Significance
Plant grafting is an ancient and agriculturally important technique. Despite its widespread use, little is known about how plants graft. Here, we perform a genome-wide transcriptome analysis of tissues above and below graft junctions. We observed a sequential activation of genes important for vascular development including cambium-, phloem-, and xylem-related genes. Massive changes in gene expression that rapidly differentiate the top of the graft from the bottom occur. These changes disappear as the graft heals and the vasculature reconnects. Many genes below the junction rapidly respond to the presence of attached tissues including genes involved in vascular differentiation and cell division. This intertissue communication process occurs independently of functional vascular connections and acts as a signal to activate vascular regeneration.
Keywords: plant grafting, regeneration, auxin, vascular tissue, wound healing
Abstract
The ability for cut tissues to join and form a chimeric organism is a remarkable property of many plants; however, grafting is poorly characterized at the molecular level. To better understand this process, we monitored genome-wide gene expression changes in grafted Arabidopsis thaliana hypocotyls. We observed a sequential activation of genes associated with cambium, phloem, and xylem formation. Tissues above and below the graft rapidly developed an asymmetry such that many genes were more highly expressed on one side than on the other. This asymmetry correlated with sugar-responsive genes, and we observed an accumulation of starch above the graft junction. This accumulation decreased along with asymmetry once the sugar-transporting vascular tissues reconnected. Despite the initial starvation response below the graft, many genes associated with vascular formation were rapidly activated in grafted tissues but not in cut and separated tissues, indicating that a recognition mechanism was activated independently of functional vascular connections. Auxin, which is transported cell to cell, had a rapidly elevated response that was symmetric, suggesting that auxin was perceived by the root within hours of tissue attachment to activate the vascular regeneration process. A subset of genes was expressed only in grafted tissues, indicating that wound healing proceeded via different mechanisms depending on the presence or absence of adjoining tissues. Such a recognition process could have broader relevance for tissue regeneration, intertissue communication, and tissue fusion events.
For millennia people have cut and rejoined plants through grafting. Generating such chimeric organisms combines desirable characteristics from two plants, such as disease resistance, dwarfing, and high yields, or can propagate plants and avoid the delays entailed by a juvenile state (1). Agriculturally, grafting is becoming more relevant as a greater number of plants and species are grafted to increase productivity and yield (2). However, our mechanistic understanding of the biological processes involved in grafting, including wound healing, tissue fusion, and vascular reconnection, remain limited.
Plants have efficient mechanisms to heal wounds and cuts, in part through the production of wound-induced pluripotent cells termed “callus.” Callus fills the gap or seals the wound and later differentiates to form epidermal, mesophyll, and vascular tissues (3). In grafted Arabidopsis hypocotyls, tissues adhere 1–2 d after grafting, and the phloem, the tissue that transports sugars and nutrients, connects after 3 d (4, 5). The xylem, tissue that transports water and minerals, connects after 7 d (4). Plant hormones are important regulators of vascular formation, and at the graft junction both auxin and cytokinin responses increase in the vascular tissue (4–6). Auxin is important for differentiation of vascular tissues whereas cytokinin promotes vascular stem cells, termed the “cambium,” to divide and proliferate in a process known as secondary growth (7, 8). Auxin is produced in the upper parts of a plant and moves toward the roots via cell-to-cell movement. Auxin exporters, including the PIN proteins, transport auxin into the apoplast, whereas auxin importers, such as the AUX and LAX proteins, assist with auxin uptake into adjacent cells (8). Disrupting this transport, such as by mutating PIN1, inhibits healing of a wounded stem (9). Blocking auxin transport with the auxin transport inhibitor TIBA (2,3,5-triiodobenzoic acid) in the shoot inhibits vascular formation and cell proliferation at the Arabidopsis graft junction (6). In addition to auxin, other compounds, including sugars, contribute to vascular formation. The localized addition of auxin to callus induces phloem and xylem but requires the presence of sugar (10, 11). In plants, sugars are produced in the leaves and transported through the phloem to the roots (12). The role of sugars in vascular formation and wound healing is not well established; however, sugars promote cell division and cell expansion (13), processes important for development including vascular formation.
The molecular and cellular mechanisms of wound healing, tissue reunion, and graft formation remain largely unknown. One emerging theme is that the top and bottom of the cut do not behave similarly. Such tissue asymmetry occurs in other plant tissues, most notably leaves. Developing leaf primordia have an inherent asymmetry that is established early to specify differences between the top and the bottom of the leaf. External signals promote early leaf polarity changes, but how asymmetry is established remains unknown (14). Auxin depletion in the upper side of the emerging leaf might be important (15), or, alternatively, a meristem-derived lipophilic molecule could activate HD-ZIPIII proteins important for asymmetry (16). Asymmetry also appears in cut Arabidopsis inflorescence stems where the transcription factor RAP2.6L expresses exclusively below the cut whereas the transcription factor ANAC071 expresses exclusively above the cut (9). Both are important for stem healing, and ANAC071 and a close homolog, ANAC096, are important for graft formation (6). Asymmetry also exists in genetic requirements, since ALF4 and AXR1, two genes involved in auxin perception, are important below but not above the graft junction for phloem connection (4). However, ANAC071 is expressed symmetrically around the hypocotyl graft junction 3 d after grafting (6) so the extent and the mechanistic basis of asymmetry during wound healing remain largely uncharacterized.
Previous efforts have characterized wound healing and tissue reunion using transcriptomic analyses. Mechanical wounding altered ∼8% of the Arabidopsis transcriptome and showed a high degree of overlap with transcriptomic changes elicited by pathogen attack and abiotic stress (17). Stem wounding and wound-induced callus formation altered the expression of hundreds or thousands of genes (9, 18, 19), whereas grafting grape vines, lychee trees, and hickory trees induced hundreds or thousands of differentially expressed genes involved in hormone response, wound response, metabolism, cell-wall synthesis, and signal transduction (5, 20–23). These grafting studies provide limited information, as tissues from above and below the graft junction were not isolated to test whether these tissues behaved differently, and controls were not performed to distinguish how grafting and tissue fusion might differ from a response associated with cut tissues that remained separated. Here, we perform an in-depth analysis to describe the spatial and temporal transcriptional dynamics that occur during healing of cut Arabidopsis tissues that are joined (grafted) or left unjoined (separated). We find that the majority of genes differentially expressed are initially asymmetrically expressed at the graft junction and that many of these genes are sugar-responsive, which correlates with severing of the phloem tissue and the accumulation of starch above the junction. However, genes associated with cell division and vascular formation activate on both sides of the graft, and, similarly, auxin responsiveness activates equally on both sides. We propose that the continuous transport of substances, including auxin, independent of functional vascular connections, promotes division and differentiation, while the enhanced auxin response and blockage of sugar transport provides a unique physiological condition to activate genes specific to graft formation that promotes wound healing.
Results
Grafting Activates Vascular Formation and Cell Division Genes.
To better understand the developmental processes that occur at the graft junction, we generated RNA deep-sequencing libraries from Arabidopsis thaliana hypocotyl tissues immediately above and immediately below the graft junction 0, 6, 12, 24, 48, 72, 120, 168, and 240 h after grafting in biological replicates for each tissue at each time point (Fig. 1A). Before RNA extraction, we separated top and bottom tissues at the graft junction. We found that the strength required to break apart the graft junction increased linearly (SI Appendix, Fig. S1) and similarly to previously reported breaking strength dynamics of grafted Solanum pennellii and Solanum lycopersicum (24, 25). When pulling apart grafts to separate top and bottom for sample preparation, grafts broke cleanly with minimal tissue from one half present in the other half (SI Appendix, Fig. S1 and Movies S1 and S2). We measured the amount of tissue from tops adherent to bottoms and vice versa and found less than 4% cross-contamination (SI Appendix, Fig. S1). In addition to grafting, we also prepared libraries from ungrafted hypocotyls (“intact” treatment) and cut plants that had not been reattached (“separated” treatment) (Fig. 1A). Here we refer to tissues harvested above the graft (the scion) or from the shoot side of separated tissue as “top” and tissues from below the graft (the rootstock) or from the root side of separated tissue as “bottom” (Fig. 1A).
Fig. 1.
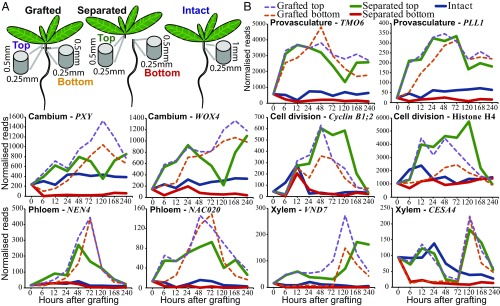
Transcriptional dynamics of genes associated with provasculature, phloem, and xylem development and cell division. (A) Separated and grafted Arabidopsis tissues were harvested ∼0.5 mm above (top) and ∼0.5 mm below (bottom) the cut site. For intact plants, ∼1-mm segments were harvested that spanned the region where cuts were made in grafted and separated plants. (B) Expression levels were plotted over time for intact, separated, and grafted samples.
To understand which developmental processes occur at the graft junction, we looked at the expression of markers associated with vascular formation and cell division. Many markers of cambium, phloem, and provascular development were activated within 6 h of grafting. Provascular markers typically showed an early peak of expression followed by a peak of cambial marker expression (Fig. 1 and SI Appendix, Fig. S2). Expression of most phloem markers peaked at 72 h (Fig. 1 and SI Appendix, Fig. S2), the time when phloem reconnections form in grafted Arabidopsis (4, 5). Notably, the early phloem marker NAC020 activated before the middevelopment phloem marker NAC086, which activated before the late-development phloem marker NEN4, consistent with the dynamics of phloem transcriptional activation during primary root development and leaf vascular induction (SI Appendix, Fig. S2) (26, 27). Certain markers associated with xylem formation, such as VND7 and BFN1, activated early in the grafted top. Other xylem development markers, such as IRX3 and CESA4, activated late in grafted samples. By 120 h after grafting, genes activated in xylem development were expressed in top and bottom, consistent with when the first xylem strands differentiate at the graft junction (4). Genes associated with cell division were activated by 12 h in the grafted top and by 24 h in the grafted bottom (Fig. 1 and SI Appendix, Fig. S2). On the other hand, control genes, the expression of which does not typically vary between tissues and treatments (28), were not differentially expressed in grafted tops or bottoms (SI Appendix, Fig. S2). The RNAseq expression data appeared to correlate well with transcriptional fluorescent reporters for both activation dynamics and the localization of expression (SI Appendix, Fig. S3).
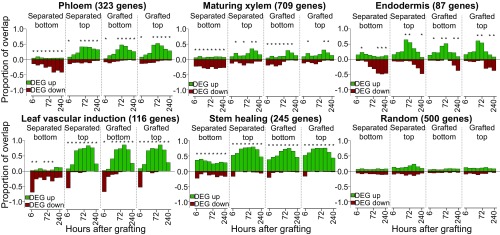
The similar activation dynamics of vascular differentiation genes between grafting and leaf vascular formation prompted us to test whether this phenomenon occurred with other known developmental processes. We obtained lists of genes, the expression of which is associated with various biological processes from previous publications (Dataset S1), and tested how many of the genes differentially expressed in our transcriptomes overlapped with the previously published lists. Differentially expressed genes in grafted samples and separated tops partially overlapped with those the expression of which is associated with phloem, xylem, and procambium formation (Fig. 2 and SI Appendix, Fig. S4). There was a high overlap between Arabidopsis inflorescence stem healing and grafting, as well as between vascular induction in leaf disk cultures and grafting (Fig. 2). Various genes expressed in a cell-type–specific manner also showed a high transcriptional overlap with graft formation, including phloem, endodermis, and protoxylem (Fig. 2 and SI Appendix, Fig. S4). In nearly all cases, the separated top, grafted top, and grafted bottom samples showed similar activation dynamics. The separated bottom samples were exceptional, however, since gene expression associated with vascular development and cell-specific processes was typically down-regulated (Fig. 2 and SI Appendix, Fig. S4). We also compared our datasets with RNAs expressed in longitudinal cross-sections of the Arabidopsis root (29). There was little overlap between grafted bottoms and sections from the root meristematic zone, whereas overlap existed between grafted tops and the root meristematic zone at early time points and between grafting and the root maturation zone (SI Appendix, Fig. S5). Our analysis also revealed that two genes expressed in the cambium, WOX4 and PXY, were induced by grafting, but the primary root marker WOX5 and the lateral root marker LBD18 were not substantially induced (Fig. 1 and SI Appendix, Fig. S2).
Fig. 2.
Transcriptional overlap between previously published vascular datasets and the grafting datasets. Genes, the transcripts of which are associated with various cell types or biological processes, were taken from previously published datasets (Dataset S1) and compared with the transcriptomic datasets generated here. The number in parentheses represents the number of cell-type–specific or process-specific genes identified in previous datasets. Overlap is presented as a ratio of 1.0 for differentially expressed genes (DEG) up- or down-regulated in our dataset relative to intact samples compared with up- and down-regulated genes in the previously published transcriptome dataset. An asterisk represents a significant overlap (P < 0.05) for a given time point.
Genes Are Asymmetrically Expressed Around the Graft.
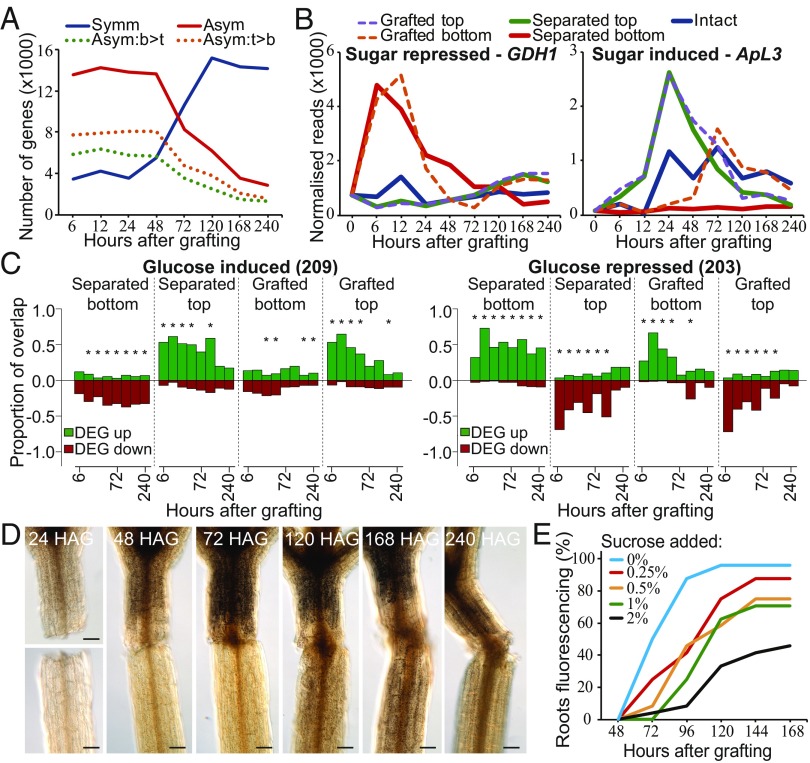
Many of the vascular development and cell-division–related genes initially activated in the grafted top whereas, in some instances, activation was delayed in the grafted bottom by up to 24 h (Fig. 1 and SI Appendix, Figs. S2 and S3). Several genes important for tissue reunion or graft formation show an asymmetric pattern of expression above and below the cut (4, 9), suggesting that asymmetry might be a common feature of grafting and tissue reunion. To investigate the extent of asymmetry at the graft junction, we identified RNAs that were differentially expressed equally in tops and bottoms of grafts (symmetrically expressed) or were more highly expressed in one tissue than in the other (asymmetrically expressed). We identified these genes by performing a pairwise comparison of the protein-coding transcriptome datasets that were differentially expressed as a consequence of grafting relative to intact hypocotyls. Several thousand RNAs were identified that fit either pattern of expression, including the transcript of the cambial markers TMO6 that was induced symmetrically and WOX4 that was induced asymmetrically (Figs. 1 and 3A). Six to 48 h after grafting, the number of graft-differentially expressed genes that were asymmetrically expressed was roughly threefold greater than those symmetrically expressed, indicating that tissues above the cut changed their expression dynamics relative to those below the cut. However, at 72 h the numbers were nearly equal, and by 120 h, the number of symmetrically differentially expressed genes was threefold greater than those asymmetrically expressed (Fig. 3A). Some of the observed asymmetry at the graft junction might have been due to a gradient of differential expression along the length of the intact hypocotyl. We reasoned that, if asymmetry was due to inherent asymmetry in intact hypocotyls, then the average expression of a gene above and below the graft junction would be similar to its expression value in intact hypocotyls. We found that, for each time point, between 141 and 1,465 genes had expression values in intact hypocotyls that were similar to the average expression between the grafted top and grafted bottom (SI Appendix, Fig. S2C), suggesting that some of these genes may be asymmetrically expressed due to inherent asymmetry in the hypocotyl. However, these numbers were a small proportion of the 13,000 genes asymmetrically expressed at early time points (Fig. 3A). As a second approach, we performed a hierarchical clustering analysis that indicated that the grafted top and grafted bottom were initially dissimilar but by 120 h had clustered together and had become highly similar (SI Appendix, Fig. S1), consistent with the symmetry analysis (Fig. 3A). Thus, graft healing promoted a shift from asymmetry to symmetry.
Fig. 3.
Asymmetric changes in accumulation of sugar-responsive RNAs and of starch occur at the graft junction. (A) A pairwise analysis between the grafted top and grafted bottom identified sets of protein-coding genes symmetrically or asymmetrically expressed at the graft junction [false discovery rate (FDR) < 0.05; likelihood of symmetric/asymmetric expression >50%]. Asymmetrically expressed genes were further divided into those the RNAs of which were higher in the top (orange dotted line) or higher in the bottom (green dotted line) (FDR < 0.05; likelihood of asymmetric expression >50%). (B) Expression profiles for transcripts of a sugar-repressed gene (GDH1) and a sugar-induced gene (ApL3) were plotted for intact, separated, and grafted samples. (C) Transcriptional overlap between previously published glucose-induced or glucose-repressed genes and our dataset. The number in parentheses represents the number of glucose-responsive genes identified in the previous dataset (Dataset S1). Overlap is presented as a ratio of 1.0 for differentially expressed genes (DEG) up- or down-regulated in our dataset relative to intact samples compared with up- and down-regulated genes in the previously published transcriptome dataset. An asterisk represents a significant overlap (P < 0.05). (D) Lugol staining of grafted plants at various time points revealed dark brown staining associated with starch accumulation. HAG, hours after grafting. (Scale bars: 100 μm.) (E) pSUC2::GFP-expressing Arabidopsis shoots were grafted to Col-0 wild-type roots and GFP movement to the roots was monitored over 7 d for phloem connection in the presence or absence of various concentrations of sucrose.
Sugar Response Correlates with Asymmetric Gene Expression.
The shift from asymmetry to symmetry could be due to phloem reconnection at 72 h (4) and the resumption of hormone, protein, and sugar transport. We tested a role for sugar by grafting in the presence of exogenous sucrose, which has previously been reported to affect grafting success (30). Low levels of exogenous sucrose lowered grafting efficiency (Fig. 3E), suggesting that differential sugar responses at the graft junction might be important for vascular reconnection. Expression of ApL3, a gene the expression of which is induced by sugar (31), was rapidly up-regulated in separated tops and grafted tops, whereas expression of DIN6, GDH1, and STP1, genes the expression of which is repressed by sugar (31–33), was rapidly up-regulated in separated bottoms and grafted bottoms (Fig. 3B and SI Appendix, Fig. S6). These observations were consistent with sugar accumulation in the grafted top and sugar depletion in the grafted bottom. The expression of these genes returned to levels similar to intact samples by 120 h and, with the exception of ApL3, the grafted samples normalized expression more rapidly than did the separated tissues. Genes associated with photosynthesis increase expression in separated bottoms 24 h after cutting, a common response to starvation (13), but likely too late to affect sugar levels before 24 h (SI Appendix, Fig. S6). A transcriptional overlap analysis with RNAs from known glucose-responsive genes (Dataset S1) revealed a substantial overlap with genes differentially expressed by grafting. RNAs from known glucose-induced genes were up-regulated in separated tops and grafted tops, whereas RNAs from known glucose-repressed genes were up-regulated in separated bottoms and grafted bottoms (Fig. 3C and SI Appendix, Fig. S6). This trend was not observed with genes differentially expressed by mannitol treatment (SI Appendix, Fig. S6), suggesting that the effect was specific to metabolically active sugars. To further investigate this effect, we stained grafted, separated, and intact plants with Lugol solution to assay for the presence of starch. Staining above the graft junction increased 48–72 h after grafting (Fig. 3D). By 120 h, staining was equal on both sides of the graft whereas in separated tops staining became stronger after 72 h (Fig. 3D and SI Appendix, Fig. S6). We concluded that starch accumulated above the cut, but after 72 h, this asymmetry disappeared only in grafted plants. To test whether the accumulation of starch and increased sugar responsiveness could explain the observed transition from asymmetry to symmetry, we compared our datasets to previously published genes that are induced by starvation or are induced by sucrose readdition (Dataset S1). At early time points, 20–31% of asymmetrically expressed genes were known to respond to sugars, whereas only 2–5% of symmetrically expressed genes were known to respond to sugars (SI Appendix, Table S1). However, at 72 h, the overlap between asymmetrically expressed genes and sugar-responsive genes reduced substantially (SI Appendix, Table S1).
Auxin Response Is Symmetric at the Graft.
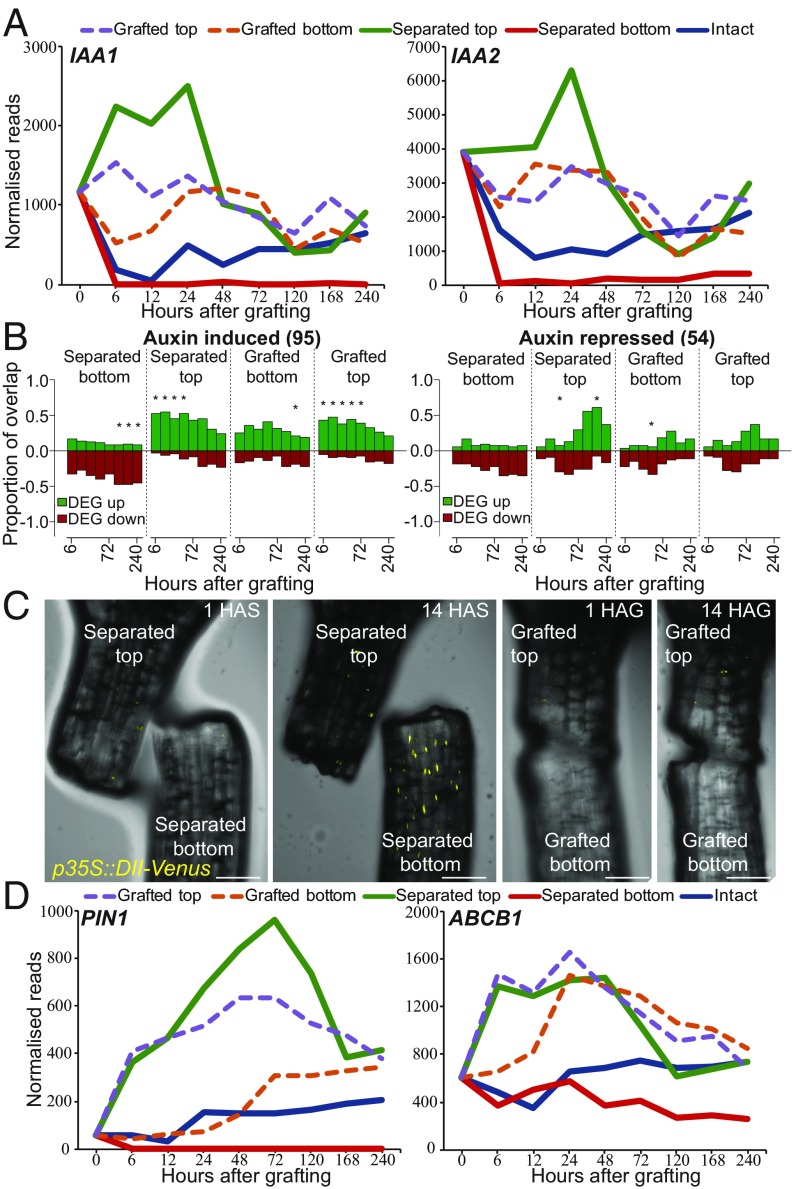
The rapid activation of many vascular markers in the grafted bottoms despite the starvation response promoted us to investigate whether other mobile substances such as phytohormones could play a role in gene activation. We compared lists of genes known to respond to cytokinin, ethylene, or methyl jasmonate (34) and found no substantial overlap between these lists and genes differentially expressed by grafting (SI Appendix, Fig. S7 and Dataset S1). Abscisic acid-responsive and brassinosteroid-responsive genes showed overlap with genes differentially expressed in our datasets, but this overlap was of a similar magnitude in both separated and grafted datasets, suggesting that the effect was not specific to grafting (SI Appendix, Fig. S7). Auxin-responsive transcripts were exceptional, however, as they showed a substantial overlap with RNAs differentially expressed by grafting (Fig. 4 A and B and SI Appendix, Fig. S7). Auxin-induced genes were up-regulated in separated tops, grafted bottoms, and grafted tops whereas they were repressed in separated bottoms (Fig. 4 A and B). Auxin-responsive genes such as IAA1 and IAA2 (35) were induced to similar levels in grafted tops and grafted bottoms by 24 h. To further investigate whether the auxin response was uniform between grafted tops and grafted bottoms, we grafted the auxin-responsive fluorescent reporter p35S:DII-Venus, the fluorescent protein of which is degraded in the presence of auxin (36). DII-Venus fluoresced in the separated bottoms but did not fluoresce in grafted bottoms 14 h after cutting (Fig. 4C), indicating that separated bottoms had a low level of auxin response but grafted tops, grafted bottoms, and separated tops had a high level of auxin response.
Fig. 4.
Auxin response is symmetric at the graft junction. (A and D) Expression profiles for various auxin-responsive genes (IAA1, IAA2) or auxin transporter genes (PIN1, ABCB1) were plotted for intact, separated, and grafted samples. (B) Overlap between previously published auxin-induced or auxin-repressed RNAs and our dataset. The number in parentheses represents the number of auxin-responsive genes identified in the previous dataset (Dataset S1). Overlap is presented as a ratio of 1.0 for differentially expressed genes (DEG) up- or down-regulated in our dataset relative to intact samples compared with up- and down-regulated genes in the previously published transcriptome dataset. An asterisk represents a significant overlap (P < 0.05). (C) Grafted and separated plants expressing the auxin-responsive p35S::DII-Venus transgene that is degraded in the presence of auxin reveal a reduction of auxin response in cut bottoms, but not in grafted bottoms. HAG, hours after grafting; HAS, hours after separation. (Scale bars: 100 μm.)
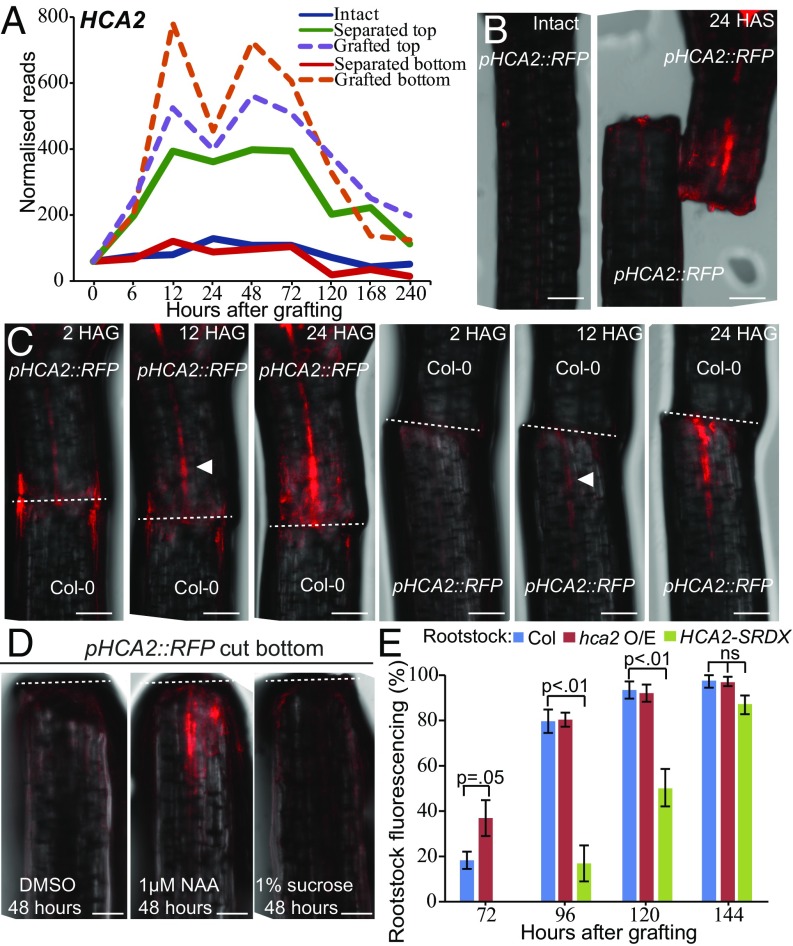
To test whether auxin contributed to activation of gene expression below the graft junction, we monitored the expression of the symmetrically expressed gene HIGH CAMBIAL ACTIVITY 2 (HCA2) (Fig. 5A). We generated a transcriptional fluorescent reporter, pHCA2::RFP, that rapidly activated in grafted bottoms, grafted tops, and separated tops (Fig. 5 B and C). Separated bottoms did not activate pHCA2::RFP expression under grafting conditions or when placed on media containing sucrose or DMSO (Fig. 5 B and D). However, 26 h of synthetic auxin [naphthaleneacetic acid (NAA)] treatment was sufficient to activate pHCA2::RFP at the cut hypocotyl bottom but was insufficient to activate pHCA2::RFP at the primary root tip of intact plants (Fig. 5D and SI Appendix, Fig. S8). We also tested whether activation of HCA2 below the graft junction was important for grafting. Enhancing HCA2 activity (hca2 mutant) in grafted roots improved phloem reconnection rates, whereas suppressing HCA2 targets (p35S::HCA2-SRDX) delayed phloem reconnection (Fig. 5E) (37).
Fig. 5.
HCA2 contributes to graft junction formation. (A) The RNAseq expression profile for HCA2 plotted for intact, separated, and grafted samples. (B and C) HCA2 transcription is up-regulated above and below the graft junction. pHCA2::RFP was grafted to Col-0 roots or Col-0 shoots to avoid ambiguity of signal origin at the junction. HCA2 was also up-regulated in separated tops but not in intact samples or in separated bottoms. HAG, hours after grafting; HAS, hours after separation. White arrowhead denotes initial fluorescent signal; dashed lines denote the cut site. (Scale bars: 100 μm.) (D) Separated hypocotyl bottoms activated pHCA2::RFP expression upon treatment of the synthetic auxin, NAA, after 48 h but did not activate pHCA2::RFP expression with DMSO or sucrose treatment. (Scale bars: 100 μm.) Dashed lines denote the cut site. (E) pSUC2::GFP-expressing Arabidopsis shoots were grafted to roots of Col-0 wild-type, hca2-overexpressing mutants, or plants expressing p35S::HCA2-SRDX. GFP movement to the roots was monitored 3–7 d after grafting for phloem connection.
Tissue Fusion Imparts a Unique Physiological Response That Differs from Tissue Separation.
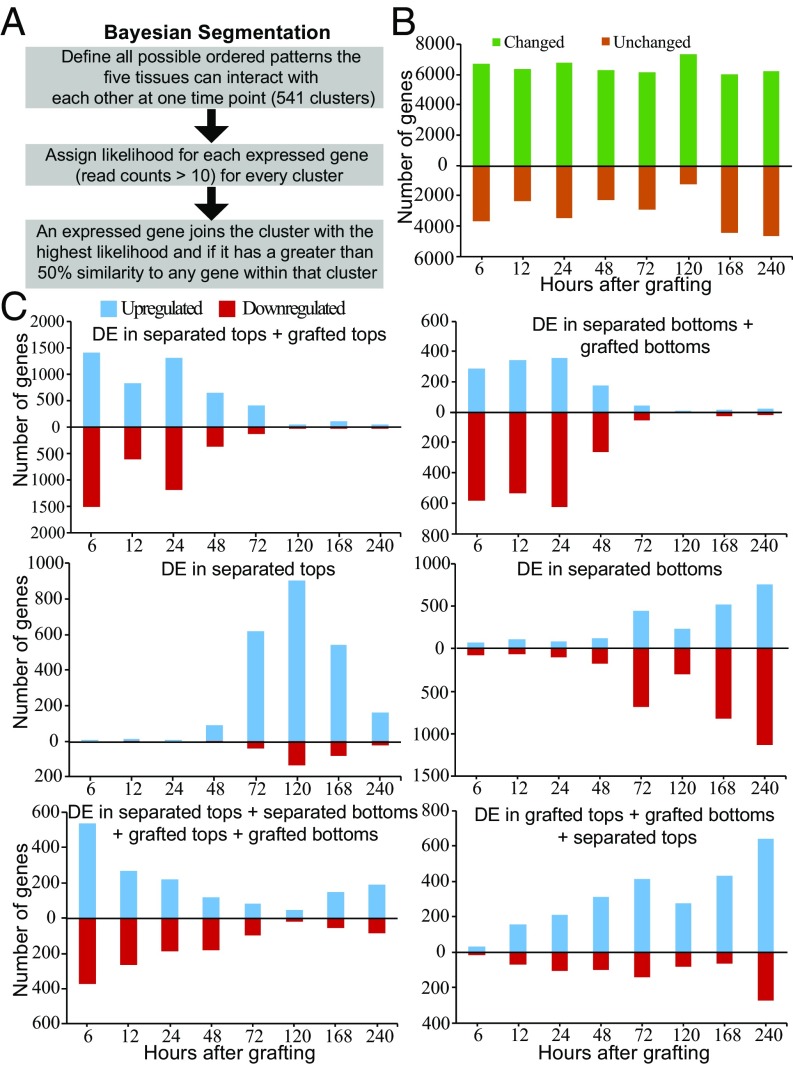
We hypothesized that the symmetric auxin response and asymmetric sugar response at the graft junction could allow a unique transcriptional response since neither separated plants nor intact plants had similar response dynamics to sugars and auxin (Figs. 3 and 4). To uncover protein-coding genes differentially expressed only by grafting, we segmented the transcriptome into groups of genes that behaved similarly and identified groups that corresponded to genes differentially expressed most highly by grafting (Dataset S2). We used an empirical Bayesian analysis (38) to define all possible patterns of differential expression between the five tissue types (intact, grafted top, grafted bottom, separated top, and separated bottom) with orderings allowed (< or >) (Fig. 6A). This analysis produced 541 ordered patterns (“clusters”) and, for each time point, posterior likelihoods on the likelihood of each pattern of expression were calculated for every gene in every tissue. A gene joined the cluster it fit best, and a gene could join only one cluster at each time point. Although there were 541 possible clusters, we found that only 113 clusters contained 10 or more genes for at least one time point whereas 28 clusters contained 200 or more genes for at least one time point (Dataset S2). In the top 113 clusters, ∼6,000 genes were differentially expressed in at least one tissue whereas between 1,000 and 4,000 genes were not differentially expressed (Fig. 6B).
Fig. 6.
Clustering the transcriptome at each time point, based on likelihoods of all possible patterns of differential expression (DE) in grafted, separated, and intact tissues. (A) Overview depicting the Bayesian segmentation. (B) Analysis of differential behavior produced 113 categories containing at least 10 genes, the expression of which was in a specific differential pattern for at least one time point (Dataset S2). One group is composed of genes the transcript levels of which are not substantially changed in the five tissues (unchanged), whereas the other group is composed of the sum of the other 112 groups (genes the transcript levels of which changed after treatment in at least one tissue) over the time points tested. (C) Major categories in the segmentation revealed RNAs the levels of which changed in all of the treatments listed relative to intact samples. Note that a gene can be represented in only one category for a given time point, that is, the category in which the transcript level changes best fit the category.
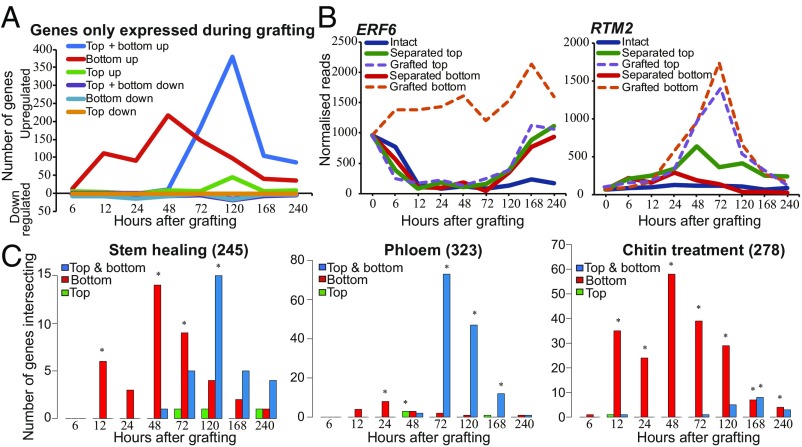
To simplify the analysis, we considered clusters in which gene expression was grouped into patterns consisting of one comparison between two groups. At early time points, the cluster containing genes with similar differential expression in both grafted tops, grafted bottoms, separated tops, and separated bottoms had high numbers that decreased with time and could represent a general wound response (Fig. 6C). A gene ontology (GO) analysis of the genes in this cluster revealed that they were highly enriched in defense, immune, and wound-responsive genes at 6 h and that this enrichment decreased as the graft healed (Dataset S3). Clusters containing genes with similar differential expression in both separated tops and grafted tops had high numbers that decreased with time, similar to the trend observed with clusters containing genes with similar differential expression in both separated bottoms and grafted bottoms. This observation indicated that the grafted top was initially transcriptionally similar to the separated top, whereas the grafted bottom was initially transcriptionally similar to the separated bottom. After the 48-h time point, clusters containing genes differentially expressed only in separated tops or differentially expressed only in separated bottoms increased in numbers, suggesting that these tissues gained a unique pattern of gene expression. The clusters containing genes with similar differential expression in both grafted tops, separated tops, and grafted bottoms increase in numbers throughout the healing process (Fig. 6C). We searched for grafting-specific cluster categories with one or more orderings in which genes were most highly differentially expressed by grafting (Dataset S2). There were very few genes down-regulated by grafting or up-regulated only in the grafted top (Fig. 7A). Instead, clusters contained several hundred differentially expressed genes up-regulated either in the grafted bottom only or up-regulated in both grafted bottom and grafted top (Fig. 7A). Genes, the expression of which changed only in the grafted bottom sample, were prevalent early during grafting and were most common at 48 h, whereas genes activated in both top and bottom became prevalent at 48 h and were most common at 120 h (Fig. 7 A and B). We performed a GO analysis and found that genes differentially expressed most highly in the grafted bottom sample were enriched in the immune response and chitin response biological process categories (Dataset S3). Previously published chitin-induced RNAs had a high proportion of overlap with differentially expressed graft bottom-specific genes (Fig. 7C). A GO analysis also revealed that grafting-specific RNAs expressed in both the grafted top and grafted bottom were enriched in vascular-related biological processes (Dataset S3). Previously published phloem-enriched, endodermal-enriched, vascular-induction, and stem-wounding associated RNAs had a high proportion of overlap with these differentially expressed graft-specific genes (Fig. 7C and SI Appendix, Fig. S9). Since few genes were grafting-specific and grafted tissues were initially transcriptionally quite similar to separated tissues (Figs. 6C and 7A), we tested whether tissues separated for short periods could be grafted with similar reconnection dynamics as tissues that had been grafted immediately. Plants were cut and grafted after 0–5 d of separation. Separation did not speed up vascular reconnection, and instead, it always took 3 d from the point of tissue attachment for phloem connections to form (SI Appendix, Fig. S9). Furthermore, the shoot lost competence to graft 2–3 d after separation whereas the root remained competent to graft up to 5 d after separation (SI Appendix, Fig. S9). Together, it appears that the grafted shoot and root have a unique physiological response that differs from the separated shoot and root and that tissue attachment is required to activate graft formation.
Fig. 7.
A subset of genes is differentially expressed only during graft formation compared with intact or separated tissues. (A) Certain genes were only differentially expressed in grafted tops, grafted bottoms, or both in grafted tops and grafted bottoms. (B) Expression profiles for a graft bottom-specific (ERF6) or a graft top and bottom differentially expressed gene (RTM2) were plotted for intact, separated, and grafted samples. (C) Grafting-specific genes are also expressed in other processes such as stem healing, phloem reconnection, and treatment with chitin. Genes, the transcripts of which are associated with these biological processes, were taken from previously published datasets (Dataset S1) and compared with our dataset to assess transcriptional overlap with genes expressed in the grafted top, in the grafted bottom, or in both grafted top and grafted bottom. The number in parentheses represents the number of process-specific genes identified in previous datasets. An asterisk represents a significant overlap (P < 0.05).
Discussion
To better understand how plants graft, we analyzed in depth an RNA deep-sequencing dataset that spatially and temporally distinguished genes activated by cutting followed by tissue attachment or continuous tissue separation. Cutting promoted a similar wound response in both grafted and separated tissues; however, by 72 h after cutting, the grafted and separated tissues became transcriptionally dissimilar (Fig. 6C), indicating that tissue fusion was mechanistically different from healing an unattached cut surface. During graft formation, tissues had a very high transcriptional overlap with genes differentially expressed by inflorescence stem healing and by vascular induction in leaves (Fig. 2 and SI Appendix, Fig. S4) (9, 26), suggesting that grafting is closely related to these processes. Graft formation had little transcriptional overlap with lateral root formation (SI Appendix, Fig. S2) (29) and appeared to follow a pathway similar to secondary root growth since the secondary growth-specific cambium markers WOX4 and PXY (39) were activated by grafting (Fig. 1 and SI Appendix, Fig. S2). Grafted tops initially showed a short-lasting and small transcriptional overlap with genes expressed during primary root formation, which may be related to the accumulation of substances activating adventitious root formation, a common response in failed grafts or in cut shoots (SI Appendix, Fig. S6C). Thus, we conclude that grafting likely proceeds via a pathway involving secondary growth with radial meristems activating in the mature cambium to heal the wound. Vascular formation genes including those specifying cambium and phloem were activated early, followed by an activation of cell division genes, suggesting that the start of cellular differentiation preceded activation of cell division. Xylem identity genes showed an early and a late activation peak (Fig. 1 and SI Appendix, Fig. S2). There is no visible xylem differentiation at the graft junction during the first peak of expression (4), and this expression could represent programmed cell death that does not lead to xylem differentiation. Alternatively, these genes might be suppressed by phloem differentiation genes that suppress protoxylem formation (40, 41). The second expression peak of xylem-related genes at 120 h occurred after the differentiation of functional phloem and coincided with the differentiation of xylem strands at the graft junction (4). Previous studies highlighted the importance of callus and pericycle cells during regeneration (18, 42), but we see little evidence that genes expressed in the pericycle or during callus formation have high transcriptional overlap with genes differentially expressed by grafting (SI Appendix, Fig. S4). Expression profiles for all protein-coding genes can be found in Datasets S4 and S5.
A high proportion of genes were initially asymmetrically expressed (Fig. 3A), and many had a delay in phloem, cambium, and cell division activation below the graft junction compared with above it (Fig. 1 and SI Appendix, Fig. S2). Several genes associated with vascular formation, such as HCA2 (37) and TMO6 (43), activated equally in both grafted top and grafted bottom at 6 h after grafting (Figs. 1 and 5 A–C). These data indicate that, at least transcriptionally, the grafted root rapidly responded to the presence of the grafted shoot and that this response was independent of functional vascular connections. This response was not present in separated roots, indicating that attachment was key for recognition and activation of graft formation (Fig. 5 and SI Appendix, Fig. S9). Sugars are known activators of cell division and cell elongation (13), and, in our datasets, a large proportion of genes asymmetrically expressed are also sugar-responsive (SI Appendix, Table S1). However, sugars are transported in the phloem (12) that is severed upon grafting, and the grafted root exhibited a sugar-starvation response and showed similar sugar-response dynamics as the separated root. Instead, we infer that some other molecular that is transported in the absence of vascular connections activated HCA2 and TMO6 as well as cell division, phloem-, and cambium-related genes in the grafted bottom.
Given auxin’s fundamental role in vascular formation (39), it is a strong candidate for an activating signal. Auxin response was largely symmetric from 12 h after grafting (Fig. 4 and SI Appendix, Fig. S7), consistent with previous findings that the auxin-inducible DR5, IAA5, and ANAC071 genes activate above and below the graft junction within 1–3 d of grafting (4–6, 44). Furthermore, exogenous auxin application combined with cutting was sufficient to activate HCA2 expression in separated root hypocotyls (Fig. 5D). One idea is that grafting caused an interruption in auxin transport, and, where opposing tissues adhered, auxin transport resumed regardless of vascular connections since auxin is transported from cell to cell through the apoplast (8). The genes encoding the auxin efflux proteins PIN1 and ABCB1 were transcriptionally activated above the graft junction (Fig. 4D), similar to the putative Pisum sativum PIN1 protein accumulating above a cut stem before vascular reconnection (45), and could reflect a role for these proteins in exporting auxin across the cut. Consistent with these observations, adding an auxin transport inhibitor to grafted Arabidopsis shoots prevented the expression of grafting-induced genes below the graft junction (6). Although auxin response was largely symmetric, our previous work demonstrated that the auxin signaling genes ALF4 and AXR1 are important for grafting only below the graft junction (4). Mutating ALF4 below the graft junction more strongly reduced auxin response than mutating ALF4 above the junction (4). Thus, proteins such as ALF4 or AXR1 might act by promoting auxin response and vascular regeneration below the graft junction, which could be particularly important when there is incomplete attachment, cellular damage, or inefficient transport. All higher plants transport auxin from shoot to root, yet not all plant species can be successfully grafted (3) so the response to auxin rather than the transport itself may be a determining factor in the ability to graft. A role for sugars is not completely ruled out, however, since the magnitude of differential expression of vascular-related genes was often lower in the grafted bottom (SI Appendix, Fig. S2). In addition, very low levels of exogenous sugars can improve graft formation under certain conditions (30). Altogether, endogenous sugars likely enhance cell division and differentiation, perhaps similar to their role in enhancing the rate of pericycle cell divisions in the hypocotyl (46).
Our analyses identified two groups of genes, the expression changes of which were unique to graft formation in our experiments (Fig. 7). One group activated shortly after grafting below the graft junction and was enriched in immune-responsive and chitin-responsive genes (Fig. 7 and SI Appendix, Fig. S9 and Dataset S3). The breakdown products of cell walls are potent elicitors of defense responses (47), so it is possible that the grafted bottom up-regulates pathways specific to wound damage response. This group was not up-regulated in separated bottoms, however, so the unique physiological state of the grafted root, indicated by the presence of the auxin response but the absence of the sugar response, could have promoted their up-regulation. The second group activated both above and below the graft junction and became highly expressed later during graft formation (Fig. 7). This group was enriched in RNAs associated with vascular development (Fig. 7 and Dataset S3), and we suggest that the products of these genes are involved in the vascular reconnection processes between the two tissues. Despite many transcriptional similarities between separated tops and grafted tissues, tissues had to be attached for at least 3 d for phloem connections to form, regardless of when cutting occurred (SI Appendix, Fig. S9). Thus, it appears that RNAs expressed in the separated top or separated bottom are insufficient to drive graft formation. Instead, genes activated uniquely by grafting or genes involved in the recognition response are those that contribute to distinguishing attached from separated plant tissues. Future work should focus on these genes to identify the pathways required for grafting that could be modified to improve graft formation, wound healing, and vascular regeneration. Likewise, the rapid transcriptional changes below the graft indicate a recognition system that promotes tissue regeneration. Identifying the cues that trigger recognition and understanding how they are perceived should be priorities, as should understanding whether this phenomenon applies more broadly to intertissue communication, tissue regeneration, or tissue fusion events, such as parasitic plant infections (48), epidermal fusions (49, 50), or petal fusions (51).
Materials and Methods
Plant Material and Microscopy.
The A. thaliana accession Columbia was used throughout except where indicated. The p35S::GFP-ER (52), pSUC2::GFP (53), pUBQ10::PM-tdTomato (54), pANT::H2B-YFP (55), pLOG4::n3GFP (56), pCASP1::NLS-GFP (57), p35S::DII-Venus (36), hca2 (37), p35S::HCA2-SRDX (37), and pDR5rev::GFP-ER (58) lines were previously published. The pARR5::GFP line (59) was previously published and is in the Ws background. For the construction of pHCA2::RFP-ER, a 2.9-kb 5′ upstream region of the HCA2 gene (At5g62940) was cloned into the pDONRp4-p1R donor vector and recombined with tagRFPer into a destination vector by the Multisite Gateway system (60). A. thaliana micrografting and grafting assays were performed according to previously published protocols (61, 62). Fluorescent images were taken on a Zeiss LSM-700 or LSM-780 confocal microscope. Black and white fluorescent images of graft junctions were taken on a Zeiss V12 dissecting microscope. FIJI software (Fiji.sc) was used to process images. Breaking force was calculated using a microextensometer (63) to apply force to either side of the graft junction until it broke.
RNAseq Sample and Library Preparation.
The grafted wild-type A. thaliana accession Col-0 was harvested at the respective time points and care was taken to separate grafts by gently pulling plants apart. Approximately 0.5 mm of tissue was taken above or below each cut site and kept separate. Intact plants had 1 mm of tissue taken from a similar location on the hypocotyl as separated or grafted plants. Grafted, separated, or intact tissues were pooled into groups of ∼24 tissues. Tissues were ground using a microcentrifuge pestle frozen in liquid nitrogen. RNA was extracted using an RNeasy Kit (Qiagen) following the manufacturer’s instructions. RNA (90–100 ng) was used to prepare RNAseq libraries using the TruSeq Stranded mRNA LT kit (Illumina) according to the manufacturer’s instructions. The final PCR was for 15 cycles, and 11–12 barcoded samples were randomly mixed to make a total of seven mixes for seven flow lanes, one mix per lane. Biological replicates of each sample were sequenced on the HiSeq 4000 platform with paired-end 75-bp transcriptome sequencing (BGI Tech Solutions). RNAseq data are available from the Gene Expression Omnibus database (GSE107203).
Iodine Staining.
Arabidopsis seedlings were placed in a fixation solution (3.7% formaldehyde, 50% ethanol, 5% acetic acid) for 1 h at room temperature and then transferred to 70% ethanol for 10 min. Afterward, plants were transferred to 96% ethanol and stored at −20 °C for up to a week. Samples were rehydrated in 50% ethanol for 1 h at room temperature, transferred to distilled water for 30 min, and then stained for 10 min in Lugol solution (Sigma) at room temperature. Plants were rinsed with water and mounted on microscope slides. Images were taken on a Zeiss Axioimager.M2 microscope.
Bioinformatic Analyses.
The reads acquired through high-throughput sequencing were quality-trimmed with sickle (64), aligned with Bowtie2, and assigned to protein-coding gene sequences acquired from TAIR10 using eXpress. Library scaling factors were inferred from the sum of the number of reads assigned to the genes in the lowest 75 percentile of expressed genes for each library (65). Analyses of the data were carried out using the R package baySeq (38), and clustering based on the posterior probabilities was acquired from this package and the clusterSeq package (66). The GO enrichment analysis on grafting-specific genes was done with a customized R script (https://github.com/AlexGa/GraftingScripts) using the package GOstats (67).
Supplementary Material
Acknowledgments
We thank Niko Geldner, Dolf Weijers, Paul Tarr, Yka Helariutta, Ruth Stadler, Li-Jia Qu, and the Nottingham Arabidopsis Stock Centre for providing seeds. Funding for this work was provided by Gatsby Charitable Trust Grants GAT3272/C and GAT3273-PR1; by Knut and Alice Wallenberg Academy Fellowship KAW2016.0274 (to C.W.M.); by a SKW Stickstoffwerke Piesteritz GmbH Research Foundation Grant (to A.G. and I.G.); by German Science Foundation (DFG) Grants GR 3526/2, GR 3526/6, and FZT 118 (to I.G.); and by Howard Hughes Medical Institute and Gordon and Betty Moore Foundation Grant GBMF3406 (to E.M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE107203).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718263115/-/DCSupplemental.
References
- 1.Goldschmidt EE. Plant grafting: New mechanisms, evolutionary implications. Front Plant Sci. 2014;5:727. doi: 10.3389/fpls.2014.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee J, et al. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci Hortic (Amsterdam) 2010;127:93–105. [Google Scholar]
- 3.Melnyk CW. Plant grafting: Insights into tissue regeneration. Regeneration (Oxf) 2016;4:3–14. doi: 10.1002/reg2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melnyk CW, Schuster C, Leyser O, Meyerowitz EM. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr Biol. 2015;25:1306–1318. doi: 10.1016/j.cub.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin H, et al. Graft-union development: A delicate process that involves cell-cell communication between scion and stock for local auxin accumulation. J Exp Bot. 2012;63:4219–4232. doi: 10.1093/jxb/ers109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka K, et al. Differential cellular control by cotyledon-derived phytohormones involved in graft reunion of Arabidopsis hypocotyls. Plant Cell Physiol. 2016;57:2620–2631. doi: 10.1093/pcp/pcw177. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto-Kitano M, et al. Cytokinins are central regulators of cambial activity. Proc Natl Acad Sci USA. 2008;105:20027–20031. doi: 10.1073/pnas.0805619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leyser O. Auxin, self-organisation, and the colonial nature of plants. Curr Biol. 2011;21:R331–R337. doi: 10.1016/j.cub.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 9.Asahina M, et al. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:16128–16132. doi: 10.1073/pnas.1110443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wetmore RH, Rier JP. Experimental induction of vascular tissues in callus of angiosperms. Am J Bot. 1963;50:418–430. [Google Scholar]
- 11.Aloni R. Role of auxin and sucrose in the differentiation of sieve and tracheary elements in plant tissue cultures. Planta. 1980;150:255–263. doi: 10.1007/BF00390835. [DOI] [PubMed] [Google Scholar]
- 12.Lough TJ, Lucas WJ. Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Ruan YL. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013;4:163. doi: 10.3389/fpls.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlemeier C, Timmermans MC. The Sussex signal: Insights into leaf dorsiventrality. Development. 2016;143:3230–3237. doi: 10.1242/dev.131888. [DOI] [PubMed] [Google Scholar]
- 15.Qi J, et al. Auxin depletion from leaf primordia contributes to organ patterning. Proc Natl Acad Sci USA. 2014;111:18769–18774. doi: 10.1073/pnas.1421878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 17.Cheong YH, et al. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwase A, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011;21:508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Ikeuchi M, et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 2017;175:1158–1174. doi: 10.1104/pp.17.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cookson SJ, et al. Graft union formation in grapevine induces transcriptional changes related to cell wall modification, wounding, hormone signalling, and secondary metabolism. J Exp Bot. 2013;64:2997–3008. doi: 10.1093/jxb/ert144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cookson SJ, et al. Heterografting with nonself rootstocks induces genes involved in stress responses at the graft interface when compared with autografted controls. J Exp Bot. 2014;65:2473–2481. doi: 10.1093/jxb/eru145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng BS, et al. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol. 2010;30:297–303. doi: 10.1093/treephys/tpp102. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, et al. Transcriptome changes between compatible and incompatible graft combination of Litchi chinensis by digital gene expression profile. Sci Rep. 2017;7:3954. doi: 10.1038/s41598-017-04328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsay DW, Yeoman MM, Brown R. An analysis of the development of the graft union in Lycopersicon esculentum. Ann Bot (Lond) 1974;38:639–646. [Google Scholar]
- 25.Moore R. Graft formation in Solanum pennellii (Solanaceae) Plant Cell Rep. 1984;3:172–175. doi: 10.1007/BF00270192. [DOI] [PubMed] [Google Scholar]
- 26.Kondo Y, et al. Vascular cell induction culture system using Arabidopsis leaves (VISUAL) reveals the sequential differentiation of sieve element-like cells. Plant Cell. 2016;28:1250–1262. doi: 10.1105/tpc.16.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta KM, et al. Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science. 2014;345:933–937. doi: 10.1126/science.1253736. [DOI] [PubMed] [Google Scholar]
- 28.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 30.Marsch-Martínez N, et al. An efficient flat-surface collar-free grafting method for Arabidopsis thaliana seedlings. Plant Methods. 2013;9:14. doi: 10.1186/1746-4811-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villadsen D, Smith SM. Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Mol Biol. 2004;55:467–477. doi: 10.1007/s11103-004-1050-0. [DOI] [PubMed] [Google Scholar]
- 32.Thum KE, Shin MJ, Palenchar PM, Kouranov A, Coruzzi GM. Genome-wide investigation of light and carbon signaling interactions in Arabidopsis. Genome Biol. 2004;5:R10. doi: 10.1186/gb-2004-5-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cordoba E, Aceves-Zamudio DL, Hernández-Bernal AF, Ramos-Vega M, León P. Sugar regulation of SUGAR TRANSPORTER PROTEIN 1 (STP1) expression in Arabidopsis thaliana. J Exp Bot. 2015;66:147–159. doi: 10.1093/jxb/eru394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- 36.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 37.Guo Y, Qin G, Gu H, Qu LJ. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell. 2009;21:3518–3534. doi: 10.1105/tpc.108.064139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardcastle TJ, Kelly KA. baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Rybel B, Mähönen AP, Helariutta Y, Weijers D. Plant vascular development: From early specification to differentiation. Nat Rev Mol Cell Biol. 2016;17:30–40. doi: 10.1038/nrm.2015.6. [DOI] [PubMed] [Google Scholar]
- 40.Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y. APL regulates vascular tissue identity in Arabidopsis. Nature. 2003;426:181–186. doi: 10.1038/nature02100. [DOI] [PubMed] [Google Scholar]
- 41.Ito Y, et al. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- 42.Sugimoto K, Jiao Y, Meyerowitz EM. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell. 2010;18:463–471. doi: 10.1016/j.devcel.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 43.Gardiner J, Sherr I, Scarpella E. Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int J Dev Biol. 2010;54:1389–1396. doi: 10.1387/ijdb.093006jg. [DOI] [PubMed] [Google Scholar]
- 44.Pitaksaringkarn W, Ishiguro S, Asahina M, Satoh S. ARF6 and ARF8 contribute to tissue reunion in incised Arabidopsis inflorescence stems. Plant Biotechnol. 2014;31:49–53. [Google Scholar]
- 45.Sauer M, et al. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006;20:2902–2911. doi: 10.1101/gad.390806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skylar A, Sung F, Hong F, Chory J, Wu X. Metabolic sugar signal promotes Arabidopsis meristematic proliferation via G2. Dev Biol. 2011;351:82–89. doi: 10.1016/j.ydbio.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Souza CA, et al. Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 2017;173:2383–2398. doi: 10.1104/pp.16.01680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musselman LJ. The biology of Striga, Orobanche, and other root-parasitic weeds. Annu Rev Phytopathol. 1980;18:463–489. [Google Scholar]
- 49.Lolle SJ, Hsu W, Pruitt RE. Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics. 1998;149:607–619. doi: 10.1093/genetics/149.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becraft PW, Stinard PS, McCarty DR. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 51.Zhong J, Preston JC. Bridging the gaps: Evolution and development of perianth fusion. New Phytol. 2015;208:330–335. doi: 10.1111/nph.13517. [DOI] [PubMed] [Google Scholar]
- 52.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 53.Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segonzac C, et al. The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell. 2012;24:3186–3192. doi: 10.1105/tpc.111.091264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Randall RS, et al. AINTEGUMENTA and the D-type cyclin CYCD3;1 regulate root secondary growth and respond to cytokinins. Biol Open. 2015;4:1229–1236. doi: 10.1242/bio.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rybel B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 57.Roppolo D, et al. A novel protein family mediates casparian strip formation in the endodermis. Nature. 2011;473:380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- 58.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 59.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 60.Siligato R, et al. MultiSite gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melnyk CW. Grafting with Arabidopsis thaliana. Methods Mol Biol. 2017;1497:9–18. doi: 10.1007/978-1-4939-6469-7_2. [DOI] [PubMed] [Google Scholar]
- 62.Melnyk CW. Monitoring vascular regeneration and xylem connectivity in Arabidopsis thaliana. Methods Mol Biol. 2017;1544:91–102. doi: 10.1007/978-1-4939-6722-3_9. [DOI] [PubMed] [Google Scholar]
- 63.Robinson S, et al. An automated confocal micro-extensometer enables in vivo quantification of mechanical properties with cellular resolution. Plant Cell. 2017;29:2959–2973. doi: 10.1105/tpc.17.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi NA, Fass JN. 2011 Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files, Version 1.33. Available at https://github.com/najoshi/sickle.
- 65.Hardcastle TJ, Kelly KA, Baulcombe DC. Identifying small interfering RNA loci from high-throughput sequencing data. Bioinformatics. 2012;28:457–463. doi: 10.1093/bioinformatics/btr687. [DOI] [PubMed] [Google Scholar]
- 66.Hardcastle TJ, Papatheodorou I. 2017. ClusterSeq: Methods for identifying co-expression in high-throughput sequencing data. bioRxiv:188581.
- 67.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.