Abstract
Hybrid sterility is a common first step in the evolution of postzygotic reproductive isolation. According to Haldane's Rule it affects predominantly the heterogametic sex. While the genetic basis of hybrid male sterility in organisms with heterogametic males has been studied for decades, the genetic basis of hybrid female sterility in organisms with heterogametic females has received much less attention. We investigated the genetic basis of reproductive isolation in two closely related avian species, the Common Nightingale (Luscinia megarhynchos) and the Thrush Nightingale (L. luscinia), that hybridize in a secondary contact zone and produce viable hybrid progeny. In accordance with Haldane's Rule, hybrid females are sterile, while hybrid males are fertile, allowing gene flow to occur between the species. Using transcriptomic data from multiple individuals of both nightingale species we identified genomic islands of high differentiation (FST) and of high divergence (Dxy), and we analyzed gene content and patterns of molecular evolution within these islands. Interestingly, we found that these islands were enriched for genes related to female meiosis and metabolism. The islands of high differentiation and divergence were also characterized by higher levels of linkage disequilibrium than the rest of the genome in both species indicating that they might be situated in genomic regions of low recombination. This study provides one of the first insights into genetic basis of hybrid female sterility in organisms with heterogametic females.
Keywords: speciation, genomic islands of differentiation, hybrid female sterility, oogenesis, birds
Introduction
Understanding the genetic basis of reproductive isolation between incipient species is a major goal of evolutionary biology. Hybrid sterility is a common first step in the evolution of postzygotic reproductive isolation (Coyne & Orr 2004). Haldane (1922) observed that hybrid sterility occurs preferentially in the heterogametic sex, i.e. males in organisms with XY sex chromosomes, such as mammals and Drosophila, and females in organisms with ZW sex chromosomes, such as birds and butterflies. While the genetic basis of hybrid male sterility in organisms with heterogametic males has been studied for decades (e.g., Storchová et al. 2004; Masly & Presgraves 2007; Mihola et al. 2009; Good et al. 2010; Presgraves 2010; Bhattacharyya et al. 2014; Zhang et al. 2015), the genetic basis of hybrid female sterility in organisms with heterogametic females has received much less attention. In fact, most studies on hybrid female sterility have been limited to anecdotal observations that hybrid females do not lay eggs (Stadie 1991; Reifová et al. 2011a), that eggs of hybrid females fail to hatch nestlings (Gelter et al. 1992), or that hybrid females have atrophied ovaries (Eroukhmanoff et al. 2016). One exception is the work on Heliconius butterflies showing that a large Z effect underlies hybrid female sterility, although specific genes have not been identified (Jiggins et al. 2001; Naisbit et al. 2002).
In the last fifteen years, technological advances in DNA sequencing have created new possibilities for studying the genetic basis of reproductive isolation in non-model organisms (Seehausen et al. 2014). One approach is to study patterns of genetic differentiation (FST) across the genome. In the presence of gene flow, genomic regions underlying reproductive isolation should show increased levels of genetic differentiation since selection will act against introgressed alleles in these regions (Wu 2001; Via & West 2008). These portions of the genome have been referred to as islands of high differentiation (Turner et al. 2005; Harr, 2006; Ellegren et al. 2012; Burri 2017; Wolf & Ellegren 2017). However, heterogeneous levels of differentiation can arise even in the absence of differential gene flow as a consequence of selection at linked sites leading to a local reduction in within-population diversity (Maynard Smith & Haigh 1974; Kaplan et al. 1989; Charlesworth et al. 1993). One way to distinguish between differential gene flow and selection at linked sites as causes for heterogeneous levels of differentiation is to take into account absolute divergence (Dxy) - the average number of pairwise differences between alleles sampled from two populations (Nei 1987). Selection at linked sites reduces variation within populations, and since FST (Wright 1951) depends on the amount of within-population variation as well as the amount of between-population variation, selection at linked sites will lead to increased values of FST (Charlesworth 1998). In contrast, absolute sequence divergence (Dxy) is not increased by selection at linked sites. In fact, Dxy may be reduced in such regions since selection in the ancestral population will reduce the coalescence time for alleles sampled in the two daughter populations. On the other hand, Dxy is expected to be higher in regions with low introgression as introgression reduces Dxy in a similar way as FST. Thus, genomic regions of high differentiation and high absolute divergence are likely to contain loci that are resistant to introgression (Noor & Bennett 2009; Nachman & Payseur 2012; Cruikshank & Hahn 2014).
Analysis of islands of high differentiation and divergence thus seems to be a promising approach for identifying genomic regions involved in reproductive isolation. For example, this method has recently been used with two flycatcher species (Ficedula hypoleuca and Ficedula albicollis) (Ellegren et al. 2012; Burri et al. 2015). These taxa have been described as conforming to Haldane's Rule (e.g. Sætre et al. 2003), although more recent work suggests that hybrids of both sexes are sterile (Alund et al. 2013). Moreover, genome-wide data to date have not revealed a single backcross individual in the hybrid zone (Kawakami et al. 2014), suggesting that postzygotic isolation is already complete in these species. Patterns of FST and Dxy across the flycatcher genome suggest that heterogeneity in levels of differentiation is due mostly to linked selection rather than to differences in levels of gene flow (Burri et al. 2015). Genome-wide comparisons of Dxy and FST have also been used in Swainson's thrush subspecies (Catharus ustulatus ustulatus and C. u. swainsoni) (Delmore et al. 2015), two cichlid ecomorphs that are in an early stage of ecological divergence (Malinsky et al. 2015), and subspecies of house mice (Phifer-Rixey et al. 2014), among others.
Here we explore the genomic landscape of differentiation and divergence in two closely related songbirds, the Common Nightingale (L. megarhynchos) and the Thrush Nightingale (L. luscinia). These two species diverged 1.8 Mya and currently hybridize in a zone of secondary contact across Central Europe (Storchová et al. 2010). Despite relatively strong assortative mating, F1 as well as backcross hybrids occur in sympatric populations (Reifová et al. 2011a, 2011b; Vokurková et al. 2013) indicating that postzygotic isolation is still not complete. Crosses of the two species in captivity as well as observations in nature show that F1 hybrid females are sterile, while F1 hybrid males are fertile, consistent with Haldane's Rule (Stadie 1991; Reifová et al. 2011a). The two nightingale species are very similar morphologically and ecologically, and for that reason, strong competition occurs between the species in sympatry (Reif et al. 2015). Interspecific competition has resulted in ecological character displacement in beak size (Reifová et al. 2011b), which might enhance reproductive isolation and thus also contribute to speciation. In previous work, using coalescent models of isolation with migration we showed that gene flow has occurred between the species and that gene flow has been higher on the autosomes than on the Z chromosome, consistent with a large Z effect (Storchová et al. 2010).
Here, we expand on that work by comparing loci across the nightingale genomes. Using transcriptomic data from multiple allopatric individuals of both nightingale species, we identified (i) genomic islands of high differentiation and (ii) genomic islands of both high differentiation and divergence. We than analyzed gene content within these islands. In addition, we compared levels of linkage disequilibrium and patterns of molecular evolution using McDonald-Kreitman tests in genomic regions inside and outside of these islands. Interestingly, we found significant enrichment of genes related to female meiosis and metabolism in islands of high differentiation. The same functional categories were also enriched in islands of high differentiation and divergence although in this case the enrichment was not significant after correction for multiple tests. Islands of high differentiation and divergence were also characterized by increased levels of linkage disequilibrium in both species, suggesting that these regions might be situated in genomic regions with reduced recombination. Our study confirms that hybrid female sterility is an important reproductive barrier between the two nightingale species and provides a first look at the genetic basis of hybrid female sterility in organisms with heterogametic males.
Materials and methods
Samples
Birds were sampled in allopatric regions close to the contact zone, in southwestern Poland for L. megarhynchos and in northeastern Poland for L. luscinia. Within each species, birds were captured at different localities 10-30 kilometers apart. The whole sampling area spanned approximately 100 km in both species. Eight male L. megarhynchos and seven male L. luscinia were collected. In addition, we sampled one male individual of the Bluethroat (L. svecica) as an outgroup. L. svecica diverged from the other two species approximately 12 Mya (Jetz et al. 2012). Sampling was done in 2010 at the beginning of the breeding season (early May). The birds were euthanized, and a piece of liver was removed and placed into RNA later. All work with live animals was done in accordance with the Local Ethic Committee for Scientific Experiments on Animals in Poznan, Poland (permission no. 36/2010).
Transcriptome sequencing
Total RNA from liver was isolated using Trizol Reagent (Ambion) according to the manufacturer's protocol. The integrity of the RNA was verified using an Experion Automated Electrophoresis system (Bio-Rad). cDNA synthesis and normalization were performed by the Evrogen Lab (Moscow, Russia). Total RNA samples were used for ds cDNA synthesis using a SMART approach (Zhu et al. 2001). SMART-prepared cDNAs were then normalized using the DSN normalization method (Zhulidov et al. 2004). The normalization procedure included cDNA denaturation/reassociation, treatment by duplex-specific nuclease (DSN) (Shagin et al. 2002) and amplification of the normalized fraction by PCR. One μg of each normalized cDNA sample was used to generate a tagged 454 sequencing library using MID adaptors following the Rapid Library Preparation Method from Roche. The libraries were sequenced on a GS FLX instrument using Titanium chemistry. Most libraries were sequenced in pairs in which two different libraries were pooled and loaded onto one sequencing region.
Transcriptome assembly and variant calling
The sequences were split according to MIDs using the sfffile tool and converted to fastq. Transcriptome assembly was performed with Newbler 2.6 (Margulies et al. 2005). Resulting contigs were mapped onto the zebra finch (Taeniopygia guttata) reference genome (taeGut1, WUSTL v3.2.4). The assembly produced 61,753 unique contigs belonging to 9,332 unique genes in the reference genome. The average length of the contigs was 1,264 bp and the median length 1,023 bp. The chromosome and position in the zebra finch genome were used to build a contig scaffold by concatenating the contigs in the order of their zebra finch homologs. All reads were mapped onto the contig scaffold with SMALT (http://www.sanger.ac.uk/science/tools/smalt-0), and variants were called using samtools mpileup (Li et al. 2009). The variant detection pipeline closely followed one used in Mořkovský et al. (2015). In brief, we applied the default filtering criteria used by vcfutils varFilter and additionally filtered for Phred quality score above 10. In addition, we excluded from the analysis all variants from contigs that spanned more than 50 kb in the zebra finch genome after the mapping since this probably reflects incorrect mapping. We identified 300,433 SNPs which met the filtering criteria. The average coverage of each variant that was used in analyses was 42.5.
Nightingales and zebra finch diverged approximately 45 million years ago (Jetz et al. 2012). However, large-scale synteny is observed among bird genomes separated by even greater evolutionary distances (Backström et al. 2008). Nonetheless, using the zebra finch genome may result in incorrect ordering of some genes in nightingales if rearrangements have occurred. This is likely to introduce some noise into downstream analyses, but should not result in systematic bias affecting the gene content or patterns of molecular evolution in islands of high differentiation and divergence.
Analysis of genetic differentiation (FST) and divergence (Dxy)
Per site unweighted FST (Wright 1951) was calculated for all SNP variants that passed our quality filters using vcftools (Danecek et al. 2011). We calculated smoothed FST values by averaging per site FST in sliding windows along chromosomes using positions in the zebra finch genome. In cases where the nightingale sequence mapped perfectly to the zebra finch genome, the exact genomic position for SNPs within this sequence was taken. In cases where there was no exact mapping, for example in less conserved introns (note that a small number of introns are captured in unspliced transcripts), the positions of SNP variants were taken within the range of the gene to which the intron belonged (this occurred in 23,819 out of 410,245 SNP variants, or 5.8 %). The size of the sliding window was 1 Mb and the step size was 100 kb.
We used a permutation analysis to identify islands of elevated FST. A distribution of potential FST values in each window was determined by shuffling the real FST values among the genomic positions of SNP variants – this kind of resampling procedure serves to establish a distribution of FST values as expected if genome position had no effect on FST. We sampled the distributions with 25,000 realizations of the permutation. Then the upper 1 % quantile of the distribution was used as a threshold to identify windows with elevated FST. To account for the different mode of inheritance and effective population size of the sex chromosomes and their effect on FST, permutations were conducted separately for the Z chromosome and for all autosomes. Windows of elevated FST less than 1Mbp apart were pooled and the whole genomic region containing these windows was designated as an island of high differentiation.
Dxy, defined as the average number of pairwise differences between alleles sampled from two populations, was calculated as follows. First, for each site we calculated the average number of pairwise differences as p1q2 + p2q1, where p and q represent the frequencies of alternative alleles and subscripts represent the species of origin. Second, the average pairwise differences for individual SNPs were summed and divided by the total length of the contig including all variable and invariant sites that passed the filtering criteria. The average Dxy in the FST islands was calculated. A subset of islands of high differentiation with Dxy higher than the genomic average was then identified for further analyses. We refer to these islands as islands of high differentiation and divergence.
Gene set enrichment analysis
To assess whether the genes in the islands of high differentiation and islands of high differentiation and divergence were associated with specific functions, we carried out a test for over-represented gene groups. We took all Ensembl (Yates et al. 2016) genes found (i) in the islands of high differentiation and (ii) in the islands of high differentiation and divergence and assigned them to KEGG pathways (Kanehisa & Goto 2000, Kanehisa et al. 2016) using the GenomeNET REST API and KEGG REST API. For each pathway we counted the genes found in the islands and calculated the enrichment score using all Ensembl genes as a background (17,488 genes). We used Fisher's exact tests and binomial tests followed by a correction for false discovery rate using the procedure of Benjamini & Hochberg (1995). To highlight the candidate genes in the Oocyte meiosis pathway (tgu04114) we used Cytoscape 3.3.0 (Shannon et al. 2003, Smoot et al. 2011).
Analysis of linkage disequilibrium
Genotypic linkage disequilibrium was assessed inside and outside of (i) islands of high differentiation and (ii) islands of high differentiation and divergence separately for the two nightingale species. We calculated the composite correlation coefficient (r2) between all pairs of SNPs within 1 Mb sliding windows using the vcftools software (Danecek et al. 2011). From these pairwise comparisons, we calculated the average value of r2 for each window. SNP positions were based on the zebra finch genome. Only SNPs having a minor allele frequency (MAF) higher than 0.15 were considered. Significance was assessed using Mann-Whitney U-tests. The analysis of linkage disequilibrium was done for the entire dataset including the Z chromosome as well as for the dataset excluding the Z chromosome. Results were nearly the same in both analyses and therefore we present results only for the entire dataset including the Z chromosome.
Patterns of molecular evolution
We conducted McDonald-Kreitman (MK) tests (McDonald & Kreitman 1991) and calculated neutrality indexes (NI; Rand & Kann 1996) inside and outside of (i) islands of high differentiation and (ii) islands of high differentiation and divergence (Supplemental Table 1). The MK test and the NI are based on comparisons of the amount of polymorphism within species and substitutions between species for synonymous and nonsynonymous mutations. Calculations were made separately for the two nightingale species. As the two nightingale species are closely related and there is only limited number of substitutions between them, we determined substitutions for each species relative to the outgroup, L. svecica. Sequences were then compared to two other more distantly related outgroups, collared flycatcher (Ficedula albicollis) and zebra finch, to assign substitutions either to L. svecica or to L. megarhynchos/L. luscinia lineage. In our analysis, we considered only those substitutions that occurred in the lineage to L. megarhynchos/L. luscinia. Analyses were performed for the entire dataset including the Z chromosome as well as for the dataset excluding the Z chromosome. Results were nearly identical for both datasets and therefore we present results only for the entire dataset including the Z chromosome.
To identify synonymous and nonsynonymous mutations we used the Exonerate program (Slater & Birney 2005) and custom Perl and Shell scripts. For gene prediction, we used protein sequences from the collared flycatcher and zebra finch obtained from Ensembl (Ver. 81, July 2015). Only protein coding sequences of genes exhibiting a one-to-one ortholog relationship between the zebra finch and flycatcher genomes were included (based on the Ensembl Compara database; Vilella et al. 2008, Herrero et al. 2016). For each gene only the best prediction of the two reference species was considered (based on the blast score provided by the Exonerate program). We also considered predictions for which only one species provided a match. Since the genomes of the flycatcher and zebra finch are fairly diverged, we aligned their coding sequences with the coding sequences created for individual nightingale samples. Regions of poor alignment containing gaps, unknown bases, stop codons or multiple substitutions/polymorphisms per codon were removed. The final dataset consisted of 4657 protein coding genes. The number of within-species polymorphisms associated with coding sequence was 3,186 for L. megarhynchos and 3,987 for L. luscinia. From these 588 were shared between the species. The number of substitutions was 3,505 for the L. megarhynchos lineage and 2,704 for the L. luscinia lineage. From these 2,129 were shared between the two lineages (i.e. occurred between the common ancestor of both nightingale species with L. svecica and the ancestor of L. megarhynchos and L. luscinia).
To compare patterns of molecular evolution inside and outside of (i) islands of high differentiation and (ii) islands of high differentiation and divergence, we constructed a generalized linear model (glm) with terms for the type of mutation (synonymous or nonsynonymous), type of variant (polymorphism or substitution), genomic location (inside or outside islands), and the three-way interaction between these terms. This three-way interaction tests for the difference in MK tests inside and outside islands. To assess its significance, we compared the full model containing this interaction to simpler model without this interaction using a Log-Likelihood ratio test (ΔLL). The analysis was carried out in R (R Core Team 2016).
Results
Patterns of differentiation and divergence along the genome
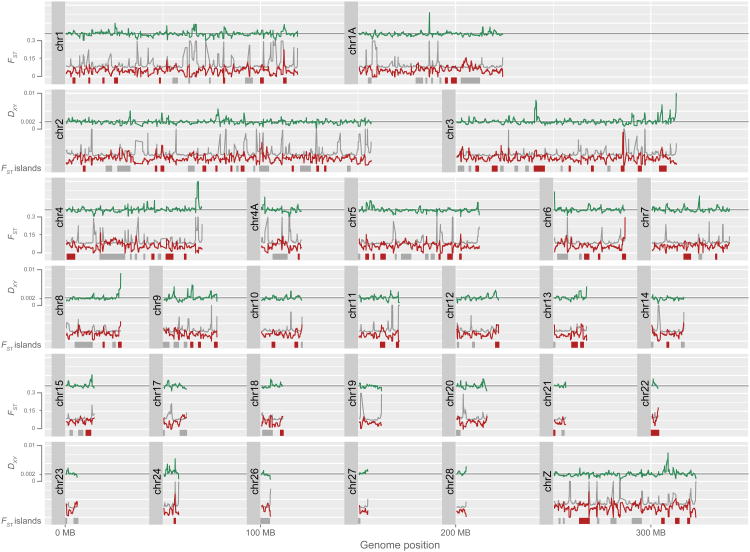
We found 136 islands of elevated differentiation (FST) covering ∼319 Mbp. The mean island size was ∼2.3 Mbp (maximum 13 Mb; minimum 1 Mbp) (Figure 1). Overall, the Z chromosome exhibited higher average FST values (0.088) than the autosomes (0.063). This is consistent with previous work that found lower levels of gene flow on the Z chromosome compared to the autosomes (average 2Nm was 0.422 on the autosomes and 0.018 on the Z chromosome; Storchová et al. 2010). However, we note that at equilibrium with respect to migration and drift FST-autosomes = 1/(4Nm + 1) and FST-Z chromosome = 1/(3Nm + 1), assuming an equal sex ratio and no sex-biased migration. The observed values of FST for the Z chromosome and for the autosomes are very close to these theoretical expectations, suggesting that these differences in FST can largely be explained by differences in the effective population size of the Z chromosome and autosomes. Within the Z chromosome, islands of high differentiation were not more common than on the autosomes. However, since the permutation tests were conducted separately for the Z chromosome and autosomes, Z-linked islands of differentiation were generally more differentiated than on the autosomes.
Figure 1.
FST and Dxy values along the chromosomes. FST is drawn by red line and Dxy by green line. Grey lines represent the genomic average for Dxy and bootstrap values for FST. Genomic position is based on the mapping of nightingale contigs to the zebra finch genome. Genomic islands with significantly elevated FST values are depicted as grey or red bars. Red bars indicate genomic islands of elevated FST with average Dxy higher than the genome average.
The average absolute divergence (Dxy) was slightly lower within islands of high differentiation (Dxy = 0.00210) than outside islands of high differentiation (Dxy = 0.00211), but the difference was not significant (p=0.6098; Welch Two Sample t-test). To identify genomic regions possibly involved in reproductive isolation, we calculated the average Dxy in islands of high differentiation, and we selected all islands of high differentiation having Dxy higher than the genomic average - we call these regions islands of high differentiation and divergence. There were 67 such islands harboring a total of 2007 genes.
Functional analysis of genes in islands of high differentiation and divergence
To better understand the nature of reproductive isolation between the nightingale species and to identify candidate speciation genes, we performed functional analyses of genes located in (i) islands of high differentiation and (ii) islands of high differentiation and divergence. We used KEGG pathways as a proxy for protein function and conducted gene set enrichment analyses. Altogether we found three KEGG pathways significantly enriched in islands of high differentiation after FDR correction (Supplemental Table 2A). These pathways included Metabolic pathway (tgu01100), Lysosome pathway (tgu04142) and Oocyte meiosis pathway (tgu04114). When we focused the analysis on islands of high differentiation and divergence, no pathway was enriched after FDR correction. However, when we considered pathways significantly enriched in these islands without FDR correction (excluding pathways with fewer than 10 genes in islands to reduce the effect of false positives), we identified four enriched pathways (Supplemental Table 2B). Interestingly these included Metabolic pathway (tgu01100), Oocyte meiosis pathway (tgu04114), Lysosome pathway (tgu04142) and Purine metabolism pathway (tgu00230). We were further interested whether any of the three pathways that were significantly enriched in islands of high differentiation as well as (without FDR correction) in islands of both high differentiation and divergence were also enriched in islands of high differentiation and low divergence (i.e. with Dxy lower than the genomic average). We found that Metabolic and Lysosome pathways were significantly (without FDR correction) enriched in these islands. Interestingly, Oocyte meiosis pathway was not enriched in these islands (Supplemental Table 2C), suggesting that the enrichment of this pathway in islands of high differentiation is driven mainly by higher density of genes related to this pathway in islands of high differentiation and divergence.
There were 17 genes belonging to the oocyte meiosis pathway in islands of high differentiation and divergence (Table 1). These genes represent candidate hybrid female sterility genes and are located on several autosomes. None of them are located on the Z chromosome. We also looked at their position in the oocyte meiosis pathway (Supplemental Figure 1). The genes function at different stages of oogenesis from prophase I of meiosis I until meiosis II.
Table 1.
List of seventeen genes of the Oocyte meiosis pathway that were found in the genomics islands of high differentiation and divergence. Location of these genes in the Oocyte meiosis pathway is indicated (see also Supplementary Figure 1). Gene information was retrieved using the Ensembl BioMart, Ensembl Version 81, July 2015. Physical location of genes in Zebra finch genome is provided.
| Ensembl Gene ID | Gene name | Gene description | Location in the pathway | Chr | Gene Start | Gene End | Strand |
|---|---|---|---|---|---|---|---|
| ENSTGUG00000009440 | CDC16 | cell division cycle 16 | APC/C-Cdc20 complex | 1 | 26694321 | 26715128 | + |
| ENSTGUG00000007142 | ADCY1 | adenylate cyclase 1 | AC | 2 | 77509308 | 77671457 | + |
| ENSTGUG00000011936 | CCNE2 | cyclin E2 | CycE-Cdk2 complex | 2 | 132836273 | 132856217 | - |
| ENSTGUG00000004938 | PPP2R5D | protein phosphatase 2 regulatory subunit B'delta | PP2A-B56 phosphatase | 3 | 19981662 | 20006850 | - |
| ENSTGUG00000005486 | SPDYA | speedy/RINGO cell cycle regulator family member A | Ringo-Cdc2 | 3 | 21135408 | 21139927 | - |
| ENSTGUG00000001762 | PPP2CB | protein phosphatase 2 catalytic subunit beta | PP2A, PP2A-B56 phosphatase | 4 | 2417725 | 2421587 | - |
| ENSTGUG00000001917 | MAD2L1 | mitotic arrest deficient 2 like 1 | Mad1/2 | 4 | 4749027 | 4751057 | - |
| ENSTGUG00000009328 | ANAPC4 | anaphase promoting complex subunit 4 | APC/C-Cdc20 complex | 4 | 52684835 | 52701909 | - |
| ENSTGUG00000009326 | INS | insulin | INS | 5 | 13622537 | 13625762 | + |
| ENSTGUG00000005982 | CCNB2 | cyclin B2 | cyclinB2/B5-Cdc2 complex (MPF) | 10 | 6469106 | 6472886 | - |
| ENSTGUG00000009363 | MAP2K1 | mitogen-activated protein kinase kinase 1 | MEK | 10 | 19125593 | 19183532 | + |
| ENSTGUG00000008504 | ADCY7 | adenylate cyclase 7 | AC | 11 | 12630250 | 12660343 | - |
| ENSTGUG00000001287 | PPP2CA | protein phosphatase 2 catalytic subunit alpha | PP2A, PP2A-B56 phosphatase | 13 | 10125175 | 10140010 | + |
| ENSTGUG00000001291 | SKP1 | S-phase kinase associated protein 1 | SCF-BTRC complex | 13 | 10154467 | 10160268 | + |
| ENSTGUG00000001417 | PTTG1 | pituitary tumor-transforming 1 | Securin | 13 | 11608459 | 11612472 | + |
| ENSTGUG00000002334 | MAD2L2 | Taeniopygia guttata MAD2 homolog (LOC100226442) | Mad1/2 | 21 | 907423 | 914246 | - |
| ENSTGUG00000003464 | CAMK2B | calcium/calmodulin dependent protein kinase II beta | CamKII | 22 | 112126 | 120186 | + |
Linkage disequilibrium and patterns of molecular evolution in the islands of high differentiation and divergence
We estimated linkage disequilibrium and performed MK tests for genomic regions inside and outside of (i) all islands of high differentiation and (ii) all islands of high differentiation and divergence separately in both nightingale species. In both species, linkage disequilibrium (measured as r2) was significantly higher inside than outside islands for both islands of high differentiation and islands of high differentiation and divergence (Table 2). All MK tests except one (L. luscinia, inside islands) were statistically significant (Supplementary Table 1). The NI revealed that the ratio of nonsynonymous to synonymous mutations was higher for polymorphisms than for substitutions. Importantly, when the amount of synonymous and nonsynonymous variation within and between species was compared inside and outside of islands using glm models (see Materials and Methods), we found a statistically significant difference in patterns of molecular evolution inside and outside of islands for all islands of high differentiation in both species (L. luscinia: Δ LL=-4.4178, p-value=0.03557; L. megarhynchos:Δ LL=-6.9914, p-value = 0.0082, Supplemental Table 1), but not for islands of high differentiation and divergence (Supplemental Table 1). Islands of high differentiation were characterized by an excess of nonsynonymous polymorphisms relative to nonsynonymous substitutions (Supplemental Table 1).
Table 2.
Linkage disequilibrium (measured as r2) inside and outside of the (i) islands of high differentiation (FST) and (ii) islands of high differentiation and divergence (FST&Dxy) in both species (LL: L. luscinia, LM: L. megarhynchos). Difference between r2 inside and outside the islands was tested by Mann-Whitney U-test.
| Type of island | Species | Position | r2 | U | p-val |
|---|---|---|---|---|---|
| FST | LL | Inside | 0.1667 | ||
| Outside | 0.1406 | 27970 | 0.0327 | ||
| LM | Inside | 0.4032 | |||
| Outside | 0.2984 | 21977 | 0.0176 | ||
| FST&Dxy | LL | Inside | 0.1667 | ||
| Outside | 0.1432 | 26106 | 0.0256 | ||
| LM | Inside | 0.4145 | |||
| Outside | 0.2954 | 22142 | 0.0002 |
Discussion
We conducted a genome-wide survey of differentiation and divergence to study the genetic basis of reproductive isolation between L. megarhynchos and L. luscinia, two closely related species that diverged 1.8 Mya (Storchová et al. 2010) and are separated by incomplete prezygotic isolation and female-limited hybrid sterility. Using transcriptomic data from multiple individuals of both species, we identified 136 regions of elevated differentiation (FST) across the genome, 67 of which had absolute divergence (Dxy) higher than the genomic average. This subset of islands with high differentiation and divergence identifies candidate genomic regions involved in maintaining reproductive isolation between the nightingale species. To identify specific candidate genes and to gain insight into the molecular mechanisms underlying reproductive isolation, we analyzed gene content and studied patterns of molecular evolution in these regions.
Three pathways (Metabolic pathway, Lysosome pathway and Oocyte meiosis pathway) were significantly enriched in islands of high differentiation (Supplemental Table 2). The same three pathways together with the Purine metabolism pathway were also enriched in islands of high differentiation and divergence, although in this case the enrichment was not significant after FDR correction for multiple comparisons. Intriguingly, the Oocyte meiosis pathway could be related to female sterility, which is the main reproductive barrier observed between the nightingale species. We identified 17 genes located in islands of high differentiation and divergence that were associated with the Oocyte meiosis pathway (Table 1). These are among the first candidate genes for female-limited hybrid sterility in taxa with heterogametic females.
Some of the candidate genes operate in the early stages of meiosis. This agrees with our current understanding of hybrid female sterility in nightingales. Experimental crosses of both species in captivity (Stadie 1991) as well as rare observations of hybrid females in nature (Reifová et al. 2011a) show that hybrid females do not lay eggs, suggesting that sterility may be caused by an early meiotic arrest. In addition, hybrid females show impaired sexual behaviour and a reduced tendency to mate which might indicate reduced levels of sex hormones produced by the ovaries (Stadie 1991, Reifová et al. 2011a). More detailed analysis of the ovaries of F1 females is needed to better understand the underlying mechanisms of hybrid female sterility in these species.
The sex chromosomes are expected to play a crucial role in hybrid female sterility and Haldane's Rule (Presgraves 2008). We previously showed that levels of between-species differentiation are higher and that levels of interspecific gene flow are lower on the Z chromosome compared to the autosomes (Storchová et al. 2010). The present results are consistent in showing higher average levels of differentiation on the Z chromosome compared to the autosomes. However, islands of high differentiation were not markedly enriched on the Z chromosome and none of the candidate genes for hybrid female sterility was located on the Z chromosome. It is difficult to draw strong inferences from this observation, since it likely reflects the conservative nature of the analysis in which significance for each genomic region on the Z chromosome was assessed only in relation to the distribution of regions within this chromosome. Since the Z chromosome showed higher levels of differentiation overall, it was more difficult to detect significant outliers. Direct mapping experiments would be useful for determining whether QTL for female sterility are located on the Z chromosome.
The two nightingale species diverged 1.8 Mya and although hybrid female sterility is an important reproductive barrier separating the two nightingale species, there are also other forms of reproductive isolation. Competition between the two species has resulted in partial habitat segregation (Reif et al. 2018) and ecological character displacement in bill size (Reifová et al. 2011b), which might enhance prezygotic as well as extrinsic postzygotic isolation in sympatric populations. There is also evidence for marked differences in sperm length between the species, which might contribute to postmating prezygotic isolation (Albrecht et al. unpublished results). In addition, the two nightingale species differ in breeding distribution; L. megarhynchos has more southern breeding distribution, while L. luscinia is distributed more northerly. In principle, this could be associated with differences in metabolic rates between the two species and could result in disrupted metabolism in hybrids (Qvarnström et al. 2016) as has been demonstrated in two flycatcher species (McFarlane et al. 2016). Possible adaptation to different climates in the two nightingale species could provide an explanation for why the Metabolic pathway is enriched in islands of high differentiation. The Lysosome pathway could be also related to different climatic adaptations as it is involved in energy metabolism (Settembre et al. 2013) and in fact there is some overlap between the Metabolic and Lysosome pathways.
Analysis of linkage disequilibrium revealed significantly higher levels of linkage disequilibrium inside than outside islands of high differentiation as well as islands of high differentiation and divergence in both nightingale species. In addition, islands of high differentiation were characterized by a significant excess of nonsynonymous polymorphism relative to nonsynonymous substitutions in both species. This suggests that purifying selection might be less efficient in these regions, although it should be noted that differences in linkage disequilibrium and effective population size in these islands could bias the results of MK tests (Eyre-Walker 2002). Nevertheless, together, these observations suggest that some loci involved in reproductive isolation may lie in regions of locally reduced recombination. In such regions selection at linked sites can lead to a locally reduced effective population size and less efficient purifying selection (Maynard Smith & Haigh 1974; Charlesworth et al. 1993). This observation is consistent with theory that predicts an association between regions of low recombination and regions contributing to reproductive isolation (Butlin 2005; Nachman & Payseur 2012). It is also consistent with recent empirical studies in other species (Carneiro et al. 2009; Geraldes et al. 2011; Janoušek et al. 2015), suggesting that variation in recombination rate plays an important role in the evolution of reproductive isolation.
Our results together with previous studies (Delmore et al. 2015; Malinsky et al. 2015) demonstrate that analysis of the genomic landscape of differentiation and divergence is a promising approach for identifying genomic regions and candidate genes involved in speciation. The two nightingale species represent an especially suitable model system for this approach. These species diverged a relatively long time ago (1.8 Mya, Storchová et al. 2010) yet show relatively high post-divergence interspecific gene flow (average 2Nm on autosomes is 0.422, Storchová et al. 2010). This situation facilitates the identification of islands of high differentiation and divergence. Nightingales also provide a useful model system for studying mechanisms that underlie female-limited hybrid sterility. Further studies, including analysis of the ovary transcriptome of both nightingale species as well as hybrids, together with a detailed inspection of the ovaries of hybrid females, will shed more insight into female-limited hybrid sterility in organisms with heterogametic females.
Supplementary Material
Acknowledgments
We thank late Marcin Antczak for the assistance in the field and Megan Phifer-Rixey for useful discussion. The study was supported by a junior grant of the Czech Science Foundation (15-10884Y) to RR and an NIH grant (RO1 GM074245) to MWN. This work was also supported by ELIXIR CZ research infrastructure project (MEYS Grant No: LM2015047) including access to computing and storage facilities. We are grateful for financial support from the Czech Academy of Sciences (grant no. RVO 67985904) and the Ministry of Education, Youth and Sports of the Czech Republic (grant no. EXCELLENCE CZ.02.1.01/0.0/0.0/15_003/0000460 OP RDE).
Footnotes
Data Accessibility: DNA sequences, demultiplexed FASTQ files, and accompanying sample identification are available from Dryad repository: doi:10.5061/dryad.2p4t3, doi:10.5061/dryad.41ng6. The analysis scripts are publicly available (https://github.com/libor-m/islands-paper).
Author Contributions: RR, LM and MWN designed the study. RR and JRe collected samples, LC and KJ prepared samples for sequencing, JRi sequenced samples, LM, VJ and JP analyzed data. RR, LM, VJ and MWN wrote the paper with contribution of all coauthors.
References
- Alund M, Immler S, Rice AM, Qvarnström A. Low fertility of wild hybrid male flycatchers despite recent divergence. Biology Letters. 2013;9:20130169. doi: 10.1098/rsbl.2013.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Karaiskou N, Leder EH, et al. A gene-based genetic linkage map of the collared flycatcher (Ficedula albicollis) reveals extensive synteny and gene-order conservation during 100 million years of avian evolution. Genetics. 2008;179:1479–1495. doi: 10.1534/genetics.108.088195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bhattacharyya T, Reifová R, Gregorová S, et al. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genetics. 2014;10:e1004088. doi: 10.1371/journal.pgen.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri R. Interpreting differentiation landscapes in the light of long-term linked selection. Evolution Letters. 2017;1:118–131. [Google Scholar]
- Burri R, Nater A, Kawakami T, et al. Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Research. 2015;25:1656–1665. doi: 10.1101/gr.196485.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK. Recombination and speciation. Molecular Ecology. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Ferrand N, Nachman MW. Recombination and speciation: loci near centromeres are more differentiated than loci near telomeres between subspecies of the European rabbit (Oryctolagus cuniculus) Genetics. 2009;181:593–606. doi: 10.1534/genetics.108.096826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Molecular Biology and Evolution. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Cruickshank TE, Hahn MW. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology. 2014;23:3133–3157. doi: 10.1111/mec.12796. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore KE, Hübner S, Kane NC, et al. Genomic analysis of a migratory divide reveals candidate genes for migration and implicates selective sweeps in generating islands of differentiation. Molecular Ecology. 2015;24:1873–1888. doi: 10.1111/mec.13150. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- Eroukhmanoff F, Rowe M, Cramer ER, et al. Experimental evidence for ovarian hypofunction in sparrow hybrids. Avian Research. 2016;7:3. [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelter HP, Tegelström H, Gustafsson L. Evidence from hatching success and DNA fingerprinting for the fertility of hybrid Pied × Collared Flycatchers Ficedula hypoleuca × albicollis. Ibis. 1992;134:62–68. [Google Scholar]
- Geraldes A, Basset P, Smith KL, Nachman MW. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Molecular Ecology. 2011;20:4722–4736. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genetics. 2010;6:e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Sex-ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Research. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, et al. Ensembl comparative genomics resources. Database (Oxford) 2016:bav096. doi: 10.1093/database/bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušek V, Munclinger P, Wang L, Teeter KC, Tucker PK. Functional organization of the genome may shape the species boundary in the house mouse. Molecular Biology and Evolution. 2015;32:1208–1220. doi: 10.1093/molbev/msv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. The global diversity of birds in space and time. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, Linares M, Naisbit RE, et al. Sex-linked hybrid sterility in a butterfly. Evolution. 2001;55:1631–1638. doi: 10.1111/j.0014-3820.2001.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Backström N, Burri R, et al. Estimation of linkage disequilibrium and interspecific gene flow in Ficedula flycatchers by a newly developed 50k single-nucleotide polymorphism array. Molecular Ecology Resources. 2014;14:1248–1260. doi: 10.1111/1755-0998.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Challis RJ, Tyers AM, et al. Genomic islands of speciation separate cichlid ecomorphs in an East African crater lake. Science. 2015;350:1493–1498. doi: 10.1126/science.aac9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biology. 2007;5:e243. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genetics Research. 1974;23:23–35. [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McFarlane SE, Sirkiä PM, Ålund M, Qvarnström A. Hybrid dysfunction expressed as elevated metabolic rate in male Ficedula Flycatchers. PLoS One. 2016;11:e0161547. doi: 10.1371/journal.pone.0161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlček C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Mořkovský L, Pačes J, Rídl J, Reifová R. Scrimer: designing primers from transcriptome data. Molecular Ecology Resources. 2015;15:1415–1420. doi: 10.1111/1755-0998.12403. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Payseur BA. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:409–421. doi: 10.1098/rstb.2011.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbit RE, Jiggins CD, Linares M, Salazar C, Mallet J. Hybrid sterility, Haldane's rule and speciation in Heliconius cydno and H. melpomene. Genetics. 2002;161:1517–1526. doi: 10.1093/genetics/161.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press; New York, NY: 1987. [Google Scholar]
- Noor MA, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bomhoff M, Nachman MW. Genome-wide patterns of differentiation among house mouse subspecies. Genetics. 2014;198:283–297. doi: 10.1534/genetics.114.166827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends in Genetics. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. The molecular evolutionary basis of species formation. Nature Reviews Genetics. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- Qvarnström A, Ålund M, McFarlane SE, Sirkiä PM. Climate adaptation and speciation: particular focus on reproductive barriers in Ficedula flycatchers. Evolutionary Applications. 2016;9:119–34. doi: 10.1111/eva.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Molecular Biology and Evolution. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Reif J, Jiran M, Reifová R, et al. Interspecific territoriality in two songbird species: potential role of song convergence in male aggressive interactions. Animal Behaviour. 2015;104:131–136. [Google Scholar]
- Reif J, Reifová R, Skoracka A, Kuczyński L. Competition-driven niche segregation on a landscape scale: evidence for escaping from syntopy toward allotopy in two coexisting sibling passerine species. Journal of Animal Ecology. 2018 doi: 10.1111/1365-2656.12808. in press. [DOI] [PubMed] [Google Scholar]
- Reifová R, Kverek P, Reif J. The first record of a female hybrid between the Common Nightingale (Luscinia megarhynchos) and the Thrush Nightingale (Luscinia luscinia) in nature. Journal of Ornithology. 2011a;152:1063–1068. [Google Scholar]
- Reifová R, Reif J, Antczak M, Nachman MW. Ecological character displacement in the face of gene flow: evidence from two species of nightingales. BMC Evolutionary Biology. 2011b;11:138. doi: 10.1186/1471-2148-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sætre GP, Borge T, Lindroos K, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proceedings of the Royal Society B: Biological Sciences. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, et al. Genomics and the origin of species. Nature Reviews Genetics. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nature Reviews Molecular Cell Biology. 2013;14:283–96. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagin DA, Rebrikov DV, Kozhemyako VB, et al. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Research. 2002;12:1935–1942. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadie C. Europaische Vogelwelt. Vol. 3. Sonderheft; 1991. Erdsanger I; Nachtigall und Sprosser; pp. 130–189. [Google Scholar]
- Storchová R, Gregorová S, Buckiová D, et al. Genetic analysis of X-linked hybrid sterility in the house mouse. Mammalian Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Storchová R, Reif J, Nachman MW. Female heterogamety and speciation: reduced introgression of the Z chromosome between two species of nightingales. Evolution. 2010;64:456–471. doi: 10.1111/j.1558-5646.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biology. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, West J. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Molecular Ecology. 2008;17:4334–4345. doi: 10.1111/j.1365-294X.2008.03921.x. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, et al. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Research. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokurková J, Petrusková T, Reifová R, et al. The causes and evolutionary consequences of mixed singing in two hybridizing songbird species (Luscinia spp.) PLoS One. 2013;8:e60172. doi: 10.1371/journal.pone.0060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Ellegren H. Making sense of genomic islands of differentiation in light of speciation. Nature Reviews Genetics. 2017;18:87–100. doi: 10.1038/nrg.2016.133. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. Journal of Evolutionary Biology. 2001;14:851–865. [Google Scholar]
- Yates A, Akanni W, Amode MR, et al. Ensembl 2016. Nucleic Acids Research. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Sun T, Woldesellassie F, Xiao H, Tao Y. Sex ratio meiotic drive as a plausible evolutionary mechanism for hybrid male sterility. PLoS Genetics. 2015;11:e1005073. doi: 10.1371/journal.pgen.1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30:892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- Zhulidov PA, Bogdanova EA, Shcheglov AS, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Research. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.