Abstract
Bacterial microcompartments are large proteinaceous structures that act as metabolic organelles in many bacterial cells. A shell or capsid, which is composed of a few thousand protein subunits, surrounds a series of sequentially acting enzymes and controls the diffusion of substrates and products into and out of the interior. The carboxysome and the propanediol utilization microcompartment represent two well-studied systems among seven or more distinct types that can be delineated presently. Recent structural studies have highlighted a number of sophisticated mechanisms that underlie the function of bacterial microcompartment shell proteins. This review updates our understanding of bacterial microcompartment shells, how they are assembled, and how they carry out their functions in molecular transport and enzyme organization.
Introduction
Many bacterial species produce giant proteinaceous compartments – polyhedral in shape and roughly 1000 Å in diameter – for the purpose of sequestering key metabolic reactions within the cell [Bobik, 2006; English et al., 1994; Kerfeld et al., 2010; Shively et al., 1973; Yeates et al., 2010; Yeates et al., 2011]. By spatially separating distinct cellular processes, they serve as metabolic organelles in bacteria. These bacterial microcompartments, referred to hereafter as MCP’s, consists of a series of sequentially acting enzymes enclosed within a thin shell or capsid, which is assembled from a few thousand shell protein subunits. MCP’s are present in approximately 20% of known bacteria, and distinct types dedicated to several different metabolic processes have been delineated; a few have been studied experimentally while others have so far only been inferred from genomic analyses.
MCP’s function to improve the flux or direct the flow of metabolites through a sequestered pathway (Figure 1). The way in which this confers a selective advantage appears to differ somewhat between different types of MCP’s. The carboxysome, the founding member of the MCP’s, encapsulates two enzymes: carbonic anhydrase and RuBisCO[Cannon et al., 2001]. Carbonic anhydrase dehydrates bicarbonate, after it has diffused across the shell from the cytosol into the lumen of the MCP, to produce CO2 [Cannon et al., 2010; Heinhorst et al., 2006]. There the CO2 is ‘fixed’ by its addition to ribulose bisphosphate in a reaction catalyzed by RuBisCO, a notoriously slow and imperfectly selective enzyme, before the nonpolar CO2 can escape back into the cytosol and across the cell membrane. Other types of MCP’s are more complex than the carboxysome, enclosing more enzymes and more complex reactions. Two types of MCP’s prevalent in enteric bacteria are the Pdu MCP, which metabolizes 1,2-propanediol, and the Eut MCP, which metabolizes ethanolamine [Bobik et al., 1999; Chen et al., 1994; Kofoid et al., 1999; Penrod and Roth, 2006; Stojiljkovic et al., 1995]. These two compartments carry out related reaction schemes (Figure 1), both producing aldehydes as intermediates – propionaldehyde in the case of Pdu and acetaldehyde in the case of Eut. These aldehyde intermediates are further metabolized to alcohols and CoA-esters within their respective MCP’s before the aldehydes can escape into the cytosol. Retaining aldehyde intermediates within the MCP prevents those chemically reactive molecules from damaging cellular DNA[Havemann et al., 2002; Rondon et al., 1995a; Rondon et al., 1995b; Sampson and Bobik, 2008]. It also prevents valuable carbon compounds from being lost by diffusion across the cell membrane, which is particularly problematic for the more volatile acetaldehyde [Penrod and Roth, 2006; Sampson and Bobik, 2008]. The Pdu and Eut MCP’s both carry out vitamin B12-dependent reactions as their first encapsulated step, with subsequent reactions requiring additional cofactors (e.g. NAD+/NADH, ATP, CoA, and iron-sulfur clusters). Although studies support the presence of active mechanisms for regenerating some cofactors internally, it appears likely that some transport of cofactors across the shell must occur [Cheng et al., 2012]. How this might be accomplished without allowing smaller aldehyde molecules to escape presents an intriguing mechanistic puzzle.
Figure 1.
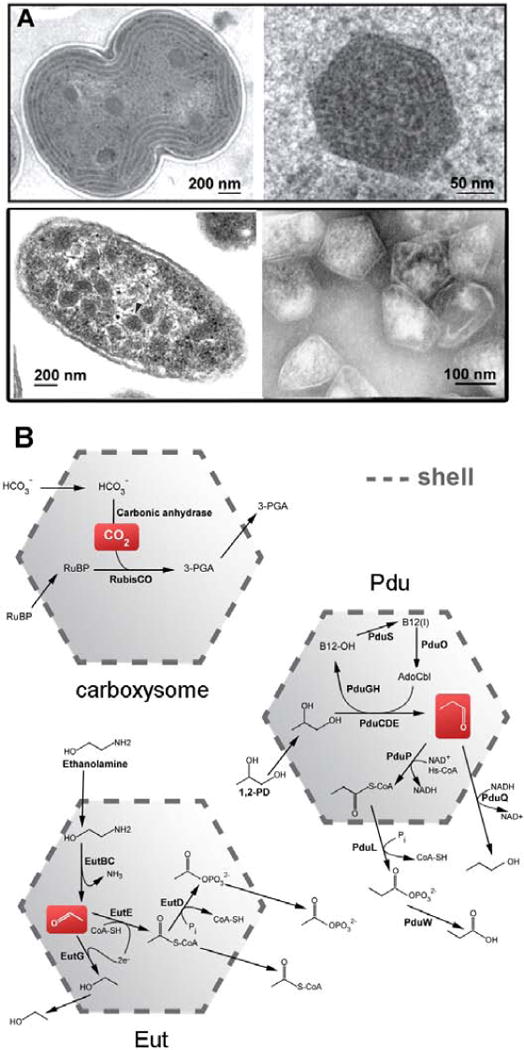
Bacterial microcompartments and models of their sequestered pathways. (A) Thin-section EM of a dividing cell of the cyanobacterium Synechocystis sp. PCC6803 (top left) along with an enlargement of a single carboxysome (top right, courtesy of Wim Vermaas). (bottom left) EM of thin-sectioned Salmonella enterica serovar Tyhpimurium LT2 (reprinted from [Crowley et al., 2008]) and (bottom right) purified Pdu microcompartments (reprinted from [Crowley et al., 2008]). (B) Models for CO2 fixation and 1,2-propanediol and ethanolamine metabolism in the carboxysome, Pdu and Eut microcompartments, respectively.
To date, the range of metabolic reactions that occur within MCP’s has only been partially explored. Aside from those described above that have been studied in some experimental detail, a few other categories have been delineated. One type, present across a wide range of bacteria, appears to metabolize 1,2-propanediol (and possibly other similar compounds such as glycerol), but with an initial reaction step different from the B12-dependent Pdu and Eut MCP’s. In this case, the key dehydration reaction involves a glycyl radical-based enzyme rather than a B12 cofactor [Jorda et al., 2013; Petit et al., 2013; Scott et al., 2006]. The name Grp (glycyl radical-based propanediol) has been proposed for this type of MCP. Gene expression data, bioinformatics analyses, and preliminary structural results suggest that the Grp MCP is similar in major respects to the other MCP’s characterized so far. Other prospective types of MCP’s, including one that has been proposed to metabolize amino alcohols in select mycobacteria [Jorda et al., 2013], and another that has been proposed to metabolize fucose and rhamnose (via 1,2-propanediol), [Petit et al., 2013; Scott et al., 2006] possibly using an encapsulated aldolase, have yet to be explored experimentally.
The shells of MCP’s play key functional roles. They form a barrier that separates the cytosol from the lumen of the MCP. They provide narrow pores for the selective diffusion of the substrates and products of the enclosed pathway. And they selectively bind and organize the enzymes that are enclosed within the MCP. In this review we focus on our current understanding of those mechanisms, with an emphasis on insights that have come from structural studies on numerous shell proteins.
MCP Shell Proteins
The shells of MCP’s are formed predominantly by a single family of proteins, referred to here as BMC shell proteins (PF00936 in the PFAM database [Finn et al., 2008]) which are typically about 100 amino acids long. It is notable that metabolically diverse MCP’s are all assembled from this conserved family of proteins. BMC genes have apparently spread across the bacterial kingdom by horizontal gene transfer events [Bobik et al., 1999]. The BMC shell proteins were first identified in seminal work by Shively et al. by the analysis of purified carboxysomes, followed by gene sequencing[English et al., 1994]. This led to the discovery of other types of MCP’s by sequence analysis [Bobik, 2006; Bobik et al., 1999; Chen et al., 1994; Stojiljkovic et al., 1995]. The shell proteins and the enzymes they encapsulate are typically encoded together in operons. In some bacteria these MCP operons encode more than 20 different shell proteins and enzymes. A remarkable property of most MCP operons is the presence of multiple BMC genes encoding a set of paralogous shell proteins – as many as seven in some cases (Figure 1). Although direct evidence is lacking on the composition of individual MCP particles, the prevailing view is that individual particles are composed of mixtures of multiple BMC paralogs, with each paralog serving a specialized function in the shell. A single shell consists of a few thousand protein subunits in total.
Crystal structures of BMC shell proteins, beginning with the first studies in 2005, have provided rich insights into mechanisms of assembly in MCP’s (Figure 2)[Crowley et al., 2010; Crowley et al., 2008; Heldt et al., 2009; Kerfeld et al., 2005; Klein et al., 2009; Pang et al., ; Sagermann et al., 2009; Takenoya et al., 2010; Tanaka et al., 2009; Tanaka et al., 2010; Tsai et al., 2007; Yeates et al., 2010]. Diverse BMC shell proteins assemble to form disk-shaped homohexamers. These are the basic building blocks of the shell. Typically, one side of the hexamer is relatively flat and polar in character. The other side often bears a bowl-shaped depression, depending on the disposition of flexible C-terminal segments of the protein. This side tends to be more hydrophobic, and is believed to represent the inward-facing or lumenal surface of the shell [Fan et al., 2012]. Each hexamer typically bears a narrow central pore, which is presumed to provide a route for molecular transport across the shell. The BMC hexamers are shaped so that they can further associate side-by-side to make an essentially solid molecular layer, about 20 Å thick. Extended layers of this type have been observed in the context of numerous crystals of different BMC shell proteins, and formation of true two-dimensional layers from purified shell proteins has been demonstrated by electron microscopy [Dryden et al., 2009]. These molecular layers are presumed to represent the nearly flat facets of the polyhedral MCP shell (Figure 2).
Figure 2.
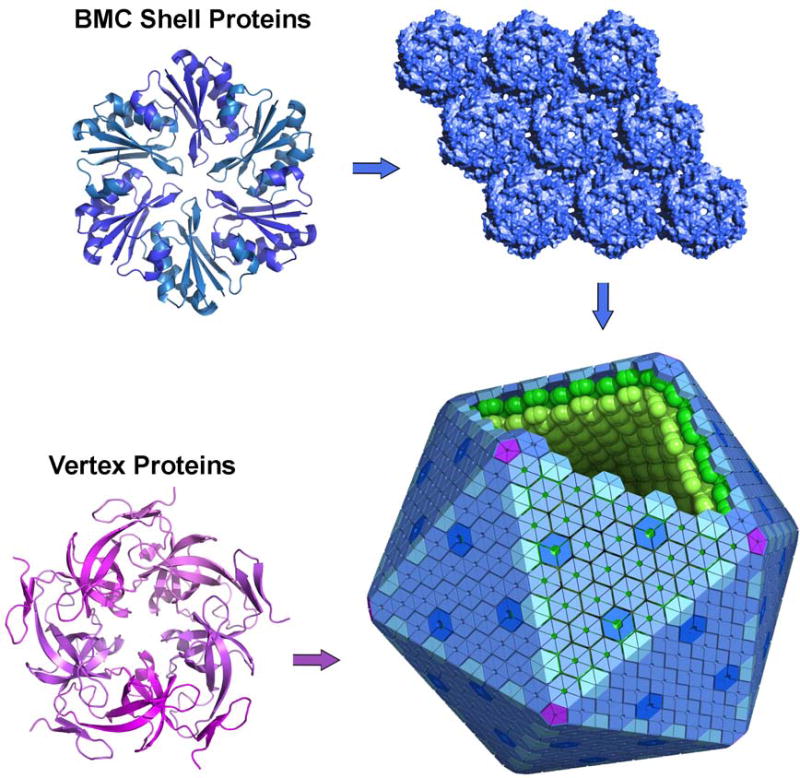
A model for assembly of bacterial microcompartments. Shell proteins from the BMC family form hexamers (or pseudohexameric trimers) that tile to make a molecular layer comprising the majority of the shell. They have central pores proposed to allow molecular transport. Different BMC paralogs (indicated in different shades) perform different roles in the shell. Special pentameric proteins, unrelated to the BMC protein family, are proposed to occupy vertex positions in the polyhedral shell, which is shown here idealized as a perfect icosahedron (bottom right). The shell proteins bind to and organize interior enzymes (arranged hypothetically in layers). Bottom right image: Creative Commons license, T. Yeates).
The relatively simple picture of identical protein subunits forming hexagons that tile to make a layer is complicated by a striking amount of structural divergence between the multiple paralogous BMC shell proteins within even a single type of MCP (Figure 3). Though the individual BMC protein domain is small, it has undergone a surprising variety of rearrangements during evolution[Yeates et al., 2010]. Some instances of the BMC shell protein reveal a circular permutation in which a highly similar tertiary structure is built from secondary structure elements occurring in a different order. This phenomenon was first identified in the shell protein PduU, where the permutation leads to protein termini in different locations, allowing for distinct elaborations at the ends of the protein chain[Crowley et al., 2008]. Gene duplication events have led to a second type of variation in the form of numerous tandem-domain BMC proteins. These assemble as trimers, which are only approximately hexameric. These trimeric structures allow the otherwise strict hexagonal symmetry of the BMC proteins to be broken. This appears to enable important conformational rearrangements in these proteins, as discussed subsequently. Yet other BMC shell proteins are both permuted and duplicated. Finally, numerous BMC shell proteins have been elaborated by fusion with other protein domains, which likely extend into the cytosol to perform other functions. Many of these fusions appear to have repetitive, charged, or otherwise atypical sequence compositions, and may be disordered. However, one fusion domain has been characterized – from the EutK shell protein – and its helix-turn-helix structure suggests a role in DNA binding[Tanaka et al., 2010].
Figure 3.
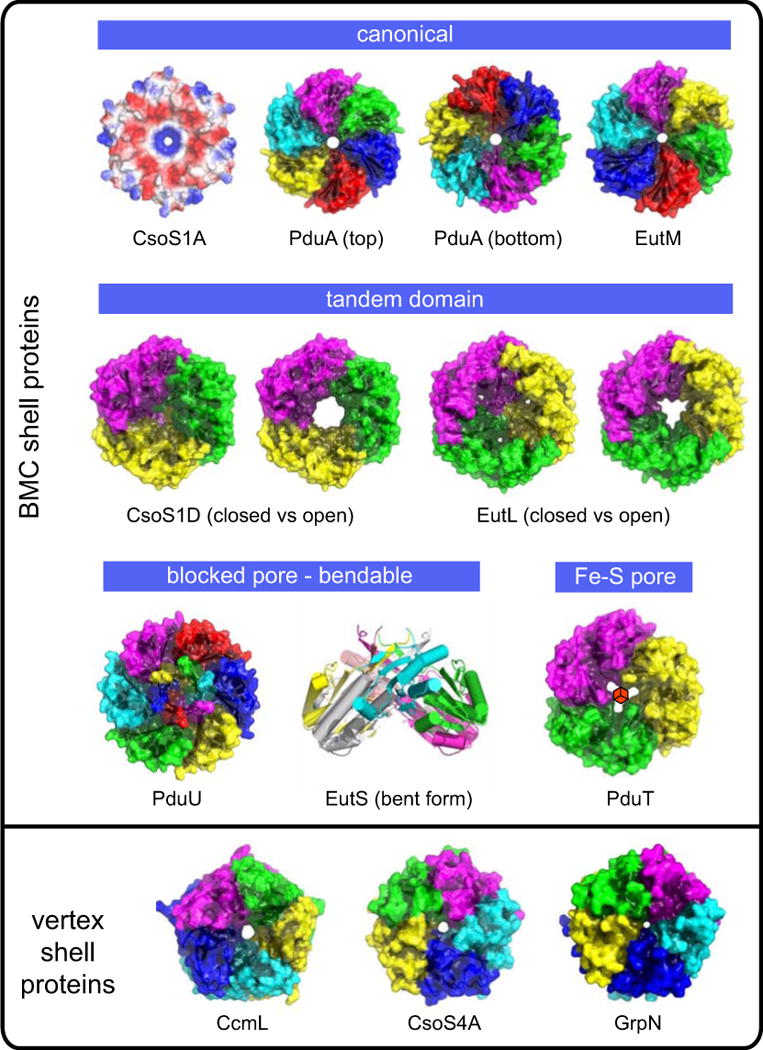
Crystal structures of representative shell proteins of different types and different presumptive functions. Except for the structure of PduA, which is shown in both orientations, the shell proteins are viewed from the outside direction. The structures are rendered for depth based on diffusion accessibility[Tsai, 2009; Yeates, 1995], with the exception of the carboxysome protein CsoS1A, which is shown colored by electrostatic potential (blue:positive, red:negative). The Fe-S cluster believed to occupy the PduT pore is indicated in its presumptive location; only the apo form of the protein has been structurally characterized. The hexameric crystal form of the presumptive vertex protein EutN is not shown.
The model of a flat molecular layer composed of hexagonal units begs the question of how a closed structure can be achieved. According to the principles of solid geometry, curvature of an otherwise flat hexagonal array requires insertion of non-hexagonal units. In particular, pentagonal units at the vertices give rise to icosahedral assemblies, as seen in many large viral capsids and geodesic domes. It was therefore illuminating when previously uncharacterized proteins (CsoS4A and CcmL) from the alpha and beta types of carboxysomes were shown by crystallography to be pentagonal, and to have edge lengths consistent with co-assembly together with BMC shell proteins[Tanaka et al., 2008]. Prior studies had failed to detect these proteins in purified carboxysomes, owing to their low abundance – 60 copies compared to a few thousand BMC shell proteins. Subsequent studies confirmed the presence of these pentameric proteins in purified carboxysomes by Western blotting[Cai et al., 2009]. The understanding that MCP’s were composed of pentameric units together with a much larger number of hexameric (BMC) units made it possible to generate a rough atomic model of an intact, essentially icosahedral carboxysome shell, based on model building and computational docking[Tanaka et al., 2008] (Figure 2). Electron cryotomography studies of isolated carboxysomes have supported the approximately icosahedral shape of the shell[Jensen and Briegel, 2007; Schmid et al., 2006], but it has not been possible yet to directly confirm the proposed arrangement of shell proteins by that method, owing to limited resolution. Nonetheless, the proposed model in which special pentameric proteins are required for vertex formation [Tanaka et al., 2008] is largely consistent with mutagenesis experiments in various systems. In experiments on the Pdu MCP and the beta-type carboxysome, deletion of the presumptive vertex protein tends to prevent formation of closed polyhedra [Cai et al., 2009; Parsons et al., 2010; Price et al., 1993; Sampson and Bobik, 2008]. In the alpha carboxysome, experiments have shown that normal looking MCP’s can be formed in deletion mutants lacking the two proteins (CsoS4A/B) that have been proposed to form vertices in that system, though carboxysomes produced by those mutants are leaky[Cai et al., 2009]. This would be consistent with the pentameric proteins being required for complete closure of the shell, but not for overall architecture.
Proteins homologous to the pentameric carboxysomal proteins (CcmL and CsoS4A) are present in other types of MCP’s, where they have retained names such as EutN, PduN, and GrpN according to the metabolic functions of their respective MCP’s. An initial crystal structure of EutN provided a confounding observation: that protein was found as a hexamer in the crystal form[Tanaka et al., 2008]. A recent crystal structure, however, showed that the GrpN protein from R. rubrum is pentameric, consistent with the proposed role of this protein family [Wheatley et al., 2013]. This prompted a reexamination of EutN, where it was found that the protein is predominantly pentameric in solution, arguing that crystallization promoted formation of a minor (hexameric) assembly form of that protein in the earlier study[Wheatley et al., 2013]. To emphasize the idea that this protein family plays a special role in forming the vertices of polyhedral MCP shells, the name BMV (bacterial microcompartment vertex) has been proposed.
Alternative models for the MCP shell have been proposed. Numerous studies have shown that hexameric or pseudo-hexameric BMC shell proteins sometimes stack to make double-layered, dodecameric species. In addition to solution and gel electrophoresis experiments, a few crystal structures have revealed two hexameric units paired with their C-terminal (bowl-shaped) surfaces arranged face-to-face[Cai et al., 2013; Klein et al., 2009; Samborska and Kimber, 2012; Tanaka et al., 2009]. This gave birth to hypotheses that MCP shells are actually composed of a double-layered shell[Klein et al., 2009; Samborska and Kimber, 2012]. So far, however, in cases where dodecameric units have appeared in crystal structures, they have not been arranged in the same layered fashion as is often seen for the single-domain BMC proteins. A double layer structure would also raise other difficult questions about how enzymes would be preferentially attached to one side, and how the shell would bend preferentially in an inside vs outside sense in order to close. But those challenges do not argue against the role of double-disk structures in limited parts of the shell. At present, it remains an open question whether the tendency of hexamers to form pairs is an artifact of the inward-facing surface being naturally evolved for protein-protein interactions (i.e. with enzymes), or whether double layer arrangements contribute to the function of MCP shells.
Molecular Transport through Shell Protein Pores
Crystal structures of most BMC shell proteins have revealed a narrow pore down the center of the hexamer. This was proposed to be the route by which molecules transit the shell in either direction [Kerfeld et al., 2005; Yeates et al., 2007]. Smaller holes have been noted at other sites within hexameric units, and at other joints between hexamers in a layer, but the central pores are the most obvious routes. Numerous crystallographic experiments have attempted to visualize key substrate or product molecules bound in the pores of BMC shell proteins. Those experiments have led to hints, but not to unambiguous models for transport, which remain somewhat speculative at present.
In the case of the carboxysome shell, a positive electrostatic potential surrounding the pore was observed in the original structural studies [Kerfeld et al., 2005; Tsai et al., 2007] (Figure 3). It was argued that a positive potential around the pore might confer an important selective advantage for passage of negatively charged molecules, particularly bicarbonate, whose rate of entry might otherwise limit the efficiency of carbon fixation. It is noteworthy that CO2, the key intermediate produced within the carboxysome, is neutral. The CO2 intermediate would therefore not experience the same electrostatic attraction to the pore as bicarbonate. Selective transport could enable a kinetic scheme in which CO2 molecules tends to encounter RuBisCO enzymes multiple times (if necessary) before having a chance to randomly diffuse out of the carboxysome. For other types of MCP’s, it is less obvious how the substrate molecule might gain an advantage in inward transport compared to the outward movement of the metabolic intermediate produced inside. However, the presence of hydrogen bond acceptors in the pore of the PduA shell protein has been offered as a potential mechanism for favoring transport of the propanediol substrate compared to the propionaldehyde intermediate in that system[Crowley et al., 2010]. The availability of structural data on a growing number of shell proteins presents new opportunities for testing hypotheses about molecular transport by structure-guided mutagenesis. In addition, as molecular dynamics methods continue to increase in power, their application to this field could add fresh insight into mechanisms of transport through the pores of shell proteins.
In certain tandem-domain BMC shell proteins, dramatic conformational changes have been observed in the pore region, with obvious implications for transport (Figure 3). Open and closed-pore forms of tandem-domain BMC shell proteins have been seen in both types of carboxysomes – CsoS1D in the alpha type [Klein et al., 2009] and CcmP in the beta type[Cai et al., 2013]. A major, open vs closed conformational change was also reported in the EutL shell protein from the Eut system [Tanaka et al., 2010]. The simplest explanation for such behavior is a gated opening in response to specific conditions or signals. What might trigger such conformational changes has not been elucidated yet. However, tandem-domain BMC shell proteins appear to sometimes contain disulfide bonds, which are essentially absent from single-domain BMC shell proteins (Michael Thompson, unpublished data), suggesting the possibility of a connection to redox state. The interpretation of open and closed pores in tandem-domain shell proteins is further complicated by the presence of very small holes in the ‘closed’ pore conformation of these proteins, not down the symmetry axis at the center of the oligomer, but through narrow openings that emerge in the individual subunits [Pang et al., 2012; Sagermann et al., 2009]. One line of argument holds that such holes arise incidentally from the conformational changes that occur during closing of the large central pore [Tanaka et al., 2010]. On the other hand, a recent structural study on the PduB shell protein revealed unexplained electron density features in these holes and elsewhere[Pang et al., 2012]. These were interpreted as glycerol, which had been included at high concentration during the crystallization process, leading to speculation that the narrow openings could support the transport of propanediol (or glycerol) in the Pdu system. For the Eut system and other complex MCP’s such as the Pdu and Grp, the need for cofactor transport might explain the existence of large gated pores. An explanation for large gated pores is harder to rationalize in the case of the carboxysome, as cofactors are not implicated in the interior reactions. It has been noted, however, that the conserved genomic proximity of CsoS1D genes to certain enzymes and proteins not evidently related to CO2 fixation leaves open the possibility of unanticipated complexity in the carboxysome[Jorda et al., 2013; Kerfeld et al., 2010].
Lastly, some shell proteins bear an iron-sulfur cluster in the pore. This was revealed first in the tandem-BMC shell protein, PduT[Crowley et al., 2010; Pang et al., 2011]. Like other tandem-domain shell proteins studied so far, PduT is a pseudohexameric homotrimer. In this case, each subunit contributes a single cysteine ligand to the 4Fe-4S cluster. This is an unusual arrangement, leaving the fourth iron atom to be bound by an unknown ligand. Other BMC shell proteins from the Grp MCP have also been discovered to bind an iron sulfur cluster at the central pore (Nicole Wheatley and Michael Thompson, unpublished data). The presence of iron-sulfur clusters in BMC shell proteins from complex MCP types – and their apparent absence from carboxysome MCP’s – resonates with the presence of redox reactions inside the MCP. A bound iron-sulfur cluster provides a potential route for electron flow to balance interior reactions, though this remains speculative. An alternate possibility for these shell proteins is to provide a route for transporting intact metal clusters needed by interior enzymes, including PduS. These hypotheses await experimental testing.
Protein-Protein Interactions in MCP’s
Various mechanisms have evolved in nature for targeting specific proteins to different organelles or different parts of the cell. Fan et al. provided a decisive explanation for how certain enzymes are targeted to the interior of Pdu MCP’s [Fan et al., 2010]. They showed that some of the interior enzymes in the Pdu system carry special N-terminal extensions that are necessary and sufficient for targeting to the MCP. Genetic truncation of an 18 residue segment at the N-terminus abrogated encapsulation of the native PduP enzyme, whereas fusion of the same sequence to green fluorescent protein caused it to be encapsulated. A bioinformatic analysis predicts the presence of N-terminal targeting sequence extensions in various enzymes from other MCP systems [Fan et al., 2010] (Figure 4). Experiments have confirmed the role of some other N-terminal sequences in targeting [Fan and Bobik, 2011], including in the Eut system[Choudhary et al., 2012]. An examination of the amino acid sequences predicted to act as targeting signals for various enzymes reveals only weak similarity between them; they follow a general pattern consistent with an amphipathic alpha helix. The variation between enzyme tails, even within a single type of MCP, admits the possibility of specific enzymes being bound to specific BMC shell protein homologs. This could provide a system for higher level organization in MCP’s. For example, prior studies showed that the PduP targeting sequence preferentially binds the PduA and PduJ shell proteins [Fan et al., 2012]. Likewise, EutC from the Eut system can only be targeted to the MCP in presence of the EutS shell protein[Choudhary et al., 2012].
Figure 4.
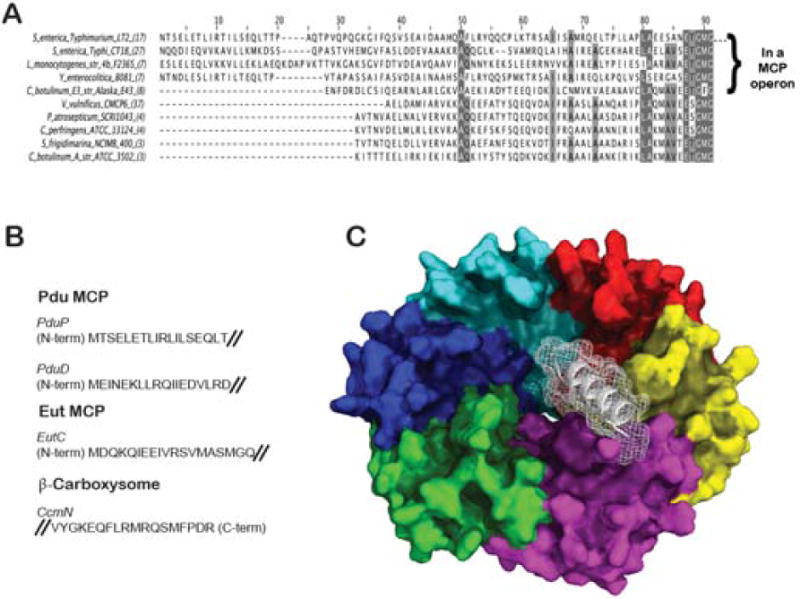
Key interior enzymes have sequence extensions that direct their association to the interior surface of the microcompartment shell. (A) An alignment of PduP sequences from various bacterial species, including a subset (top group) in which that enzyme is compartmentalized in a Pdu MCP. The sequence extension has been shown to be necessary and sufficient to encapsulate enzymes (adapted from [Fan et al., 2010]) (B) Representative sequences that have been shown experimentally to be involved in binding to MCP shells [Choudhary et al., 2012; Fan and Bobik, 2011; Fan et al., 2010; Kinney et al., 2012]. (C) A computational prediction of the PduP tail (white) bound to the interior surface of the hexameric PduA shell protein. The binding mode illustrated is based on a more exhaustive computational search compared to an earlier proposed model[Fan et al., 2012]).
Attempts to visualize the detailed atomic interactions between shell proteins and interior enzymes (or their terminal sequences) by crystallographic methods have so far been unsuccessful. Some predictions about those interactions have been generated from mutagenesis and preliminary computer modeling [Fan et al., 2012]. The N-terminal enzyme tails are predicted to be alpha helical. Mutagenesis studies show that enzyme targeting (e.g. of PduP) is eliminated by mutations in its N-terminus, or in the C-terminal helix of the PduA shell protein. That finding supports an earlier prediction that the concave side of BMC shell proteins would represent the inward facing surface [Tanaka et al., 2009]. Based on the known structure of shell proteins such as PduA, and using a helical model for the N-terminal tail of PduP that binds to the inward facing surface of PduA, tentative docking models can be produced computationally. One such docking model is shown in Figure 4.
Some of the mechanisms for enzyme and shell organization have also been illuminated in the carboxysome. In the β-type carboxysome, a peculiar protein, CcmM, has been shown to be important in binding other interior proteins. Remarkably, the CcmM sequence bears in its C-terminal region three or four tandem segments that are homologous to the small subunit of RuBisCO[Ludwig et al., 2000]. These repeated domains from CcmM are presumed to bind RuBisCO by mimicking the ring of four small subunits that ordinarily forms the end cap of the native RuBisCO oligomer. The N-terminal domain of CcmM is a redox sensitive carbonic anhydrase [Pena et al., 2010]. CcmM has been shown to interact with additional carboxysomal proteins, including CcmN [Cot et al., 2008; Long et al., 2007]. Recent experiments on CcmN show that its C-terminal region binds to the CcmK shell proteins and is essential for carboxysome assembly [Kinney et al., 2012]. Based on that finding, additional sequences important for MCP assembly were proposed.
An emerging understanding of how enzymes are targeted to MCP’s opens up prospects for engineering novel enzymatic containers. Preliminary experiments have already provided proof of concept. Green fluorescent protein was successfully directed to Pdu MCP’s in their native Salmonella host[Fan et al., 2010]. A similar experiment demonstrated the targeting of GFP and beta galactosidase to MCP’s constructed from only EutS shell subunits in E. coli[Choudhary et al., 2012]. And carboxysomes have been engineered to encapsulate RuBisCO molecules derived from heterologous species [Menon et al., 2008]. The ability to express MCP shells in heterologous hosts offers additional possibilities. Pdu-type shells having less regular morphologies than native shells have been produced in E. coli [Parsons et al., 2010], and carboxysomes of the alpha type have been produced in E. coli with morphologies approximating native carboxysomes. Efforts are underway to encapsulate reaction pathways for producing high value compounds within MCP’s.
Concluding Remarks
Considerable insights about function have come from crystal structure studies on individual shell proteins from various MCP systems. However, in the absence of crystal structures of complexes between shell proteins and interior enzymes, a deeper understanding of how different elements are organized will depend on other experimental approaches. So far, electron microscopy studies have not revealed additional clues about how various paralogous shell proteins might be arranged in MCP’s. With improvements in resolution and image processing methods, electron cryotomography might provide new information along those lines. In the meantime, biophysical experiments are providing some clues about interactions between shell proteins [Parsons et al., 2010; Samborska and Kimber, 2012], while the advent of atomic structures for MCP shell proteins has enabled structure-guided mutagenesis experiments aimed at identifying important interactions between shell proteins and interior enzymes [Fan et al., 2012; Kinney et al., 2012]. Numerous hypotheses regarding molecular transport through the pores of BMC shell proteins have been put forward, and these are now amenable to testing by mutagenesis as well. Ultimately, these studies may reveal new paradigms by which protein-based systems can control metabolite movement. BMC shell proteins are also ripe for molecular dynamics simulations aimed at testing hypotheses of selective permeability. Computational studies could also be important in identifying novel compounds for inhibiting or modulating MCP function. With regard to engineering applications, the possibilities are vast, and the successful demonstration of a designed container with practical utility would establish MCP’s as a general framework for future work in synthetic biology with potential applications in the production of biofuels/pharmaceuticals and as drug delivery vehicles.
Acknowledgments
The authors thank members of the Yeates lab for discussions and unpublished data.
Contributor Information
Todd O. Yeates, Department of Chemistry and Biochemistry, University of California, Los Angeles, California
Julien Jorda, UCLA-DOE Institute for Genomics and Proteomics, University of California, Los Angeles, California.
Thomas A. Bobik, Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, Iowa
References
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar typhimurium Lt2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One. 2009;4:e7521. doi: 10.1371/journal.pone.0007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of ccmp: A tandem bacterial microcompartment-domain protein from the beta-carboxysome forms a subcompartment within a microcompartment. J Biol Chem. 2013 doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: Carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon GC, Heinhorst S, Kerfeld CA. Carboxysomal carbonic anhydrases: Structure and role in microbial co2 fixation. Biochim Biophys Acta. 2010;1804:382–392. doi: 10.1016/j.bbapap.2009.09.026. [DOI] [PubMed] [Google Scholar]
- Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Fan C, Sinha S, Bobik TA. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the pdu microcompartment of Salmonella enterica. PLoS One. 2012;7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Structural insight into the mechanisms of transport across the Salmonella enterica pdu microcompartment shell. J Biol Chem. 2010;285:37838–37846. doi: 10.1074/jbc.M110.160580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci. 2009;18:2629–2635. doi: 10.1002/pro.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- Fan C, Bobik TA. The n-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the pdu microcompartment. J Bacteriol. 2011;193:5623–5628. doi: 10.1128/JB.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci U S A. 2012;109:14995–15000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA. Pdua is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium lt2. J Bacteriol. 2002;184:1253–1262. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst S, Williams EB, Cai F, Murin CD, Shively JM, Cannon GC. Characterization of the carboxysomal carbonic anhydrase CsoSCA from Halothiobacillus neapolitanus. J Bacteriol. 2006;188:8087–8094. doi: 10.1128/JB.00990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J. 2009;423:199–207. doi: 10.1042/BJ20090780. [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 2013;22:179–195. doi: 10.1002/pro.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein ccmn reveals common feature of bacterial microcompartment assembly. J Biol Chem. 2012;287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- Kofoid E, Rappleye C, Stojiljkovic I, Roth JR. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Sultemeyer D, Price GD. Isolation of ccmklmn genes from the marine cyanobacterium, Synechococcus sp pcc7002 (cyanobacteria), and evidence that CcmM is essential for carboxysome assembly. Journal of Phycology. 2000;36:1109–1118. [Google Scholar]
- Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric rubisco species. PLoS One. 2008;3:e3570. doi: 10.1371/journal.pone.0003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang A, Liang M, Prentice MB, Pickersgill RW. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr. 2012;68:1642–1652. doi: 10.1107/S0907444912039315. [DOI] [PubMed] [Google Scholar]
- Pang A, Warren MJ, Pickersgill RW. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr. 67:91–96. doi: 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- Pang A, Warren MJ, Pickersgill RW. Structure of PduT, a trimeric bacterial microcompartment protein with a 4fe-4s cluster-binding site. Acta Crystallogr D Biol Crystallogr. 2011;67:91–96. doi: 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell. 2010;38:305–315. doi: 10.1016/j.molcel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Pena KL, Castel SE, de Araujo C, Espie GS, Kimber MS. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, ccmm. Proc Natl Acad Sci U S A. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penrod JT, Roth JR. Conserving a volatile metabolite: A role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by clostridium phytofermentans. PLoS One. 8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by clostridium phytofermentans. PLoS One. 2013;8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Howitt SM, Harrison K, Badger MR. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. Strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, Horswill AR, Escalante-Semerena JC. DNA polymerase i function is required for the utilization of ethanolamine, 1,2-propanediol, and propionate by Salmonella typhimurium lt2. J Bacteriol. 1995a;177:7119–7124. doi: 10.1128/jb.177.24.7119-7124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, Kazmierczak R, Escalante-Semerena JC. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium lt2. J Bacteriol. 1995b;177:5434–5439. doi: 10.1128/jb.177.19.5434-5439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–8887. doi: 10.1073/pnas.0902324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samborska B, Kimber MS. A dodecameric CcmK2 structure suggests beta-carboxysomal shell facets have a double-layered organization. Structure. 2012;20:1353–1362. doi: 10.1016/j.str.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: Polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Baeumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the ccha cchb eute eutj eutg euth gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenoya M, Nikolakakis K, Sagermann M. Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J Bacteriol. 2010;192:6056–6063. doi: 10.1128/JB.00652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Sawaya MR, Yeates TO. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science. 2010;327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- Tsai Y. Diffusion accessibility. 2009 http://nihserver.mbi.ucla.edu/diff_acc/
- Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci. 2013 doi: 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO. Algorithms for evaluating the long-range accessibility of protein surfaces. J Mol Biol. 1995;249:804–815. doi: 10.1006/jmbi.1995.0339. [DOI] [PubMed] [Google Scholar]
- Yeates TO, Crowley CS, Tanaka S. Bacterial microcompartment organelles: Protein shell structure and evolution. Annu Rev Biophys. 2010;39:185–205. doi: 10.1146/annurev.biophys.093008.131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol. 21:223–231. doi: 10.1016/j.sbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: A viral capsid-like protein shell in bacterial cells. Biochem Soc Trans. 2007;35:508–511. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]


