Summary
The Pdu microcompartment is a proteinaceous, subcellular structure that serves as an organelle for the metabolism of 1,2-propanediol in Salmonella enterica. It encapsulates several related enzymes within a shell composed of a few thousand protein subunits. Recent structural studies on the carboxysome, a related microcompartment involved in CO2 fixation, have concluded that the major shell proteins from that microcompartment form hexamers that pack into layers comprising the facets of the shell. Here we report the crystal structure of PduU, a protein from the Pdu microcompartment, representing the first structure of a shell protein from a non-carboxysome microcompartment. While PduU is a hexamer like other characterized shell proteins, it has undergone a circular permutation leading to dramatic differences in the hexamer pore. In view of the hypothesis that microcompartment metabolites diffuse across the outer shell through these pores, the unique structure of PduU suggests the possibility of a special functional role.
Keywords: bacterial microcompartment; protein assembly; organelles; 1,2-propanediol; molecular transport
Introduction
Bacterial microcompartments are large, virion-sized protein assemblies comprised of specific enzymes encased within a protein shell. They function as organelles by sequestering particular metabolic processes within the cell. Microcompartments have been observed directly by electron microscopy in several species of bacteria (Fig. 1c, d) (Bobik et al., 1999; Brinsmade et al., 2005; Drews and Niklowitz, 1956; Gantt and Conti, 1969; Jensen and Bowen, 1961; Shively et al., 1970), and their relatively widespread existence across the eubacteria has been inferred by the presence of protein sequences homologous to the proteins that comprise the outer shell of diverse microcompartments (Bobik, 2006; Chen et al., 1994; Shively et al., 1998; Stojiljkovic et al., 1995). In particular, the shells of varied microcompartments are built primarily from small proteins belonging to the bacterial microcompartment (BMC) domain family of proteins (Havemann et al., 2002; Kerfeld et al., 2005; Shively et al., 1998; Tsai et al., 2007). A few thousand copies of these proteins are present in the shell of a single microcompartment. In any given organism, these major shell proteins are typically encoded in multiple homologous copies, arranged on the bacterial chromosome near genes for metabolic enzymes, at least some of which are either known or presumed to be encapsulated by the shell. Some characterization has been performed on representatives from three distinct metabolic classes of microcompartments, and up to seven distinct functional classes are predicted based on genomic sequence analysis (Bobik, 2006). To date, detailed structural studies have been conducted only on the carboxysome shell (Iancu et al., 2007; Kerfeld et al., 2005; Schmid et al., 2006; Tanaka et al., 2008; Tsai et al., 2007), a microcompartment that enhances carbon fixation in cyanobacteria and some chemoautotrophs by encapsulating essentially all of the ribulose-bisphosphate carboxylase-oxygenase (RuBisCO) enzymes in the cell, together with carbonic anhydrase (Price et al., 1992; Shively et al., 1973).
Figure 1.
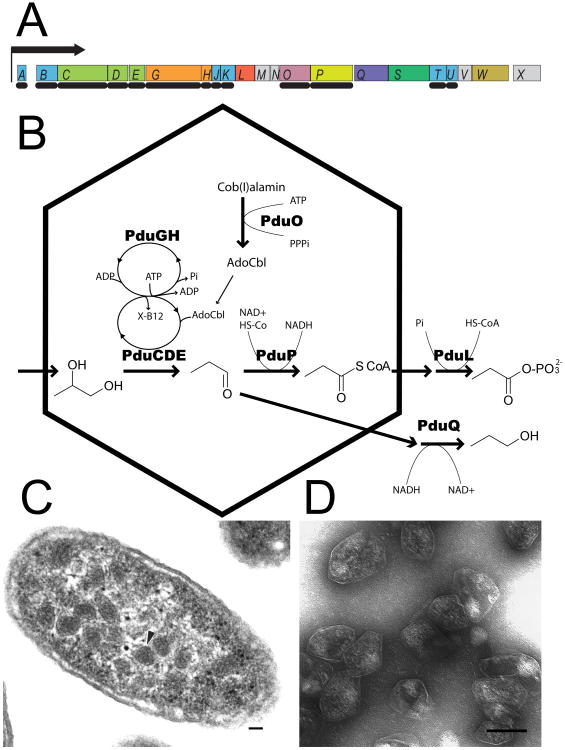
The Pdu microcompartment of Salmonella enterica. (a) Representation of the ∼17 kb pdu operon, whose genes are transcribed from a single promoter upstream from pduA (Bobik et al., 1992; Bobik et al., 1999; Bobik et al., 1997; Chen et al., 1994). The pdu- genes are labeled with their respective pdu- suffix designations, and drawn in proportion to their lengths and positions in the pdu- operon (Bobik et al., 1999). PduB and PduB' arise from alternate start sites. Dark shaded arrows indicate genes whose encoded proteins are detected in isolated microcompartments (Bobik et al., 1999; Bobik et al., 1997; Havemann and Bobik, 2003; Havemann et al., 2002; Johnson et al., 2001; Leal et al., 2003). Additionally, the genes marked by an asterisk (*) encode proteins homologous to the BMC shell proteins. (b) A current model of the function of the Pdu microcompartment. The Pdu microcompartment performs the initial steps in the metabolism of 1,2-propanediol (1,2-PD) to propionaldehyde (PA) and propionyl-CoA (PR-CoA) by the B12–dependent diol dehydratase (PduCDE) and the aldehyde dehydrogenase (PduP), respectively (Bobik et al., 1997; Leal et al., 2003). The Pdu microcompartment also contains the enzymatic reactions to generate (biologically active) adenosyl-cobalamin (Ado-B12) from its precursor cob(I)alamin, and to displace inactive de-adenosylated B12 derivatives (represented by X-B12) from the diol dehydratase. These reactions are carried out by cob(I)alamin adenosyltransferase (PduO) (Johnson et al., 2004; Johnson et al., 2001), and diol dehydratase reactivase (PduGH), respectively (Bobik et al., 1999). Given this model, the shell most likely allows passage of several relatively bulky substrates: 1,2-PD, PA, PR-CoA and HS-CoA (coenzyme A), nicotinamide adenine dinucleotide (NAD+) species, adenine nucleotides, and B12 derivatives. (c, d) Transmission electron (TEM) micrographs of Pdu microcompartments (arrows) within S. enterica (c) and of purified Pdu microcompartment organelles (d) (scale bar 100 nm).
The pdu operon in Salmonella enterica serovar Typhimurium LT2 (referred to hereafter as S. enterica) as was recently determined to encode proteins necessary for forming a microcompartment for the B12-dependent metabolism of 1,2-propanediol (1,2-PD) (Fig. 1) (Bobik et al., 1999; Chen et al., 1994; Havemann and Bobik, 2003; Shively et al., 1998). The compound 1,2-PD is proposed to serve as an important carbon and energy source for aerobic and anaerobic metabolism in S. enterica and other bacterial species (Bobik, 2006). Several studies implicate the metabolism of 1,2-PD in bacterial pathogenesis (Adkins et al., 2006; Conner et al., 1998; Heithoff et al., 1999; Klumpp and Fuchs, 2007), and demonstrate that microcompartment formation is required for its efficient metabolic utilization as a sole carbon source (Havemann et al., 2002; Sampson and Bobik, 2008).
Havemann and Bobik (Havemann and Bobik, 2003) recently characterized the total protein content of purified Pdu microcompartments from S. enterica, and determined that all but one of its fifteen detectable proteins are encoded by the 21-gene pdu operon. Seven of these proteins (PduA, -B, -B′, -J, -K,-T, and –U) have a BMC domain. Immunogold labeling of microcompartments confirmed that PduA is present in the outer shell (Havemann et al., 2002); the other homologous BMC domain proteins (PduB, -B′, -J, -K, -T and –U) are also presumed to be involved in formation of the shell. The Pdu microcompartments also incorporate four enzymes involved in 1,2-PD metabolism (Fig. 1b) (Bobik et al., 1999; Bobik et al., 1997; Johnson et al., 2001; Leal et al., 2003). The complexity of the Pdu microcompartment and its role in host metabolic interactions make it an attractive target for structural and functional studies.
High-resolution structural investigations of microcompartment shell proteins have been initiated on the carboxysome (Kerfeld et al., 2005; Tanaka et al., 2008; Tsai et al., 2007). Crystal structures of four different BMC domain proteins showed that they form cyclic hexamers with a tendency to pack into two dimensional layers representing the outer microcompartment shell. In addition, all of the carboxysome shell proteins have a narrow pore, with a diameter in the 4 Å – 7 Å range, running through the center of the hexamer. These pores are predicted to allow transport of the carboxysome substrates and products across the protein shell (Kerfeld et al., 2005; Tsai et al., 2007).
It is likely that microcompartments of various types will follow some of the same principles that have emerged from carboxysome studies, but with key differences accommodating their unique metabolic functions. Pdu microcompartments are slightly larger than typical carboxysomes and appear to be somewhat less geometrically regular in shape (Fig. 1c, d) (Bobik et al., 1999). In addition, compared to the carboxysome, the Pdu microcompartment organizes different and more varied enzymes in its interior, and must allow the diffusion of more diverse metabolites and cofactors (Fig. 1b) (Bobik et al., 1999). Structural studies on Pdu proteins are likely to shed light on these elements of function.
We report here the crystal structure of the PduU shell protein from S. enterica, the first BMC shell protein from a non-carboxysome microcompartment. PduU is one of the less abundant proteins in the Pdu microcompartment (Havemann and Bobik, 2003), and does not appear to be required for proper microcompartment assembly, but its presence is required for optimal biochemical function of the microcompartment (unpublished data). We reveal here that PduU is a hexamer as expected, but its three-dimensional topology reveals a surprising circularly-permuted variation on the typical BMC fold. A likely evolutionary mechanism for the origin of the observed permutation is presented. The resulting, novel characteristics of the PduU hexamer are discussed in light of their potential implications for function in the Pdu shell.
Results
Structure and Assembly Features
The crystal structure of PduU was determined by molecular replacement and refined at a resolution of 1.8 Å, with final R and Rfree values of 0.20 and 0.24 (see Materials and Methods) (Supp. Material, Table 1)). The crystal unit cell revealed two cyclic hexamers in the R3 space group, each sitting on the three-fold axis of symmetry. There are therefore four independent protein chains (-A, -B, -C and –D), two from each hexamer, in the asymmetric unit. The final atomic model is nearly complete: the first six residues were disordered in chains A and B from hexamer 1, the first three residues were disordered in chain C, and the first five residues were disordered in chain D from hexamer 2. There were no significant discrepancies between the two independent hexamers, and the protein backbones of chains A, B, C and D could be overlaid with an r.m.s.d. of < 0.23 Å. The qualities of the models were checked using the ERRAT program (Colovos and Yeates, 1993) and Ramachandran plots; 94% of the residues were within the favored regions and 99% of the residues were within the favored or allowed regions.
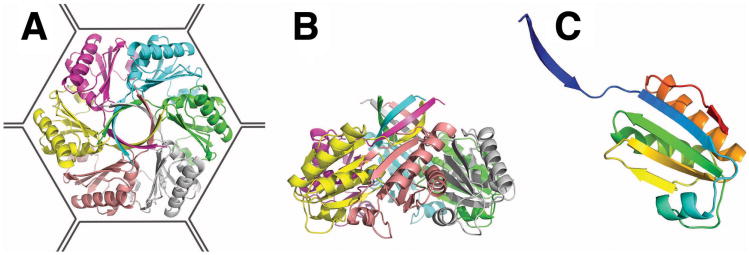
The PduU monomer adopts a compact, wedge-shaped fold representing a variation on the previously described BMC fold (Fig. 2c) (Kerfeld et al., 2005). Six subunits form a cyclic hexamer roughly 70 Å in diameter (Fig. 2a, b). Within the hexamers, individual PduU subunits form extensive interactions with neighboring subunits, burying an average surface area of 2626 Å2 for each monomer. The natural hexameric state of PduU was experimentally supported by native gel electrophoresis experiments using a Ferguson plot (Ferguson, 1964) (not shown). While some of the basic features of the PduU hexamer match those reported for homologous carboxysome shell proteins, there are some dramatic differences in folding and assembly.
Figure 2.
Crystal structure of the PduU shell protein. (a) The PduU hexamer viewed along the sixfold axis and (b) perpendicular to the sixfold axis with distinct protein chains colored separately. The outline drawn around the hexamer (a) illustrates its presumed packing in the microcompartment shell amongst the other (more abundant) homologous shell protein hexamers (e.g. PduA, PduB, PduB', and PduJ). A prominent feature of the PduU hexamer is the six-stranded, parallel β-barrel formed by one N-terminal β-strand from each monomer. Whether this feature at the top of the hexamer faces out towards the cytosol or towards the interior of the microcompartment has not been established. (c) Ribbon diagram depicting the PduU monomer, colored from blue (N-terminal) to red (C-terminal). Over residue positions 19 to 109, the chain adopts a variation of the typical bacterial microcompartment (BMC) fold (Kerfeld et al., 2005). The eighteen amino-terminal, and eight carboxy-terminal residues form novel secondary structural elements.
Three of the four hexameric carboxysome shell proteins whose structures have been determined (i.e. all but CcmK4) formed extended, two-dimensional molecular layers that are believed to represent the outer shell of the microcompartment (Kerfeld et al., 2005; Tsai et al., 2007). In contrast, the two independent PduU hexamers observed in the present crystal are arranged in loosely packed arrays (not shown) that are probably not reflective of the arrangement of BMC protein hexamers in native Pdu shells. In one of the crystalline lattice layers, (annotated as hexamer 1 in the crystal), the hexamers are touching at their corners. In the hexamer 2 layer, which must be spaced equally to the first layer, the edges of neighboring hexamers are parallel, but the spacing is too large to permit intimate contact; contacts between hexamers in the crystal involve the histidine-tagged C-terminal tails. The transverse center-to-center distance between adjacent PduU hexamers in the crystal is 74 Å, compared to the range of 67 Å – 70 Å for previously reported carboxysome hexamers in closely packed arrangements.
Structural Rearrangements in the BMC Fold
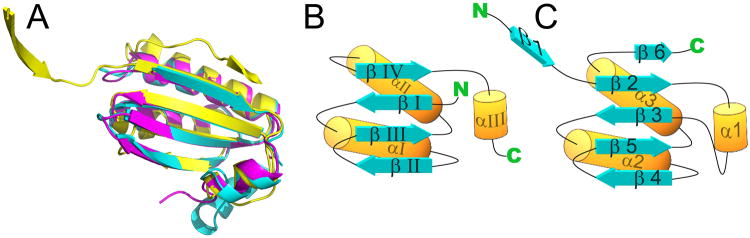
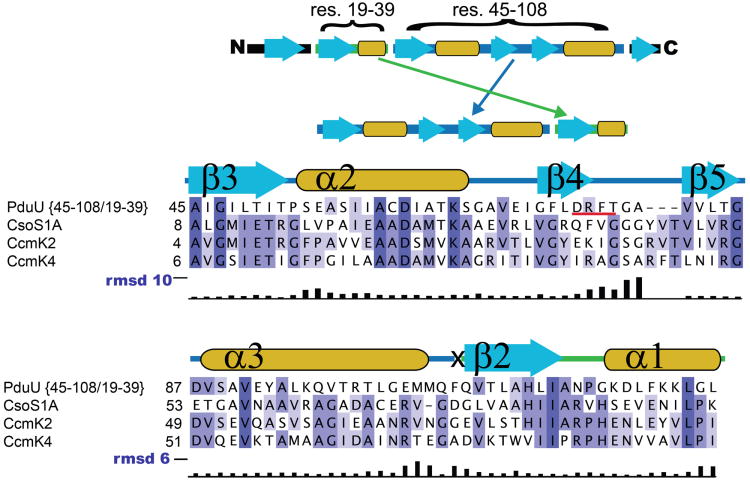
The structure of PduU reveals a core that is similar to the typical BMC fold, but the sequential ordering of structurally aligned secondary structure elements is different (Fig. 3b, c). The secondary structure elements contributed by the C-terminal region of the typical BMC fold are instead contributed by the N-terminal region of PduU. The relationship between the two cores can therefore be described as a circular permutation (Cunningham et al., 1979). This arrangement is unique amongst the carboxysome and other Pdu BMC proteins studied thus far. Circular permutation has been noted before as a mechanism for protein fold evolution (Grishin, 2001; Hahn et al., 1994; Ponting and Russell, 1995). To examine the sequence similarity between spatially analogous residues, a permuted version of the PduU sequence was generated based on its three-dimensional alignment with the typical BMC core (Figs. 3b, 3c, and 4). Sequence alignments between carboxysome shell proteins of known structure and permuted PduU showed higher amino acid sequence identity than alignments with native PduU. The sequence identity in an alignment to carboxysome shell protein CsoS1A, for example, increased to 21% for permuted PduU, compared to only 14% for native PduU (see Figure 4 for details). In addition, multiple sequence alignments (Edgar, 2004) against diverse BMC protein sequences reveal the conservation of key residues that cannot be aligned using native PduU (Fig. 4). For instance, using the permuted PduU sequence, Ile26 and Pro29 (numbered as in the native PduU sequence) align with like residues in the C-terminal regions of other BMC proteins (e.g. Ile80 and Pro83 in CcmK4); using the native PduU sequence ordering, those residues could not be aligned to conserved residues in the BMC domain (Fig. 4). The improvement in sequence identity, including key residues widely conserved in BMC domain proteins, suggests that PduU is related to the typical BMC domain by an effective permutation of the encoding gene sequence.
Figure 3.
Structural comparison of PduU to carboxysome shell proteins. (a) Ribbon diagrams of superimposed protein backbones of PduU (yellow), CcmK4 (cyan), and CsoS1A (magenta). The deviations were low overall, but substantially elevated for PduU residues 76-79, which form a loop lining the central cavity. Simplified diagrams of the arrangement of secondary structure elements are shown for (b) a typical BMC domain protein and (c) PduU. Additionally, the secondary structure elements are labeled with respect to their occurrence in the respective sequences. An apparent circular permutation is evident.
Figure 4.
Sequence comparison of typical BMC domain proteins to a permuted version of PduU. The sequence alignment represents the four structurally characterized BMC proteins. To perform this alignment, a hypothetical PduU sequence was constructed by permuting the native PduU sequence according to a structural alignment between PduU and carboxysome shell proteins of known structure (schematic, top). PduU sequence segments were shuffled as follows (numbered according to parent sequence position): residues 45-108, then residues 19-39. The “X” above marks the position between adjoining segments. Non-homologous terminal sequence regions are omitted here for clarity. The blue colored boxes represent aligned, conserved residues, and the corresponding PduU secondary structural elements are diagrammed above the alignment following the labeling presented in Fig 3c. The PduU loop region containing residues D76 –T79 (underlined with red) is shorter than the corresponding loops in other BMC proteins of known structure. Deviations between corresponding Cα positions after superimposing CcmK2 and PduU are graphed below in black bars (scale markers in blue).
In addition to the permutation in the core, there are novel features in both termini of PduU, one of which is particularly significant. A minor elaboration is present at the C-terminus, where residues 109-116 contribute an extra strand to the edge of the core β-sheet (Fig. 3), whereas at the N-terminus an extension in PduU creates an entirely new motif at the center of the hexamer, as discussed below. Owing to these variations, both ends of the protein chain terminate on the side of the hexamer opposite from that seen in other BMC domain proteins.
The Central Pore
The N-terminus of PduU extends toward the middle of the hexamer, where residues 8-15 from each subunit contribute to a parallel six-stranded β-barrel, which is unique among BMC domain proteins studied so far (Fig. 5). Beginning at residue number eight, the barrel is comprised by the amino acid sequence DRMIQEYV. The β-barrel is broken by a subsequent proline residue at position 16. The barrel has an average diameter of ∼16 Å, measured between equivalent C-alpha positions on opposite sides of the barrel, and a height of ∼12 Å. The shear value, which describes the degree of slanting of β-strands in a barrel, is 12, which has been noted to be a low-energy configuration for six-stranded β-barrels (Murzin et al., 1994). Interestingly, a similar homo-oligomeric β-barrel, formed by seven β-strands instead of six, is found in the extramembrane domain of the heptameric mechanosensitive channel of small conductance (MscS) (Bass et al., 2002). Hereafter we refer to the side of the PduU hexamer with the novel β-barrel as the top; it is not known yet which side of the layer formed by assembled BMC proteins represents the inside vs. the outside of the microcompartment.
Figure 5.
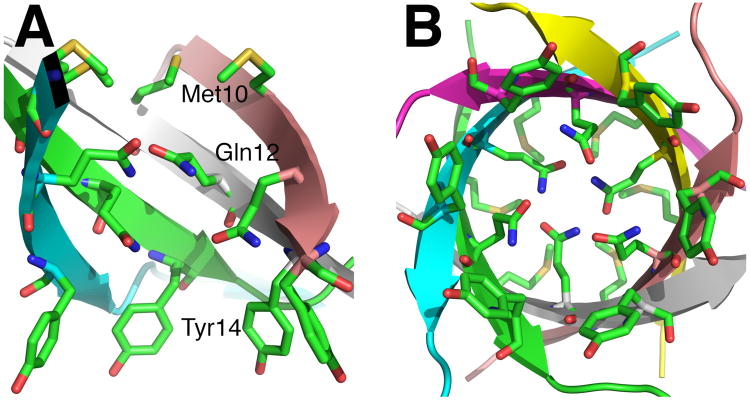
Structure of the β-barrel cap in PduU. (a, b) Cut-away views showing the side chain packing within the hexamer β-barrel. The side chains (sticks) of Met10, Gln12, and Tyr14 are buried within the β-barrel. Owing to steric restrictions, all Gln12 side chains were modeled in two alternating conformations: “up” towards the N-terminus and “down” towards the C-terminus. Inter-chain hydrogen bonding between upward-oriented Gln12 side chains is present in the configuration shown in (a) (dashed line). (c) Stereo view of the PduU β-barrel viewed from its C-terminal end. Gln12 and other interior side chains are illustrated. The configurations shown in both panels are from the hexamer labeled “1” in the crystal asymmetric unit.
Although the central β-barrel in PduU sits on the axis believed to serve as a pore for transport in related BMC proteins, the barrel appears to be too tightly packed with large amino acid side chains to allow efficient diffusion (Fig. 5). The side chains from residues Met10, Gln12, and Tyr14 are oriented toward the β-barrel interior. Beginning at the N-terminal opening of the barrel, the side chains from Met10 pack symmetrically about the six-fold axis, and nearly occlude the entire width of the β-barrel interior (Fig. 5). The six Gln12 side chains cannot be accommodated in equivalent configurations inside the β-barrel due to steric collisions. Instead, the electron density supports modeling two side chain conformations, which alternate in sequence around the barrel (Fig. 5a, b). In the “up” conformation, the side chains are oriented toward the N-terminal pole of the β-barrel, while in the “down” conformation the side chain amides are oriented toward the C-terminal pole. There are some minor conformational differences in Gln12 packing between the two independent hexamers visualized in the crystal. In one hexamer, down-oriented Gln12 side chains are oriented favorably for intermolecular hydrogen bonding between alternating side chain amide groups; in the other hexamer, up-oriented Gln12 side chains are oriented for hydrogen bonding between the amide groups (Fig. 5a). The tight packing of methionine and glutamine side chains suggests that efficient molecular transport down the center of the PduU hexamer would not be possible, at least not with the N-terminus in the configuration observed here. We note that, notwithstanding the minor differences in amino acid side chain orientations, the N-terminus of PduU adopts very nearly equivalent configurations in the two independent hexamers in the crystal, arguing against the β-barrel being a spurious structure.
The opposite side of the PduU hexamer is likewise unique. Whereas the top side is blocked by the N-terminal β-barrel, the bottom side of the PduU hexamer is opened much wider at the site of the central pore than other structurally characterized BMC domain proteins (i.e. those from the carboxysome) (Fig. 6). Viewed in cross-section with the central β-barrel at the top, the result is a roughly bell-shaped cavity or deep depression, with Glu12, which is near the C-terminal end of the p barrel, forming the apex. Whereas the analogous central pores of the carboxysome hexamers are narrow (4-7 Å in diameter), the central cavity on the bottom side of the PduU hexamer is approximately 17 Å across (Fig. 6b). The widened opening in PduU is due mainly to a loop truncation, rather than differences in amino acid side-chains. In PduU, the loop between residues 76 and 79 replaces a loop that has three more residues in homologous carboxysome shell proteins of known structure, and 3 to 10 additional residues in other Pdu BMC proteins (Fig. 4). The cavity on the bottom side of the PduU hexamer appears to present an unusually hydrophobic surface. A high percentage of the surface of the cavity is comprised by the exposed aromatic faces of side chains from Tyr14 and Phe78, which contribute an average of 74 Å 2 and 125 Å 2 per subunit, respectively (Fig. 6b). The Tyr14 residues at the C-terminal end of the central β-barrel protrude from the barrel in a ring, with the aromatic plane normals oriented nearly perpendicular to the central axis of symmetry. The aromatic faces of Phe78 side chains are arranged similarly in a ring near the bottom opening of the cavity. Sequence alignments with other Pdu proteins suggest that this aromatic surface is unique to PduU. Less substantial, but significant, contributions to the surface are made by Ser52, Thr54, and Glu55; these contribute an average of 12 Å2, 17 Å2 and 7 Å2 per subunit, respectively (Fig. 6b). The relatively hydrophobic character of this surface suggests the likelihood of native interactions with other molecules– either other proteins or smaller molecules – that have not been visualized yet.
Figure 6.
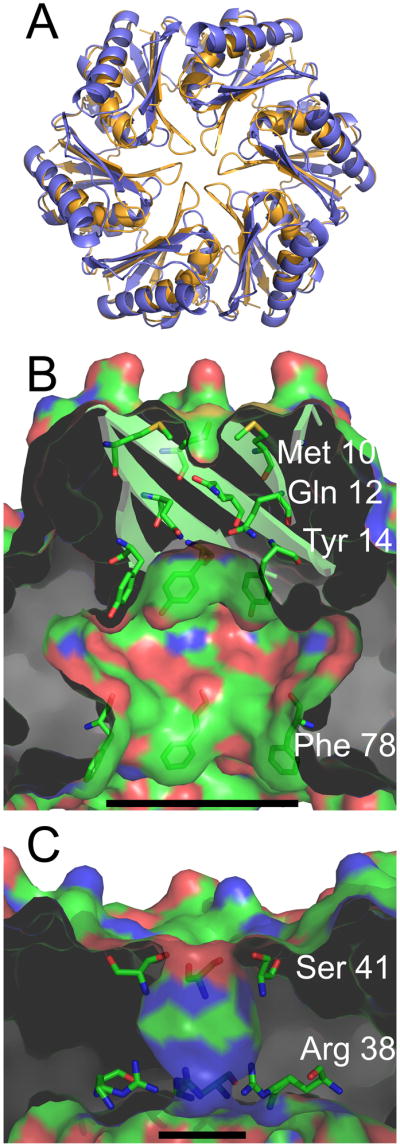
Properties of the central cavity in the PduU hexamer. (a) Comparison of a carboxysome shell protein hexamer, CcmK4 (orange) (Kerfeld et al., 2005), with the PduU hexamer (blue). To illustrate differences at the PduU central pore region, the N-termini are truncated so that only residues 19-116 are shown. (b) A cut-away view of the central cavity of the PduU hexamer, which is suggested here to be oriented with the open cavity facing the lumen of the Pdu microcompartment. The image is colored by atom type, with carbon (green), nitrogen (blue), and oxygen (red). Side chains from residues Met10, Gln12, Tyr14, and Phe78 are shown in order to illustrate their contributions to the surface. The substantial surface contributions by aromatic side chains are a distinction from carboxysome shell proteins whose pores are lined mainly by charged and polar side chains (Kerfeld et al., 2005; Yeates et al., 2007); CcmK4 is shown in panel (c). Measurement bars are added for scale, representing approximately 17 Å (b) and 8 Å (c). They indicate the distance between the Cβ atoms of Phe78 from opposing protein chains in panel (b) and between Arg38 N3 side chain atoms from opposing chains in panel (c) (17.1 Å and 7.8 Å, respectively). Note that the pore that is present in the carboxysome shell protein hexamer (panel c) is blocked by the β-barrel in PduU (panel b).
Discussion
The structure of PduU recapitulates some of the features that have been observed in structures of the major shell proteins of the carboxysome; the core of PduU is related to the typical BMC domain, and its natural oligomeric state is a cyclic hexamer (Kerfeld et al., 2005; Tsai et al., 2007). Beyond this, however, PduU reveals unique features that are likely to relate to its function. At least three functional properties would seem to be important for the major shell proteins of bacterial microcompartments. The hexameric proteins must interact with each other in order to form a layer that surrounds the organelle. Additionally, they must make critical interactions with the interior components of the microcompartment. And they must allow movement of small molecules across the shell. The structure of PduU provides functional insights into each of these issues.
By itself, PduU does not appear to form well-packed hexagonal layers as easily as the carboxysome proteins that represent major constituents of that shell. This property is shared by PduU and one particular carboxysome shell protein, CcmK4 (Heinhorst et al., 2006; Kerfeld et al., 2005). Neither PduU nor CcmK4 is a major constituent of its respective shell (Havemann et al., 2002; Heinhorst et al., 2006). The low abundance of these proteins is consistent with their failure to form layers on their own, since they presumably interact with homologous but distinct hexamers that are more abundant in native shells. The need for multiple homologues of the BMC domain proteins – and this is the case for every genome in which they have been found – is not fully understood yet. In the case of CcmK4, the tendency of that protein to form strips instead of layers has led to the speculative suggestion that it could occur at edges of the icosahedral shell (Kerfeld et al., 2005). In the case of PduU, the inability to form homogeneous PduU hexamer layers might be a mechanism for limiting its incorporation into Pdu microcompartment shells.
One of the major structural differences seen in PduU is the deep cavity or depression on one side of the hexamer (Fig. 6a, b). This open cavity takes the place of a much narrower pore observed at the hexameric axis in the BMC domain proteins from the carboxysome shell (Fig. 6a, c). The surface of the cavity in PduU presents several hydrophobic residues (Fig. 6b), which suggests the likelihood of yet unknown molecular interactions. If the purpose of the hydrophobic surface is to establish specific interactions with interior components of the microcompartment, then that side of the hexameric layer would have to face the microcompartment interior. Which side of the hexameric layer of BMC domain proteins represents the outside of a microcompartment and which side faces inward is a question that has not been resolved yet for the carboxysome or the Pdu microcompartment.
Another unique feature in PduU is the intermolecular six-stranded β-barrel that appears to block the central pore that exists in other BMC domain proteins (Fig. 4). Beta-barrels occur often in transmembrane proteins, where they create pores for molecular transport (Schulz, 2002). However, β-barrels in structurally characterized transport proteins are comprised of at least 12 strands, making them considerably larger in diameter than the PduU β-barrel. Given its narrow diameter, one hypothesis is that instead of serving a transport function, PduU might serve to organize a particular enzymatic component in the interior of the microcompartment. An alternative hypothesis is that the β-barrel in PduU could open up under certain conditions to allow transport. The β-barrel in PduU is similar in certain respects to the C-terminal, symmetric seven-stranded β-barrel from the MscS mechanosensitive channel (Bass et al., 2002), which has likewise been proposed to undergo structural transitions (Koprowski and Kubalski, 2003). If the β-barrel in PduU is capable of opening, then the unusually wide entrance on the other side of the PduU hexamer (discussed above) would allow the diffusion of larger molecules than those thought to pass through the pores of corresponding carboxysome shell proteins. Although it is not entirely clear yet which compounds must cross into and out of the Pdu microcompartment, some of the bulkier substrate might include nucleotides, nicotinamide adenine dinucleotides, coenzyme A species, and coenzyme B12 derivatives (Fig. 1b) (Bobik, 2006; Chen et al., 1994). The thermal displacement parameters (B-factors) were examined in the region of the β-barrel, but were not found to be significantly elevated compared to the rest of the protein. Further studies will be required to test hypotheses about the potential roles of PduU in binding and transport.
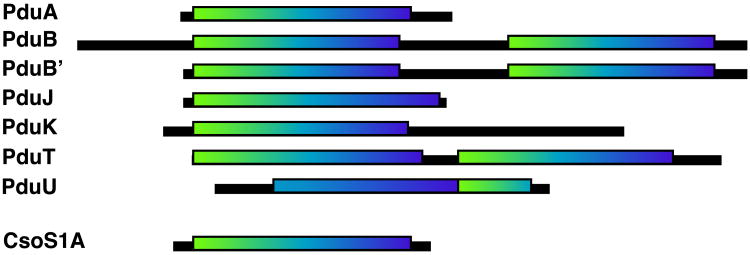
Some of the key structural differences between PduU and other BMC domain proteins arise from the apparent circular permutation in PduU (Figs. 3 and 4). What evolutionary sequence of events might have led to this genetic rearrangement? The apparent circular permutation could have arisen by tandem gene duplications of a more typical BMC-type protein in the PduU operon, followed by truncation at the ends of the duplicated gene to yield a permuted single domain. This scenario is consistent with a model first described in the saposin family (Ponting and Russell, 1995), and is supported by the frequent appearance of multiple BMC protein genes encoded adjacent to each other (Bobik et al., 1999), as well as the existence of individual microcompartment shell proteins apparently containing two tandem BMC domains. PduB and PduT illustrate two such cases (Fig. 7).
Figure 7.
Cartoon representations of the seven S. enterica Pdu shell proteins containing BMC domains recognizable by sequence homology. The boxes outline the sequence segments containing predicted BMC domains. The typical BMC domains are shaded from white at the N-terminus to black at the C-terminus. PduU can be aligned to parts of two consecutive BMC domains, or can alternatively be described as a circular permutation of a single BMC domain.
As a model system for studying bacterial microcompartments, Salmonella offers two key features. Protocols have been developed for isolating Pdu microcompartments in intact form (Havemann and Bobik, 2003), and genetic manipulation in Salmonella is straightforward. This opens up the possibility of isolating mutant microcompartments for in vitro experiments; this has not been possible yet for carboxysomes, which have been difficult to purify from bacterial species that are amenable to genetic manipulation (Heinhorst et al., 2006). S. enterica also produces a so-called Eut microcompartment for the metabolism of ethanolamine, which might be amenable as well to combined biochemical and genetic studies (Stojiljkovic et al., 1995). Structure-guided mutagenesis in the Pdu and Eut systems should make it possible to answer unresolved questions about bacterial microcompartments.
Experimental Procedures
Transmission electron microscopy
Transmission electron microscopy imaging of Salmonella enterica serovar Typhimurium LT2 sections and purified Pdu microcompartments was performed as previously described (Bobik et al., 1999; Havemann and Bobik, 2003)using a Zeiss EM-10CA transmission electron microscope (Zeiss, Co., Germany).
Cloning and protein purification
Briefly, the gene pduU from S. enterica was amplified from the plasmid construct MGS2 (Bobik et al., 1999) for cloning into the plasmid pET22b (Novagen, Darmstadt, Germany). The resulting construct encodes full-length PduU (116 amino acids) plus a C-terminal Leu-Glu-His6 affinity sequence. Protein expression from sequence-verified PduU-pET22b constructs (hereafter PduU) was carried out in BL21 (DE3) RIL+ E. coli cells (Stratagene, La Jolla, CA, USA) in 1 L cultures of Luria-Bertani (LB) media + 1% glucose. Protein was purified from the soluble cell fraction by HisTrap HP affinity chromatography (GE Healthcare, Waukesha, WI, USA) followed by Q Sepharose HP anion-exchange chromatography (GE Healthcare). All steps were carried out in the presence of 0.2 mM PMSF and 1 mM DTT. 12% SDS-PAGE was performed to monitor the purity of the sample.
Crystallization and Data Collection
Protein for crystallization was concentrated to 9 mg/ml in 25mM Tris pH 8.0 and 25mM NaCl. A Mosquito robotic instrument (TTP Labtech Ltd., Melbourn, UK) was used to set up crystallization trials by the hanging drop vapor diffusion method. The protein crystallized in 25 of 480 initial screening conditions, in most cases forming crystals with hexagonal plate morphologies. Crystal conditions typically contained 20% (v/v) low molecular weight polyethylene glycol (PEG), overall ionic strength <500 mM, and pH range 6.0-8.0. An optimized condition with 50mM HEPES, pH 8.0, 300mM LiSO4, and 20% PEG-3350 produced good single crystals with the morphology of hexagonal plates.
Single crystals were harvested in nylon loops and cryoprotected by the addition of 30% (v/v) ethylene glycol, and then frozen for subsequent data collection in a nitrogen stream at −176°C. A full data set was collected to a resolution limit of 1.8 Å at the Advanced Light Source beamline 8.2.2 (Berkeley, CA, USA) (Supp. Material, Table 1).
Structure Determination And Analysis
Diffraction data were indexed and processed using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). The program PHASER (McCoy et al., 2005) was used to solve the structure by molecular replacement using as a search model the crystal structure of CsoS1A, a hexameric carboxysome shell protein from H. neapolitanus (PDB ID, 2EWH) (Tsai et al., 2007). The model was built using the graphics program COOT (Emsley and Cowtan, 2004) and refined using the program REFMAC (CCP4, 1994; Murshudov et al., 1997). NCS restraints were applied at “medium” strength setting to the four molecules in the asymmetric unit. Restraints were released for the last five rounds of the refinement. The final R and Rfree values were 20.3% and 24.2% respectively (Supp. Materials, Table 1).
The Pymol molecular graphics program (http://www.pymol.org/) was used for visual investigation, molecular measurements, and generation of cartoon structure images of the PduU structure. Measurement bars and labels were added to exported images in the image-rendering program Photoshop (Adobe Systems, Inc., San Jose, CA, USA). Surface area measurements were calculated by the program AREAIMOL (Lee and Richards, 1971). Superpositioning of Cα backbone atoms was performed by the program SUPERPOSE (Krissinel and Henrick, 2004). Root mean squared deviations between pairs of structures were calculated over the following chain segments: CcmK2 residues 71-90, CcmK4 residues 73-92, and CsoS1A residues 74-93, over PduU residues 19-38; CcmK2 residues 4-37, CcmK4 residues 6-39, and CsoS1A residues 8-41, over PduU residues 45-78; and CcmK2 residues 41-69, CcmK4 residues 43-71, and CsoS1A residues 45-73, over PduU residues 79-107.
Supplementary Material
Acknowledgments
This work was supported primarily by the BER program of the DOE Office of Science (TOY) and by a grant from the National Science Foundation (no. MCB0616008) (TAB). The authors thank the staff at ALS synchrotron beamline 8.2.2 for technical assistance, Duilio Cascio for crystallographic advice, Morgan Beeby for gel electrophoresis analysis, James Stroud, Christopher Miller, Shiho Tanaka and Yingssu Tsai for helpful comments, and Cheryl Kerfeld for a critical reading of the manuscript.
References
- Adkins JN, Mottaz HM, Norbeck AD, Gustin JK, Rue J, Clauss TR, Purvine SO, Rodland KD, Heffron F, Smith RD. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol Cell Proteomics. 2006;5:1450–1461. doi: 10.1074/mcp.M600139-MCP200. [DOI] [PubMed] [Google Scholar]
- Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- Bobik TA, Ailion M, Roth JR. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci U S A. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: Circularly permuted amino acid sequences. Proc Natl Acad Sci U S A. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G, Niklowitz W. Cytology of Cyanophycea. II. Centroplasm and granular inclusions of Phormidium uncinatum. Arch Mikrobiol. 1956;24:147–162. [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferguson KA. Starch-Gel Electrophoresis--Application to the Classification of Pituitary Proteins and Polypeptides. Metabolism. 1964;13(1):985–1002. doi: 10.1016/s0026-0495(64)80018-4. [DOI] [PubMed] [Google Scholar]
- Gantt E, Conti SF. Ultrastructure of blue-green algae. J Bacteriol. 1969;97:1486–1493. doi: 10.1128/jb.97.3.1486-1493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- Hahn M, Piotukh K, Borriss R, Heinemann U. Native-like in vivo folding of a circularly permuted jellyroll protein shown by crystal structure analysis. Proc Natl Acad Sci U S A. 1994;91:10417–10421. doi: 10.1073/pnas.91.22.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B(12)-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinhorst S, Cannon GC, Shively JM. Carboxysomes and Carboxysome-like Inclusions. In: Shively JM, editor. Complex Intracellular Structures in Prokaryotes. Berlin: Springer-Verlag; 2006. pp. 141–165. [Google Scholar]
- Heithoff DM, Conner CP, Hentschel U, Govantes F, Hanna PC, Mahan MJ. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The Structure of Isolated Synechococcus Strain WH8102 Carboxysomes as Revealed by Electron Cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TE, Bowen CC. Organization of the centroplasm in Nostoc pruniforme. Iowa Acad Sci Proc. 1961;68:86–89. [Google Scholar]
- Johnson CL, Buszko ML, Bobik TA. Purification and initial characterization of the Salmonella enterica PduO ATP:Cob(I)alamin adenosyltransferase. J Bacteriol. 2004;186:7881–7887. doi: 10.1128/JB.186.23.7881-7887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Pechonick E, Park SD, Havemann GD, Leal NA, Bobik TA. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J Bacteriol. 2001;183:1577–1584. doi: 10.1128/JB.183.5.1577-1584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- Klumpp J, Fuchs TM. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology. 2007;153:1207–1220. doi: 10.1099/mic.0.2006/004747-0. [DOI] [PubMed] [Google Scholar]
- Koprowski P, Kubalski A. C termini of the Escherichia coli mechanosensitive ion channel (MscS) move apart upon the channel opening. J Biol Chem. 2003;278:11237–11245. doi: 10.1074/jbc.M212073200. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. Arch Microbiol. 2003;180:353–361. doi: 10.1007/s00203-003-0601-0. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweeet RM, editors. Methods in Enzymology, volume 276: Molecular Crystallography, Part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Ponting CP, Russell RB. Swaposins: circular permutations within genes encoding saposin homologues. Trends Biochem Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- Shively JM, Bradburne CE, Aldrich HC, Bobik TA, Mehlman JL, Jin S, Baker SH. Sequence homologs of the carboxysomal polypepide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboysomes-polyhedral bodies. Can J Bot. 1998;76:906–916. [Google Scholar]
- Shively JM, Decker GL, Greenawalt JW. Comparative ultrastructure of the Thiobacilli. J Bacteriol. 1970;101:618–627. doi: 10.1128/jb.101.2.618-627.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells. Biochem Soc Trans. 2007;35:508–511. doi: 10.1042/BST0350508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.