Abstract
Antagonizing MDM2 and MDMX to activate the tumor suppressor protein p53 is an attractive therapeutic paradigm for the treatment of glioblastoma multiforme (GBM). However, challenges remain with respect to the poor ability of p53 activators to efficiently cross the blood–brain barrier and/or blood–brain tumor barrier and to specifically target tumor cells. To circumvent these problems, we developed a cyclic RGD peptide-conjugated poly(-ethylene glycol)-co-poly(lactic acid) polymeric micelle (RGD-M) that carried a stapled peptide antagonist of both MDM2 and MDMX (sPMI). The peptide-carrying micelle RGD-M/sPMI was prepared via film-hydration method with high encapsulation efficiency and loading capacity as well as ideal size distribution. Micelle encapsulation dramatically increased the solubility of sPMI, thus alleviating its serum sequestration. In vitro studies showed that RGD-M/sPMI efficiently inhibited the proliferation of glioma cells in the presence of serum by activating the p53 signaling pathway. Further, RGD-M/sPMI exerted potent tumor growth inhibitory activity against human glioblastoma in nude mouse xenograft models. Importantly, the combination of RGD-M/sPMI and temozolomide — a standard chemotherapy drug for GBM increased antitumor efficacy against glioblastoma in experimental animals. Our results validate a combination therapy using p53 activators with temozolomide as a more effective treatment for GBM.
Keywords: Stapled peptide, p53, Micelle, Glioma-targeting delivery, Chemosensitization, Temozolomide
1. Introduction
Despite intensive therapy, glioblastoma multiforme (GBM) remains difficult to treat and of poor prognosis with a medium-term survival of approximately 14.6 months and a 5-year overall survival rate below 4% [1–3]. Considering various physiological barriers, surgery is still the main treatment option for glioma. However, complete surgical resection of glioma is almost impossible to achieve because of its extremely infiltrative nature, which always leads to recurrence [4]. On the other hand, chemo/radiation therapy is often of low efficiency due to the integrity of the blood–brain barrier (BBB) and blood–brain tumor barrier (BBTB) [5–7]. New therapeutic paradigms are needed for improved GBM treatment.
The tumor suppressor protein p53 is a transcription factor that transactivates, in response to cellular stresses, the expression of various target genes that mediate cell cycle arrest, senescence, or apoptosis [8–10]. Dubbed the “guardian of the genome”, p53 is critical for maintaining genetic stability and preventing tumor development [11]. Not surprisingly, the p53 signaling pathway is impaired in almost all human tumors due to either debilitating mutations or functional inhibition by its negative regulators MDM2 and MDMX [12]. MDM2 and MDMX cooperate to block p53 transactivation activity and target p53 for degradation, and are often amplified or overexpressed in tumors harboring wild type p53 [13]. Numerous reports have shown that antagonizing MDM2/MDMX to activate p53 can kill tumor cells in vitro and in vivo in a p53-dependent manner, promising a novel therapeutic approach to the treatment of many tumors with wild type (WT) p53 status and elevated levels of MDM2/MDMX [14–16].
Activation of p53 by MDM2/MDMX antagonists has been proven to be effective in treating glioma where approximately 70% of primary tumors harbor WT p53 [17–20]. Different structural classes of MDM2/MDMX antagonists have been reported, including low molecular weight compounds [14,21–23], small peptides [24–27], and miniature proteins [28–30], among others. Compared with small molecule drugs, peptide antagonists, in principle, can be more efficacious due to enhanced affinity and specificity for their protein targets, thus low toxicity. However, a major drawback of peptide antagonists is their poor druggability on account of their poor in vivo stability and low intracellular delivery efficiency.
Recently, stapled peptide technology was introduced to stabilize α-helical peptides via the addition of an all-hydrocarbon cross-link between two α-methyl-substituted amino acids using Grubbs ruthenium catalyst, which could increase their affinity, in vivo stability and cellular uptake [32–34]. Brown et al. recently reported the synthesis of a stapled peptide, sPMI [34], based on a dual-specificity inhibitor of MDM2 and MDMX termed PMI discovered by Lu and colleagues [24]. sPMI potently antagonized both MDM2 and MDMX and activated p53 in vitro without apparent off-target cellular toxicity. However, several factors still limit the therapeutic efficacy of stapled PMI and its in vivo application, including (1) fast renal clearance due to its small size, (2) lack of tumor-targeting specificity, and (3) low solubility and potential absorption by serum proteins. Therefore we were inspired to design advanced peptide drug carriers to improve the therapeutic efficiency of sPMI by endowing it with the ability to specifically target tumor cells to activate p53 in glioma while minimizing its systemic doses and potential toxicity.
Therapeutic agents for malignant gliomas have been particularly inefficient for the existence of BBTB, which hampers the accumulation and uptake in tumors [35]. Moreover, even though BBB is compromised to some extent under the situation of malignant gliomas, it remains to be the obstacle influencing the therapeutic efficacies via systemic administration. Fortunately, there are many receptors over-expressed on the BBTB (tumor microvessels) and/or glioma cells, such as integrins, that can mediate RGD modified drug delivery systems targeting gliomas and enhance tumor uptake. We have previously developed a polymeric micelle modified with a cyclic RGD peptide (c(RGDyK)) for systemic delivery of hydrophobic drugs, such as paclitaxel [36–37]. The RGD peptide, which recognizes αvβ3 and αvβ5 integrin overexpressed on glioma cells and neovasculature [38–39], enables the drug carrier to circumvent the BBTB and target the glioma [40]. Here we prepared a c(RGDyK)-conjugated poly(ethylene glycol)-block-poly(lactic acid) micelle carrying the sPMI peptide (RGD-M/sPMI), and demonstrated its anti-glioma effect through activation of the p53 pathway in vitro and in vivo. Importantly, the combination of the RGD-M/sPMI micelle and temozolomide (TMZ) increased antitumor efficacy against glioblastoma in experimental animals. Our results validate a combination therapy using p53 activators with standard TMZ chemotherapy as a more effective treatment for GBM.
2. Materials and methods
2.1. Materials
The olefinic amino acids Fmoc-S5-OH and Fmoc-R8-OH were supplied by OKeanos Tech Co. Ltd. (Beijing, China). Fmoc-protected α-amino acids (other than the olefinic amino acids Fmoc-S5-OH and Fmoc-R8-OH) were purchased from GL Biochem Ltd. (Shanghai, China). Rink amide MBHA resin was from Xi'an Innovision Bioscience Co., Ltd. O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluorophosphate (HBTU) was purchased from American Bioanalytical Co. Ltd. (Natick, MA). Nutlin, diisopropylethylamine (DIEA) and Grubbs catalyst first-generation were supplied by Sigma-Aldrich. MPEG2000–PLA2000 and mal–PEG3000–PLA2000 were obtained from Advanced Polymer Materials Inc., Canada. Temozolomide was purchased from Dalian Meilun Biology Technology Co. Ltd. DiR (1,1′-dioctadecyl-3,3,3′,3′-tetramethyl indotricarbocyanine iodide) was from Invitrogen, USA. Rabbit anti-p21 was purchased from Abcam, USA. Mouse anti-p53 was from CST. Rabbit anti-actin was from Bioworld Technology Inc. Mouse anti-MDM2 was supplied by Santa Cruz. DAPI was supplied by Roche, Switzerland. Integrin αvβ3 was supplied by R&D System, USA. Biacore series S sensor chips CM5 and HBS-EP buffer were purchased from GE Healthcare Bio-Sciences AB (Sweden). Methanol, acetonitrile, and other HPLC grade reagents were obtained from Fisher Scientific, USA. All chemicals were analytic reagent grades.
U87MG glioblastoma cells were obtained from Shanghai Institute of Cell Biology. It was cultured in special Dulbecco's modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 °C in a 5% CO2 humidified atmosphere.
Male BALB/c Nu/Nu mice 4–6 weeks of age were purchased from Shanghai Laboratory Animal Center, CAS (SLACCAS) (Shanghai, China) and kept under SPF conditions. All animal experiments were carried out in accordance with guidelines evaluated and approved by the ethics committee of Fudan University.
2.2. Synthesis and characterization of sPMI
2.2.1. Synthesis of sPMI
sPMI was synthesized via Fmoc chemistry with solid phase peptide synthesis protocol as described in detail in the supplementary materials. The sequence of sPMI is Ac-1Thr-2Ser-3Phe-cyclo(4-R8-5Glu-6Tyr-7Trp-8Ala-9Leu-10Leu-11S5)-NH2. The addition of an all-hydrocarbon cross link across positions 4 and 11 by two α-methyl-substituted amino acids using Grubbs ruthenium catalyst. Crude products were precipitated with cold ether and purified to homogeneity by preparative C18 reversed-phase HPLC. After purification, sPMI and linear PMI were labeled with fluorescein isothiocyanate via N-terminal amino of peptide in phosphate buffer (1:1.2 M ratio, pH 8.0) to yield sPMI-FAM and PMI-FAM. The products were purified by C18 RP-HPLC and molecular masses were ascertained by electrospray ionization mass spectrometry (ESI-MS).
2.2.2. Characterization of sPMI
The solution conformational properties of sPMI and the linear peptide PMI were evaluated at 50 µM in 10% (v/v) TFE (2,2,2-trifluoroethanol) and 90% 10 mM phosphate buffer, pH 7.2. The spectra were collected on a spectropolarimeter (e.g., Jasco J-710) using standard measurement parameters (e.g., temperature, 20 °C; wavelength, 190–260 nm; step resolution, 0.5 nm; speed, 20 nm/s; accumulations, 10; response, 1 s; bandwidth, 1 nm; path length, 0.1 cm).
2.3. Cellular uptake of sPMI
Human glioblastoma cell line U87MG cells were seeded at 60,000 cells per well in four-well chambered cover-glass (In vitro Scientific) in normal medium [DMEM with 10% (vol/vol) FBS] and allowed to grow overnight. Cells were then incubated with 10 µM FAM-sPMI or FAM-PMI for 3 h in DMEM with 10% FBS. Cells were washed with Dulbecco's phosphate buffered saline, fixed with 4% (wt/vol) paraformaldehyde, finally incubated by DAPI to stain the cell nucleus. Imaged using an LSM 510 Zeiss Axiovert 200M (v4.0) confocal microscope. Images were analyzed using an LSM image browser.
2.4. Preparation and characterization of RGD-M/sPMI micelle
2.4.1. Preparation of RGD-M/sPMI micelle
An RGD-M micelle was synthesized as previously reported [36]. A mixture of c(RGDyK)–PEG3000–PLA2000 and mPEG2000–PLA2000 was used for the preparation of micelles so that the RGD ligand could straighten out, recognize and bind to integrins. In brief, the RGD-M micelle was prepared as follows: 20 mg micelle materials (containing 1 mg c(RGDyK)–PEG3000–PLA2000 and 19 mg mPEG2000–PLA2000) and 1 mg sPMI were co-dissolved in 3 ml methanol, and rotary evaporated to form a thin film at 40 °C. The RGD-M/sPMI micelle was obtained by hydrating the thin film with physiological saline. The unloaded sPMI was removed by gel filtration over a Sephadex G-50 column with physiological saline.
2.4.2. Characterization of RGD-M/sPMI micelle
The RGD-M/sPMI micelle was analyzed by dynamic light scattering and TEM. To investigate the encapsulation efficiency (E.E.) and loading capacity (L.C.), different amounts of sPMI were co-dissolved with RGD-M in the preparation process of micelles. The drug-loaded micelle was diluted by acetonitrile and the concentration of sPMI was measured via HPLC. E.E. was defined as the ratio between actual sPMI amount detected by HPLC and theoretic feeding sPMI amount. L.C. was defined as the ratio between sPMI loaded in MIE detected by HPLC and the weight of lyophilized micelle.
2.4.3. Surface plasmon resonance assay
SPR-based direct binding assays were carried out at 25 °C on a Biacore T200 instrument, using a CM5 sensor chip to which integrin αvβ3 is covalently attached. The RGD-PEG-PLA/sPMI micelle or RGD peptide was prepared in PBS buffer, pH 7.4, in a 2-fold serial dilution and was injected onto the integrin αvβ3-immobilized CM5 sensor chip at a flow rate of 20 µl/min for 2 min, followed by 4 min dissociation. Nonlinear regression analysis was performed using GraphPad Prism 5.0 to give rise to KD values according to the equation RU = RUmax·C / (KD +C).
2.5. In vitro activity of RGD-M/sPMI micelle
2.5.1. Western blot analysis
Western blotting assay was performed to determine the protein expression level. U87MG cells (six-well plates, 2 × 105 cells per well) were treated with various drugs in normal medium [DMEM with 10% (vol/vol) FBS] for 24 h. Total cellular lysates were collected and then clarified by centrifugation. Protein concentrations were quantified using BSA kits. Equal amounts of proteins were subjected to Western blotting analysis using an NuPAGE electrophoresis system as described previously [41].
2.5.2. Anti-proliferation assay
U87MG cells were seeded to 96-well tissue culture plates in 200 µl of normal medium [DMEM with 10% (vol/vol) FBS] to obtain a concentration of 3 × 103 cells per well, and then the plates were incubated for 24 h at 37 °C in a 5% CO2 atmosphere. The medium in each well was refreshed with 200 µl normal medium containing the RGD-M/sPMI micelle with various sPMI concentrations or Nutlin at 37 °C in a 5% CO2 atmosphere for 72 h. Experiments were repeated at least three times. The in vitro activity of sPMI was determined by the MTT assay (PowerWave XS, Bio-TEK, USA).
2.5.3. Annexin V assay
U87MG cells were seeded in 12-well tissue culture plates (1 × 105 cells per well) in normal medium [DMEM with 10% (vol/vol) FBS] for 24 h before drug treatment and incubated with the drug for an additional 24 h. No treatment controls were established in parallel. Attached cells were trypsinized and collected, while culture medium that may contain detached cells was also collected. Both parts of cells were combined and collected by centrifugation at 1000 rpm/min for 5 min. Annexin V positive cells were quantified using an Annexin V-FITC kit (Beyotime Institute of Biotechnology).
2.5.4. Cell-cycle analysis
For cell-cycle analysis, the procedure is similar with the Annexin V assay except that cells were fixed in 70% ethanol at 4 °C for 12 h after collection. Then cells were washed twice with PBS and propidium iodide staining solution and RNase A solution were added before analysis by flow cytometry.
2.6. Biodistribution test of RGD-M micelle
Male Balb/c nude mice 4–6 weeks of age were purchased from Shanghai Laboratory Animal Center, CAS (SLACCAS) (Shanghai, China) and kept under SPF conditions. All animal experiments were carried out in accordance with guidelines evaluated and approved by the ethics committee of Fudan University. The U87MG glioblastoma model was established by inoculation of 5 × 105 cells suspended in 5 µl PBS into mouse brain. To investigate the intracranial U87MG tumor xenograft targeting efficiency, DiR (Invitrogen, USA) was encapsulated in the RGD-M micelle. In brief, DiR was co-dissolved with the micelle materials in acetonitrile. After forming micelles, the free DiR was removed via the CL4B column. The intracranial U87MG tumor bearing mice 12 days after implantation were injected with 100 µl of the DiR encapsulated RGD-M micelle via the tail vein. After 2 h of injection, the fluorescent images were detected using an in-vivo image system, therefore tissues were harvested and imaged.
2.7. Anti-glioblastoma study of RGD-M/sPMI
2.7.1. Anti-glioblastoma study in subcutaneous tumor models
To evaluate the therapeutic efficiency of the RGD-M/sPMI micelle and in combination with TMZ on U87MG xenograft in vivo, we established two different brain tumor models based on sites of inoculation — subcutaneous and intracranial. The first subcutaneous animal models were established by inoculation of 4 × 106 U87MG cells (cells suspended in 100 µl PBS) into the subcutaneous tissue of the right shoulder blade of male Balb/c nude mice. After two weeks when the tumor volume was about 50–100 mm3, the treatment began. Forty eight mice were randomly assigned to six groups (n = 8) to receive 100 µl of saline, sPMI (10 mg/kg), RGD-M/sPMI micelle (4 mg/kg and 10 mg/kg respectively), TMZ (50 mg/kg), and TMZ plus RGD-M/sPMI sPMI (50 mg/kg and 10 mg/kg) via the tail vein except TMZ by oral administration on the initial day and every 2 days until the 9th day. Tumor size was monitored via serial caliper (GuangLu®, China) measurement every 2 days and the volume was counted as length × (width)2 / 2. The serial measurements of tumor weight were followed when the animals were sacrificed.
2.7.2. Anti-glioblastoma study in intracranial tumor models
In the second animal models, U87MG cells (5 × 105 cells suspended in 5 µl PBS) were implanted into the right striatum (1.8 mm lateral, 0.6 mm anterior to the bregma and 3 mm of depth) of male Balb/c nude mice by using a stereotactic fixation device with a mouse adaptor. The mice were randomly divided into five groups (n = 10) and treated with 100 µl of saline, RGD-M/sPMI micelle (10 mg/kg), TMZ (50 mg/kg), TMZ plus RGD-M/sPMI (50 mg/kg and 10 mg/kg respectively) and TMZ plus RGD-M/sPMI (10 mg/kg and 10 mg/kg respectively) via tail vein injection except TMZ by oral administration on the 10th, 12th, 14th, 16th and 18th day after implantation. The survival times were recorded.
2.8. Statistical analysis
The IC50 values were calculated by nonlinear regression analysis with the GraphPad Prism® 5.0 version program. Differences between treatment groups in the subcutaneous anti-glioblastoma effect were assessed using a two-way ANOVA analysis. Survival data were presented using Kaplan–Meier plots and were analyzed using a log-rank test. p < 0.05, p < 0.01 and p < 0.001were considered significant respectively.
3. Results
3.1. Characterization of sPMI peptide
The sPMI peptide was synthesized via the Fmoc-protected solid phase peptide synthesis method. The crude sPMI peptide was analyzed and purified by HPLC. The purity and molecular mass were confirmed by HPLC and ESI-MS as shown in Fig. S1. The determined molecular mass of 1462.4 Da is within experimental error of the expected value of 1463.2 Da calculated on the basis of the average isotopic composition of sPMI.
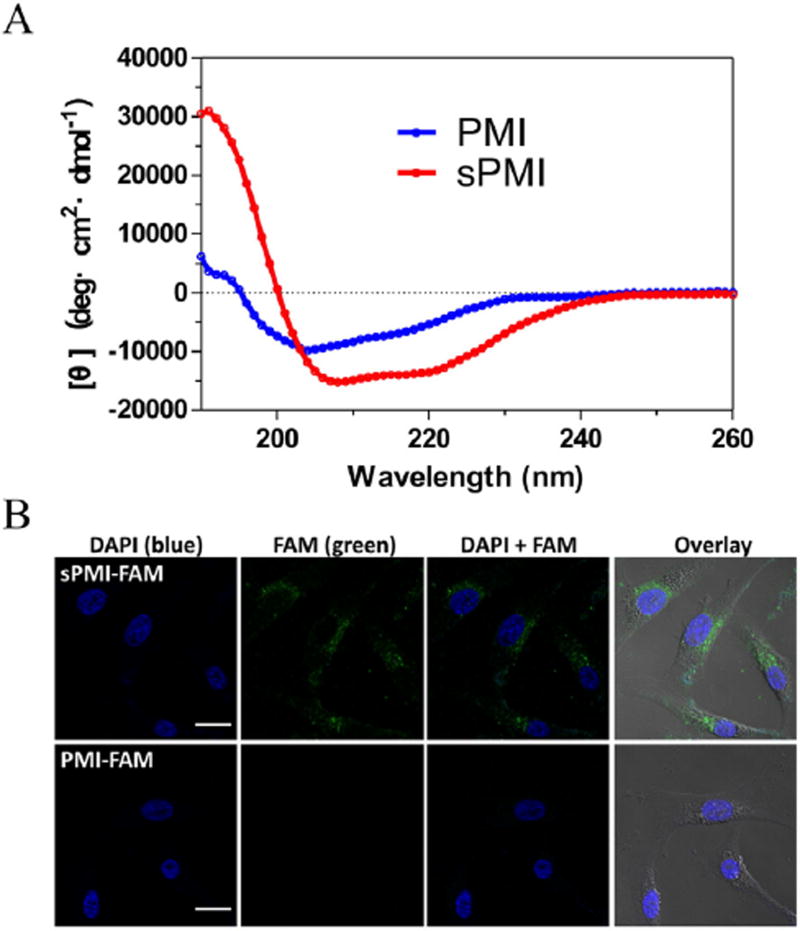
Binding studies using surface plasmon resonance techniques showed that compared with its unstapled parent peptide, sPMI exhibited no significantly decreased binding to MDM2 and MDMX (Fig. S2). The solution conformation of sPMI was determined by CD spectroscopy, and the spectra data revealed that sPMI adopted a largely α-helical conformation characterized by the double negative peaks at 208 and 222 nm and the single positive peak at 195 nm, whereas the corresponding linear PMI peptide was weakly α-helical (Fig. 1A).
Fig. 1.
sPMI adopts an α-helical structure and traverses cell membranes. (A) Comparative CD spectroscopy of sPMI and parent PMI peptide in aqueous buffer, pH 7.0, illustrates their intrinsic α-helical properties. (B) FAM-labeled sPMI showed a diffused intracellular localization, demonstrating efficient cellular uptake.
To investigate cellular uptake of sPMI, a fluorescent moiety (FAM), was introduced to the N terminus of the stapled peptide sequence. U87MG cells treated with either 10 µM FAM-sPMI or FAM-PMI were imaged by a confocal microscope (Figs. 1B and S3). FAM-sPMI showed a diffused intracellular localization, confirming efficient cellular penetration of the FAM labeled sPMI peptide. It was evident that PMI demonstrated fast degradation and most intact peptides disappeared after 24 h of incubation with 50% rat serum. In conspicuous contrast, sPMI displayed nearly no degradation under the same condition (Fig. S4).
3.2. Characterization of RGD-M/sPMI micelle
The particle size of the RGD-M/sPMI micelle was 22.4 nm and the polydispersity index is 0.099. Particle size distribution of a TEM image was shown in Fig. 2A. The drug encapsulation efficiency and loading capacity were 99% and 4.9% respectively. The particle sizes were not changed while encapsulating sPMI or DiR (Table 1 in supplementary material). The function of the c(RGDyK) peptide on the surface of a micelle binding to its target protein integrin αvβ3 receptor was evidenced by SPR assay. The modification of the c(RGDyK) peptide on the micelle's surface somehow increased its receptor binding affinity compared to that of peptide alone, yielding a KD value of 4.05 µM (Fig. 2B) for the RGD-M micelle while 45.4 µM for the c(RGDyK) peptide under the same condition (data not shown). The in vitro release profile of RGD-M/sPMI in different media was studied and showed in Figs. S5 and S6. The release of RGD-M/sPMI in PBS 7.4 was lower than in PBS 6.5, which indicated that RGD-M/sPMI released sPMI more quickly in the acidic environment. The lowest release that occurred in DMEM medium with 10% FBS, which simulated the extracellular environment in vivo, demonstrated that RGD-M/sPMI would be stable and would avoid drug release at the extracellular environment.
Fig. 2.
Characterization of the RGD-PEG-PLA/sPMI micelle. (A) Size distribution of the RGD-PEG-PLA/sPMI micelle was measured by TEM. (B) The RGD-PEG-PLA/sPMI micelle retains the binding ability to integrin α3βv via surface plasmon resonance.
3.3. RGD-M/sPMI micelle activates p53 signaling and main cellular functions
3.3.1. Western blotting
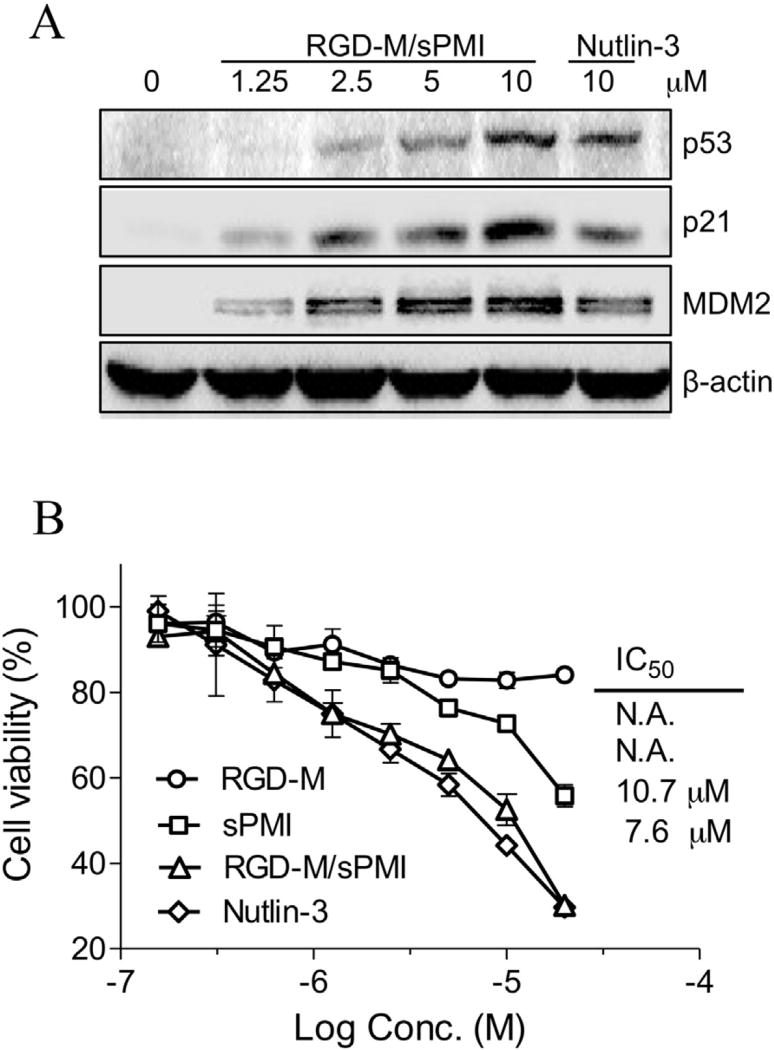
When an antagonist targets the p53-binding domains of MDM2 and MDMX, its negative regulators, should stabilize p53 and activate the pathway. P53 pathway activation should happen in cells expressing WT p53. In the presence of serum, a dose-dependent increase of p53 protein levels in U87MG cells was examined by the treatment of the RGD-M/sPMI micelle. Accompanied elevation of the p53 transcriptional targets p21 and MDM2 were also detected as shown in Fig. 3A.
Fig. 3.
RGD-M/sPMI micelle activates p53 signaling in U87MG cells. (A) RGD-M/sPMI micelle stabilizes p53 and elevates protein levels of p53 targets p21 and MDM2. U87MG cells were incubated with 1.25, 2.5, 5.0, or 10 µM RGD-M/sPMI or 10 µM Nutlin-3 for 24 h, and cell lysates were analyzed by Western blotting. (B) The viability of U87MG cells was determined after incubation with the RGD-M/sPMI micelle and expressed as a percentage of controls ± SEM.
3.3.2. Anti-proliferation
The anti-proliferative effect of sPMI, RGD-M, RGD-M/sPMI and Nutlin-3 on U87MG cells was evaluated using the MTT method. As shown in Fig. 3B, RGD-M/sPMI showed a submicromolar anti-proliferation ability, even in the presence of serum, with IC50 of 10.7 µM closed to 7.6 µM for Nutlin-3, while control empty micelle showed no significant cytotoxity. However, the anti-U87MG cell activity of sPMI was dramatically decreased in the presence of serum, while showing significantly improved potency in the absence of serum (Fig. S7).
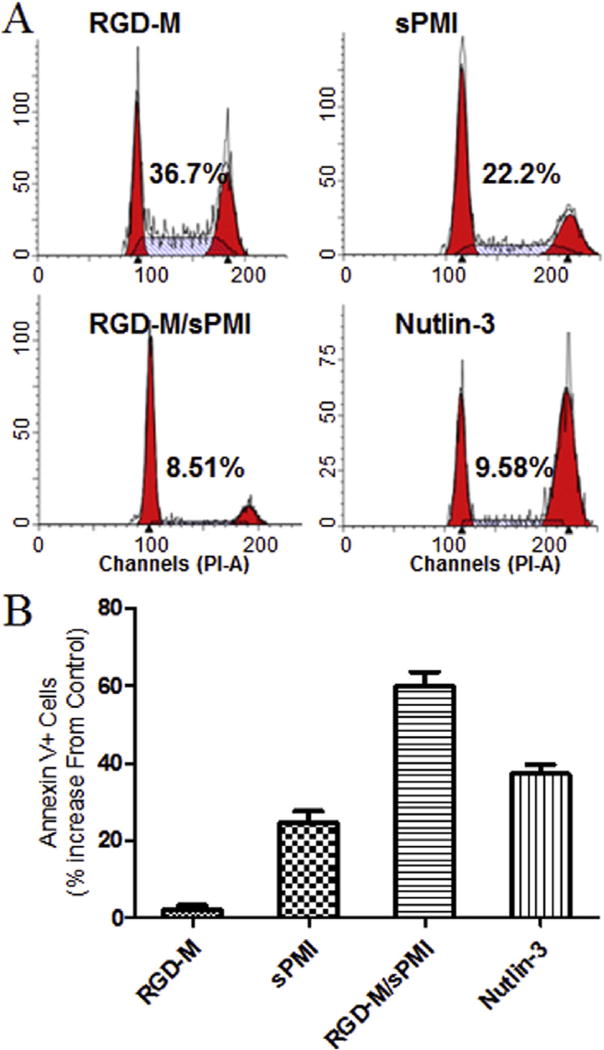
3.3.3. Cell-cycle arrest and apoptosis
The most important functions for p53 tumor suppressor protein are inductions of cell-cycle arrest and apoptosis. The RGD-M/sPMI micelle showed effective cell-cycle arrest in the G1 and G2 phases in U87MG cells as well as Nutlin-3, leading to depletion of the S phase (Fig. 4A). Next, we examined the Annexin V assay to quantify the apoptotic effect of the RGD-M/sPMI micelle (Fig. 4B). The apoptosis effect of sPMI and Nutlin-3 were similar in the U87MG cells. Encapsulation of sPMI by the micelle increased the apoptosis effect compared with sPMI alone, while control empty micelle showed no significant apoptosis effect.
Fig. 4.
RGD-M/sPMI micelle activates main p53 cellular functions in U87MG cells. (A) The RGD-M/sPMI micelle arrests cell-cycle progression in U87MG cells. Exponentially growing U87MG cells were incubated with 10 µM of the RGD-M/sPMI micelle for 24 h, and cell cycle distribution was determined by PI labeling and cell-cycle analysis. Numbers indicate the percentage of S-phase cells. (B) Apoptotic response to 10 µM of Nutlin-3, sPMI, RGD-M/sPMI and RGD-M micelle in U87MG cells for 24 h, and the percentage of apoptotic cells (mean ± SEM) were determined by the Annexin V assay.
3.4. Anti-glioblastoma activity of RGD-M/sPMI
3.4.1. RGD-M/sPMI micelle suppresses tumor growth
In the first subcutaneous animal model, the volumes of tumors were measured every other day. As shown in Fig. 5A, the tumor size in animals of the saline group increased by 8.3-fold on day 14 compared to tumor size at the start of treatment. sPMI at 10 mg/kg treatment was effective in slowing tumor growth (5.8-fold increase) compared with mock treatment. Remarkably, the RGD-M/sPMI micelle at low and high doses both showed tumor inhibition (4.3-fold and 1.8-fold increase respectively). As the first-line chemotherapeutic agent for glioma, the TMZ treatment group showed dramatic tumor inhibition (0.3-fold increase). Furthermore, combination of RGD-M/sPMI (10 mg/kg) and TMZ (50 mg/kg) displayed a better tumor growth inhibitory effect during the same period (0.4-fold decrease). The tumor weight measurements after sacrifice confirm the tumor volume results (Fig. 5B). The p53 activation in subcutaneous glioma-bearing models was examined by immunohistochemistry after sacrifice. As shown in Fig. S8, the expression of p53, p21 and MDM2 was examined to evaluate p53 activation efficiency. The group of glioma-bearing mice treated with RGD-M/sPMI at 10 mg/kg showed more expression of p53, p21 and MDM2 compared with the saline, sPMI (10 mg/kg) and RGD-M/sPMI (4 mg/kg) groups, which are related with in vitro results.
Fig. 5.
RGD-M/sPMI micelle suppresses tumor growth in vivo of U87MG subcutaneous xenografts in nude mice. (A) Tumor volumes were callipered throughout the study, and data were plotted as mean ± SEM. (B) Tumor weights were measured after the mice were sacrificed. *p < 0.05, **p < 0.01, ***p < 0.001.
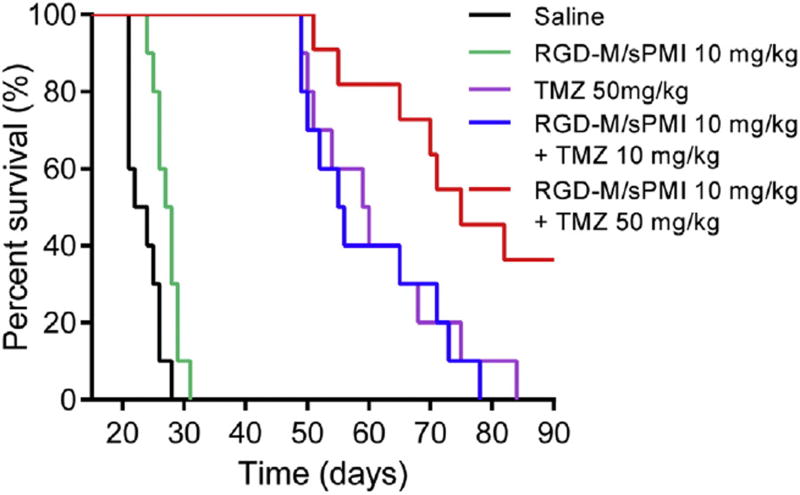
3.4.2. RGD-M/sPMI micelle prolongs animal survival
In the second animal model, non-invasive NIR fluorescence imaging showed that much stronger fluorescence intensity of RGD-PEG-PLA was observed in the glioma site than that of PEG-PLA at 2 h following administration (Fig. S9A). This result was confirmed by imaging those brains collected from mice bearing an intracranial glioma 2 h post-injection (Fig. S9B). In fact, in our previous published study [37], we had confirmed that the 125I radiolabeled RGD-M micelle could target and accumulate in tumor sites, while these could be blocked by the non-radiolabeled RGD-M micelle, indicating the accumulation of RGD-M in the tumor mainly via receptor active targeting and a possible enhanced permeability and retention (EPR) effect.
The therapeutic efficiency was evaluated by measuring the survival time of 5 groups of mice bearing an intracranial U87MG glioma following the treatment respectively. As shown in Fig. 6, mice treated with the RGD-M/sPMI micelle survived significantly longer than the untreated mice. The medium survival time of untreated mice was 22 days, and prolonged to 27.5 days upon RGD-M/sPMI micelle treatment (P < 0.05). The medium survival time of TMZ-treated mice at 50 mg/kg was significantly prolonged to 59 days. Of note, a combination of RGD-M/sPMI (10 mg/kg) and TMZ (50 mg/kg) dramatically prolonged the survival time to 70 days (p < 0.001). Interestingly, decreasing the dosage of TMZ to 10 mg/kg while in combination with RGD-M/sPMI (10 mg/kg) prolonged the survival time to 55.5 days which was similar to a high dosage of TMZ (50 mg/kg, 59 days). This result, along with the fact that the RGD-M/sPMI micelle alone only slightly prolonged the survival time, suggested the synergistic effect of a combination regimen, while prolonging the survival time or decreasing the dosage of TMZ.
Fig. 6.
RGD-M/sPMI micelle suppresses tumor growth in vivo of U87MG intracranial glioblastoma-implanted nude mice. Kaplan–Meier survival curves for intracranial glioblastoma-implanted nude mice treated with RGD-M/sPMI and TMZ.
4. Discussion and conclusion
Recent studies showed that stapled peptide antagonists of MDM2 and MDMX with high affinity and specificity could be developed as a novel class of therapeutic agents for the treatment of many tumors with WT p53 status though p53 activation [31–34]. These peptide sequences conserve an amphipathic α-helix with functional hydrophobic amino acids which are essential to dock side a hydrophobic cavity of the oncoprotein. Thus, the addition of a stapled cross-linked all hydrocarbon side-chain to amphipathic α-helical peptides extremely increases its hydrophobicity resulting in low solubility in physiological conditions. Indeed, the studies with sPMI were somehow limited by its solubility, as evidenced by peptide precipitation at 50 µM. In fact, CD analysis of sPMI could not be performed at physiological solution without the addition of TFE. In addition, anti-proliferation activity of sPMI was limited in the presence of fetal bovine serum as evidenced in Fig. S7. Besides, fast renal clearance due to its small size and lack of tumor-targeting specificity will limit therapeutic efficacy of stapled PMI. Of note, a biocompatible PEG-PLA micelle has been widely used as drug delivery systems for hydrophobic drugs. Compared with other materials, its main advantages included the higher drug encapsulation efficiency and loading capacity as well as safety and non-toxicity, which has been approved by FDA for human use. Therefore, we chose PEG-PLA for the incorporation of a stapled peptide.
In the present work, we used a PEG-PLA micelle as an advanced peptide carrier that dramatically increased the solubility of sPMI, thus alleviating serum sequestration and effectively delivering to GBM in vivo. As expected, the RGD-M/sPMI micelle exerted potent p53-dependent anti-proliferation activity against U87MG cells in the presence of 10% FBS, while sPMI alone showed little activity. Cell-cycle arrest and apoptosis assay confirmed the restriction of activity by serum. With respect to GBM, the integrity of BBB and BBTB severely restricts p53 therapy. We took the advantage of the c(RGDyK) peptide with high affinity and specificity with integrin αvβ3 and αvβ5, which is widely overexpressed on brain tumor neovasculature and glioma cell membrane. In nude mouse xenograft models, the RGD-M/sPMI micelle suppressed tumor growth and prolonged animal survival. In vitro mechanistic studies suggest that the RGD-M/sPMI micelle inhibits tumor growth by activating the p53 pathway.
As the first-line chemotherapeutic agent for GBM since 1999, TMZ standard treatment has benefited numerous glioma patients. However, more and more reports have showed that the overall survival benefit from TMZ treatment is only moderate, in part because of glioma cell resistance to TMZ and tumor recurrence [42–43]. However, activation of p53was reported to sensitize the glioma to TMZ [44–45]. In the present work, the combination of the RGD-M/sPMI micelle and TMZ increased antitumor efficacy against glioblastoma in experimental animals. Besides, as shown in Fig. S10, the H&E staining images of heart, liver, spleen, lung, and kidney revealed that all of the samples did not cause any damage in main organs, which suggested that all of the sPMI formulation had low toxicity. Overall, our results validate a combination therapy using p53 activators with TMZ standard treatment as a more effective treatment for GBM.
Although patients' compliance of TMZ is good, dose-dependent side-effects including severe myelosuppression had restricted its treatment dosage. Overdoses of TMZ treatment also could result in lymphocytopenia and increase risk of infection [46]. Our intracranial U87MG nude model experiments showed that combined administration of the RGD-M/sPMI micelle at 10 mg/kg with a low dose of TMZ at 10 mg/kg showed a medium survival time of 55.5 days, which was similar with 59.5 days at high dose of TMZ at 50 mg/kg. Our work validated an additional advantage of combination therapy using p53 activators with TMZ standard treatment, which has the potential to lower the effective therapeutic dose of TMZ, resulting in reducing the side effects and improving glioma patients' compliance. Together above, this combination therapy regimen might be a more effective and safe therapy for GBM.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program, No. 2013CB932500), the National Natural Science Foundation of China (No. 81273458 & No. 81473149), the National Science & Technology Major Project (2012ZX09304004), Development Project of Shanghai Peak Disciplines-Integrated Chinese and Western Medicine and the National Institutes of Health Grant CA 167296 (to Wuyuan Lu).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2015.09.061.
References
- 1.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Wagner M, Bigner DD, Friedman AH, Friedman HS. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin. Cancer Res. 2007;13:1253–1259. doi: 10.1158/1078-0432.CCR-06-2309. [DOI] [PubMed] [Google Scholar]
- 2.Hou LC, Veeravagu A, Hsu AR, Tse VC. Recurrent glioblastoma multiforme: a review of natural history and management options. Neurosurg. Focus. 2006;20:E5. doi: 10.3171/foc.2006.20.4.2. [DOI] [PubMed] [Google Scholar]
- 3.Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat. Rev. Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton HB. Advances in strategies to improve drug delivery to brain tumors. Expert Rev. Neurother. 2006;6:1495–1509. doi: 10.1586/14737175.6.10.1495. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Lu W. Recent advances in brain tumor-targeted nano-drug delivery systems. Expert Opin. Drug Deliv. 2012;9:671–686. doi: 10.1517/17425247.2012.682726. [DOI] [PubMed] [Google Scholar]
- 6.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 7.Ding H, Inoue S, Ljubimov AV, Patil R, Portilla-Arias J, Hu J, Konda B, Wawrowsky KA, Fujita M, Karabalin N, Sasaki T, Black KL, Holler E, Ljubimova JY. Inhibition of brain tumor growth by intravenous poly (beta-l-malic acid) nanobioconjugate with pH-dependent drug release[corrected] Proc. Natl. Acad. Sci. U. S. A. 2010;107:18143–18148. doi: 10.1073/pnas.1003919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Lane DP, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 9.Vousden KH, Lane DP. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 10.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 11.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 12.Picksley SM, Lane DP. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays. 1993;15:689–690. doi: 10.1002/bies.950151008. [DOI] [PubMed] [Google Scholar]
- 13.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 14.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Aguilar A, Bernard D, Wang S. Small-molecule inhibitors of the MDM2-p53 protein–protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Zeng SX, Lu H. Targeting p53–MDM2–MDMX loop for cancer therapy. Subcell. Biochem. 2014;85:281–319. doi: 10.1007/978-94-017-9211-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagpal J, Jamoona A, Gulati ND, Mohan A, Braun A, Murali R, Jhanwar-Uniyal M. Revisiting the role of p53 in primary and secondary glioblastomas. Anticancer Res. 2006;26:4633–4639. [PubMed] [Google Scholar]
- 18.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, Gu B, Wei G, Yuan W, Zhan C, Lu WY, Lu W. D-peptide inhibitors of the p53–MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Shen J, Wei X, Xie C, Lu W. Targeted delivery of a novel palmitylated D-peptide for antiglioblastoma molecular therapy. J. Drug Target. 2012;20:264–271. doi: 10.3109/1061186X.2011.645162. [DOI] [PubMed] [Google Scholar]
- 21.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 22.Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, Bernard D, Zhang J, Lu Y, Gu Q, Shah RB, Pienta KJ, Ling X, Kang S, Guo M, Sun Y, Yang D, Wang S. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Di Lello P, Fry D, Garvie C, Huang KS, Gao L, Tovar C, Lovey A, Wanner J, Vassilev LT. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan C, Zhao L, Wei X, Wu X, Chen X, Yuan W, Lu WY, Pazgier M, Lu W. An ultrahigh affinity d-peptide antagonist of MDM2. J. Med. Chem. 2012;55:6237–6241. doi: 10.1021/jm3005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 27.Czarna A, Popowicz GM, Pecak A, Wolf S, Dubin G, Holak TA. High affinity interaction of the p53 peptide-analogue with human Mdm2 and Mdmx. Cell Cycle. 2009;8:1176–1184. doi: 10.4161/cc.8.8.8185. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Liu M, Monbo J, Zou G, Li C, Yuan W, Zella D, Lu WY, Lu W. Turning a scorpion toxin into an antitumor miniprotein. J. Am. Chem. Soc. 2008;130:13546–13548. doi: 10.1021/ja8042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, Pazgier M, Liu M, Lu WY, Lu W. Apamin as a template for structure-based rational design of potent peptide activators of p53. Angew. Chem. Int. Ed. Engl. 2009;48:8712–8715. doi: 10.1002/anie.200904550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kritzer JA, Zutshi R, Cheah M, Ran FA, Webman R, Wongjirad TM, Schepartz A. Miniature protein inhibitors of the p53-hDM2 interaction. Chembiochem. 2006;7:29–31. doi: 10.1002/cbic.200500324. [DOI] [PubMed] [Google Scholar]
- 31.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To KH, Olson KA, Kesavan K, Gangurde P, Mukherjee A, Baker T, Darlak K, Elkin C, Filipovic Z, Qureshi FZ, Cai H, Berry P, Feyfant E, Shi XE, Horstick J, Annis DA, Manning AM, Fotouhi N, Nash H, Vassilev LT, Sawyer TK. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3445–3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown CJ, Quah ST, Jong J, Goh AM, Chiam PC, Khoo KH, Choong ML, Lee MA, Yurlova L, Zolghadr K, Joseph TL, Verma CS, Lane DP. Stapled peptides with improved potency and specificity that activate p53. ACS Chem. Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- 33.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernal F, Wade M, Godes M, Davis TN, Whitehead DG, Kung AL, Wahl GM, Walensky LD. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan C, Lu W. The blood–brain/tumor barriers: challenges and chances for malignant gliomas targeted drug delivery. Curr. Pharm. Biotechnol. 2012;13:2380–2387. doi: 10.2174/138920112803341798. [DOI] [PubMed] [Google Scholar]
- 36.Zhan C, Wei X, Qian J, Feng L, Zhu J, Lu W. Co-delivery of TRAIL gene enhances the anti-glioblastoma effect of paclitaxel in vitro and in vivo. J. Control. Release. 2012;160:630–636. doi: 10.1016/j.jconrel.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Zhan C, Gu B, Xie C, Li J, Liu Y, Lu W. Cyclic RGD conjugated poly(ethylene glycol)-co-poly(lactic acid) micelle enhances paclitaxel anti-glioblastoma effect. J. Control. Release. 2010;143:136–142. doi: 10.1016/j.jconrel.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 38.Bello L, Francolini M, Marthyn P, Zhang J, Carroll RS, Nikas DC, Strasser JF, Villani R, Cheresh DA, Black PM. Alpha(v)beta3 and alpha(v)beta5 integrin expression in glioma periphery. Neurosurgery. 2001;49:380–389. doi: 10.1097/00006123-200108000-00022. discussion 390. [DOI] [PubMed] [Google Scholar]
- 39.Gladson CL, Cheresh DA. Glioblastoma expression of vitronectin and the alpha v beta 3 integrin. Adhesion mechanism for transformed glial cells. J. Clin. Invest. 1991;88:1924–1932. doi: 10.1172/JCI115516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Park R, Shahinian AH, Tohme M, Khankaldyyan V, Bozorgzadeh MH, Bading JR, Moats R, Laug WE, Conti PS. 18F-labeled RGD peptide: initial evaluation for imaging brain tumor angiogenesis. Nucl. Med. Biol. 2004;31:179–189. doi: 10.1016/j.nucmedbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Jiang G, Wei ZP, Pei DS, Xin Y, Liu YQ, Zheng JN. A novel approach to overcome temozolomide resistance in glioma and melanoma: inactivation of MGMT by gene therapy. Biochem. Biophys. Res. Commun. 2011;406:311–314. doi: 10.1016/j.bbrc.2011.02.042. [DOI] [PubMed] [Google Scholar]
- 42.Martin S, Janouskova H, Dontenwill M. Integrins and p53 pathways in glioblastoma resistance to temozolomide. Front. Oncol. 2012;2:157. doi: 10.3389/fonc.2012.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimoto K, Mizoguchi M, Hata N, Murata H, Hatae R, Amano T, Nakamizo A, Sasaki T. Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma. Front. Oncol. 2012;2:186. doi: 10.3389/fonc.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 45.Kim SS, Rait A, Kim E, Pirollo KF, Nishida M, Farkas N, Dagata JA, Chang EH. A nanoparticle carrying the p53 gene targets tumors including cancer stem cells, sensitizes glioblastoma to chemotherapy and improves survival. ACS Nano. 2014;8:5494–5514. doi: 10.1021/nn5014484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tentori L, Graziani G. Recent approaches to improve the antitumor efficacy of temozolomide. Curr. Med. Chem. 2009;16:245–257. doi: 10.2174/092986709787002718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.