2004;5:463–474. [Google Scholar]
- 51.Shapiro LH. Asymmetric assortative mating between two hybridizing Orchelimum Katydids (Orthoptera: Tettigoniidae) Am Midl Nat. 2001;145:423–427. [Google Scholar]
- 52.Abernethy K. The establishment of a hybrid zone between red and sika deer (genus Cervus) Mol Ecol. 1994;3:551–562. doi: 10.1111/j.1365-294x.1994.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 53.Krebs RA. The mating behavior of Papilio glaucus (Papilionidae) J Res Lepid. 1988;26:27–31. [Google Scholar]
- 54.Régnière J, Powell J, Bentz B, Nealis V. Effects of temperature on development, survival and reproduction of insects: Experimental design, data analysis and modeling. J Insect Physiol. 2012;58:634–647. doi: 10.1016/j.jinsphys.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 55.Lactin DJ, Holliday NJ, Johnson DL, Craigen R. Improved rate model of temperature-dependent development by arthropods. Environ Entomol. 1995;24:68–75. [Google Scholar]
- 56.Schoolfield RM, Sharpe PJH, Magnuson CE. Non-linear regression of biological temperature-dependent rate models based on absolute reaction-rate theory. J Theor Biol. 1981;88:719–731. doi: 10.1016/0022-5193(81)90246-0. [DOI] [PubMed] [Google Scholar]
- 57.Campbell GS. Soil physics with BASIC: Transport models for soil-plant systems. Elsevier; Amsterdam: 1985. [Google Scholar]
- 58.Thornton PE, et al. 2016 Daymet: Daily Surface Weather Data on a 1-km Grid for North America (ORNL DAAC, Oak Ridge, TN), Version 2. Available at https://daac.ornl.gov/cgi-bin/dsviewer.pl?ds_id=1328. Accessed December 2, 2016.
- 59.Scriber JM. Climate-driven reshuffling of species and genes: Potential conservation roles for species translocations and recombinant hybrid genotypes. Insects. 2013;5:1–61. doi: 10.3390/insects5010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sperling FA. Heredity–Abstract of article: Mitochondrial DNA variation and Haldane’s rule in the Papilio glaucus and P. troilus species groups. Heredity. 1993;71:227–233. [Google Scholar]
- 61.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Derryberry EP, Derryberry GE, Maley JM, Brumfield RT. HZAR: Hybrid zone analysis using an R software package. Mol Ecol Resour. 2014;14:652–663. doi: 10.1111/1755-0998.12209. [DOI] [PubMed] [Google Scholar]
- 67.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 68.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.