Abstract
Purpose of review
The long-lived HIV reservoir remains a major obstacle for a HIV cure. Current techniques to analyze this reservoir are generally population-based. We highlight recent developments in methods visualizing HIV, which offer a different, complementary view; and provide indespensable information for cure strategy development.
Recent findings
Recent advances in fluoresence in situ hybridization techniques enabled key developments in reservoir visualization. Flow cytometric detection of HIV mRNAs, concurrently with proteins, provides a high-throughput approach to study the reservoir on a single-cell level. On a tissue-level, key spatial information can be obtained detecting viral RNA and DNA in situ by fluorescence microscopy. At total-body level, advancements in non-invasive immuno-positron emission tomography (PET) detection of HIV proteins may allow an encompassing view of HIV reservoir sites.
Summary
HIV imaging approaches provide important, complementary, information regarding the size, phenotype and localization of the HIV reservoir. Visualizing the reservoir may contribute to the design, assessment and monitoring of HIV cure strategies in vitro and in vivo.
Keywords: HIV cure, in situ hybridization (ISH), flow cytometry, microscopy, RNA flow, viral reservoir, positron-emission tomography (PET)
Introduction
Despite the tremendous success of antiretroviral therapy (ART), HIV persists in infected subjects predominantly in rare latently-infected CD4+ T cells [1–3]. Life-long ART is necessary, as treatment interruption would allow virus to rebound from this pool of cells and plasma viral load to return to pre-ART levels [4]. Broadly, a HIV cure may be achieved either by complete eradication of the HIV reservoir from the body (known as a sterilizing cure) or a long-term control of HIV replication without ART (functional cure) [5]. Recent cure strategies have mainly focused on the “shock/kick and kill” strategy whereby HIV is reactivated from its latent state using latency-reversing agents (LRAs). This is followed by the death of the infected cell, due to the cytopathic effects of HIV itself or killing by the host’s own immune system. However, despite initial promise, LRA-based strategies have shown limited success in clinical trials [7], suggesting that additional information regarding the HIV reservoir and alternative approaches are required to help rationally design effective cure strategies.
In this context a more detailed understanding of such viral reservoirs is required. Firstly, a better understanding of the distribution of the HIV reservoir at both the cellular and anatomic level is essential. This will help guide treatment agents that can efficiently target the various cell types and penetrate the different organs that harbor HIV. Secondly, although a substantial fraction of CD4+ T cells of HIV-infected subjects on ART harbor integrated HIV DNA (~600 copies/million resting CD4+ T cells [8]), the majority of integrated proviral sequences (~90–95%) are defective and not able to produce infectious viral particles (defined as replication-competent virus) [9,10]. Thus, the specific detection of the HIV-infected cells containing a provirus that is able to transcribe viral RNA (vRNA, transcription-competent reservoir), translate proteins (translation-competent reservoir) and/or is replication-competent is of critical importance. Lastly, potential cure therapies need to be monitored for their effectiveness in decreasing the size of the viral reservoir in vitro and in vivo. Therefore, the size of these HIV reservoirs need to be evaluated longitudinally by techniques that are scalable in the context of clinical trials for cure strategies.
Looking beyond classical reservoir measures
Multiple techniques have been developed to quantify and characterize the HIV reservoir. PCR-based approaches represent simple and rapid methods to measure, for example, the total and integrated forms of HIV DNA [11–14], and have been widely used to study both circulating cellular populations and also disrupted tissue biopsies [15,16]. Such techniques have identified specific anatomic locations, including the gut-associated lymphoid tissues, and cell populations, such as transitional memory (TTM) and central memory (TCM) CD4+ T cells, as preferential sites of HIV integration [15,16]. In contrast to measures of integated DNA, inducible infectious virus was detected after reactivation from the TCM, but rarely the TTM, by the quantitative viral outgrowth assay (qVOA) [17]. While further sudies are necessary, these apparent discrepancies may be explained by the nature of the reservoir measured by these different techniques. Assays quantifying HIV DNA, such as PCR, can not discriminate between intact or defective proviral sequences and, therefore, may vastly overestimate the size of the replication-competent reservoir in these populations [18]. At the other end of the scale, results obtained using qVOA represents a minimal estimate, as multiple rounds of stimulations might be needed for reactivation [9] and replication-competent viruses in vivo may not be able to spread in vitro. For the evaluation of cure strategies, these techniques could thus either mask an achieved cure by detecting defective HIV sequences, or miss replication-competent reservoirs that could be reactivated in vivo, but may not respond to stimulation in vitro. These subtleties illustrate the need for tools that can identify and distinguish different forms of HIV reservoirs to better estimate replication-competence.
To determine the contribution of different cell populations to the reservoir, the techniques described above require cell suspensions to be first sorted, based on phenotypic markers, before analysis. The measured HIV reservoir size therefore represents a population-level value; it does not provide information about the heterogeneity within the studied cell pool and the specific anatomic location. Furthermore, this requirement for sorting limits the number of populations that can be analyzed concurrently and is not practical for rare subsets. Therefore, approaches that can detect and characterize the HIV reservoir at a single-cell level, either in heterogeneous, mixed cell suspensions or in situ, are required.
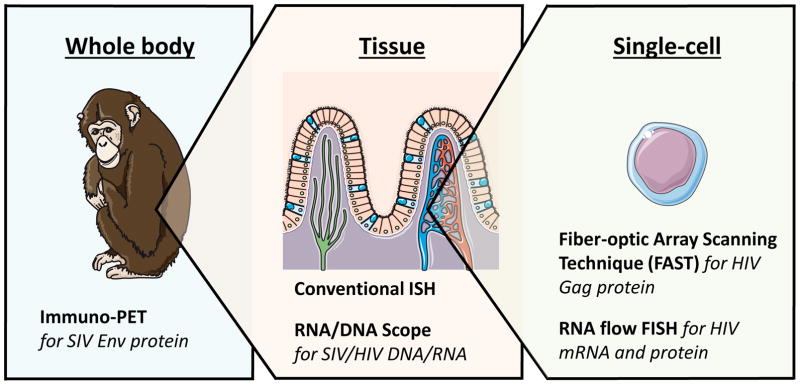
Given these caveats, imaging techniques, including flow cytometric and microscopic approaches, have been pursued as powerful strategies for the detection and characterization of HIV reservoirs in the context of cure research. Here, we detail techniques that allow the visualization of HIV at the proviral DNA, RNA or protein level in single cells, individual tissues or throughout the body (see Figure 1 and Table 1 for overview). Each of these approaches provide different and often complementary information on the size, type and distribution of the viral reservoir. Much of the work discussed here illustrates the study of baseline expression of vRNAs and proteins in infected cells during ART without any further stimulation, in vitro or in vivo. Although this has revealed important information about phenotype and localization of transcription- and translation-competent reservoir cells that are spontaneously producing viral products in the absence of stimulation, this provides an understanding of only one aspect of the reservoir. Further studies must build upon this work to investigate latently infected cells, which are transcriptionally/translationally silent but are able to produce infectious viral particles upon stimulation. Understanding the reservoir on this level will help develop, evaluate and monitor HIV cure strategies in vitro and in vivo.
Figure 1.
Schematic representation of levels of HIV visualization visualization including corresponding assays
Table 1.
Overview of HIV imaging techniques
| Level | Assay | Detection of | Assay overview | Advantages | Limitations | Key references |
|---|---|---|---|---|---|---|
| Single-cell | Fiber-optic Array Scanning Technology (FAST) | HIV Gag protein | Microscopic detection of HIV Gag protein combined with CD4 downregulation, a cellular hallmark of HIV infection | Relatively high-throughput | Limited characterization possible, limited access to technology | [21] |
| RNA flow FISH | HIV RNA (with or without HIV Gag protein) | Flow cytometric detection of RNA using multiple probes for hybridization followed by signal amplification with branched DNA amplifiers | High-throughput, multi-parametric characterization, flexible and easily adaptable | Single-cell suspension required, large cell numbers requirement, 2–3 day assay | [19,22,37,38] | |
| Tissue | Convention al in situ hybridization | HIV/SI V RNA or DNA | Microscopic detection of RNA using radiolabeled probes for hybridization or enzymatic approaches, detection of DNA using prior sequence amplification (in situ PCR) | Detection and localization of HIV RNA/DNA in anatomically intact tissue sections | Long assay duration | [46,52] |
| DNA/RNA Scope | Microscopic detection of using multiple probes for hybridization followed by signal amplification with branched DNA amplifiers RNA/DNA | Short assay duration, concurrent detection of HIV/SIV RNA and DNA | Limited multi-parametric characterization of reservoir | [50,51] | ||
| Whole body | Immuno-PET (positron-emission-tomography) | SIV Env protein | Detection of Env expression using radio-nucleotide-labeled gp120-specific antibodies followed by PET analysis | Non-invasive, in vivo imaging of SIV protein | Reduced sensitivity, bias towards tissues with higher antibody penetration | [61] |
Visualization of HIV at the single-cell level
The detection of viral proteins, such as Gag, by specific antibody staining followed by flow cytometric analysis has been a valuable tool to study HIV infection at the single-cell level in vitro, where the frequencies of cells actively translating HIV protein is high. In contrast, HIV-infected cells in virally-suppressed ART-treated subjects are exceedingly rare (minimal estimated frequency of 1 replication-competent cell in a million resting CD4+ T cells by qVOA [8]). Due to a high false positive rate, Gag protein staining alone can not be used to accurately quantify the translation-competent reservoir at frequencies lower than 1000 cells per million, masking the true HIV-infected cell pool [19]. Indeed, proviral HIV DNA was detected in only 10% of HIV Gag protein+ cells sorted from ART-treated subjects [20]. Therefore, new approaches have been developed recently that combine the detection of Gag protein with a second marker for HIV infection, and thus can overcome this false-positive rate [19,21,22].
One such approach combines the detection of cells expressing Gag protein with the downregulation of CD4, a well-characterized feature of HIV infection [21]. For this approach, the O’Doherty lab used a microscopy technique originally developed for the detection of rare circulating tumor cells [23,24] known as FAST (Fiber-optic Array Scanning Technology). Briefly, cells isolated from ART-treated HIV-infected individuals and analysed directly ex vivo without stimulation, are stained with antibodies, placed on microscopy slides and automatically laser scanned for Gag expression. Gag-expressing cells are then re-imaged by automated digital microscopy for CD4 internalization. This two step process limits the false positive detection rate [21], and enabled the quantification of rare circulating cells (0.33 to 2.7 cells per million PBMCs) spontaneously producing Gag protein in the periphery of HIV-infected donors on ART [21]. Further work is required to determine if this approach can be applied to the quantification of latent translation-competent HIV reservoir cells after LRA treatment in vitro and in vivo.
As CD4 downregulation requires the function of viral proteins Nef, Vpu and Env [21,25–30] the loss of surface CD4 provides indirect information about the integrity of the infecting proviruses. Thus, this approach quantifies infected cells that are able to translate multiple HIV proteins. We suggest that such cells are substantially more likely to contain intact replication-competent proviruses, when compared to cells identified based on detection of integrated HIV DNA only. Therefore, results obtained with this technique will be - despite a slight overestimation - a closer estimate to the true HIV reservoir size. While powerful, this approach enables only a limited characterization of reservoir cells as a maximum of four markers (in addition to Gag and CD4) can be analyzed. Furthermore, access to the FAST microscopy technology is limited, which constrains the use of this technique.
Building on this concept of single-cell detection of the reservoir, flow cytometry-based approaches also allow single-cell analysis. However, the technology has the additional advantages of being widely available and of further enabling high-throughput, multi-parametric characterization of the reservoir. One such flow cytometry-based approach utilizes Gag protein staining combined with the detection of vRNA by RNA flow FISH (fluorescent in situ hybridization) [19,22]. Original versions of this approach were able to identify cells containing HIV mRNAs by flow cytometry for the first time, but were limited by high false-positive detection rates [31–34]. A more recent iteration, developed by our group in collaboration with Affymetrix (commercially available as the PrimeFlow™ assay from ThermoFisher) attempts to overcome this by using multiple oligomeric probes that hybridize in pairs to the target RNA of interest [35]. Only two adjacently bound probes can be subsequently recognized by the branched-DNA amplification system, which reduces the probability of off-target binding. Finally, this amplified, branched scaffold is labelled with fluorescent probes. This approach was previously developed for the detection of cellular RNAs [36], but is easily customizable for the identification of cells transcribing HIV mRNAs [19,22,37,38]. In studies by our laboratory, GagPol vRNA was targeted because it is highly abundant within an infected cell and its sequence is relatively well conserved between primary isolates [39]. Furthermore, its length enables binding of a high number of probe pairs, increasing the probability of vRNA detection despite possible sequence variation that might prevent some pairs from recognition. The flow-cytometric detection of GagPol vRNA+ cells allowed HIV transcription in response to latency reversal to be studied in in vitro infection models [37]. However, we and others found that the false-positive rate observed with GagPol probes precluded the identification of the low frequencies of events (<1000 events/million), that would be predicted in clinical samples [19,37]. In an attempt to overcome this, a recent study utilized a different probe set, with reported higher specificity than previously used sets [38]; nevertheless, false-positive GagPol vRNA+ events were still identified in HIV-negative individuals. Therefore, in this study, the frequency of false-positive events detected in HIV-negative individuals was subtracted from the total number of GagPol RNA+ events detected in HIV-infected subjects [38]. Although this provided an estimate of the true frequency of the transcription-competent cells - assuming a constant background rate throughout all HIV-negative and HIV-infected donors tested - this approach limits the characterization of the infected cell population, due to the contamination with false-positive events.
As an alternative approach, we and others combined the detection of both HIV GagPol RNA and Gag protein. This dual-staining reduced the number of false-positive events to 1 HIV GagPol RNA+/Gag protein+ (HIVRNA+/Gag+) cell in 8 million total analyzed CD4+ T cells from HIV-uninfected donors [19]. The detection of HIVRNA+/Gag+ cells was linear and specific down to 0.5–1 events per million CD4+ T cells and therefore did not require mathematical corrections in our hands [19,22]. This strategy allowed the detection of rare circulating HIVRNA+/Gag+ CD4+ T cells in samples from untreated and ART-treated HIV-infected donors without in vitro stimulation, but also enabled the study of both the size and cellular distribution of the latent, reactivated translation-competent reservoir in in vitro-stimulated CD4+ T cells from HIV-infected subjects on ART [19]. Following in vitro PMA/Ionomycin stimulation, ~4.7 HIVRNA+/Gag+ per million CD4+ T cells were detected. This frequency is 2 log-fold lower than the reservoir size measured by PCR for HIV DNA in the same cohort, presumably as cells containing a provirus deficient for GagPol transcription and Gag translation would not be detected in the HIVRNA/Gag assay [19]. However, it should be noted that the cellular reservoirs detected with the HIVRNA/Gag assay could include cells containing a provirus with point mutations/deletions in Gag that do not prevent translation, or defects in other viral proteins. Nevertheless, despite this possible overestimation, the frequency of HIVRNA+/Gag+ cells was only slightly higher than that compared to the minimal estimate obtained with qVOA (~1.4 infectious units/million CD4+ T cells detected in the same cohort, a 3.6 fold difference) [19]. Thus, the detection of the translation-competent reservoir through detection of both vRNA and HIV proteins, provides an alternative approach to narrow down estimates of the true size of the reservoir.
Importantly, as these approaches are flow cytometry based, they can be used to identify and phenotype in detail single cells supporting viral replication or containing a translation-competent provirus that respond to LRA-stimulation in vitro in clinical samples. Flow-based techniques were used to assess the ability of well-known LRAs such as romidepsin [38] (a histone deacetylase inhibitor [40]) and bryostatin and ingenol [19] (both protein kinase C (PKC) agonists [41,42]) to reactivate the latent HIV reservoir in vitro. While both PKC agonists reactivated proviruses in similar frequency of CD4+ T cells, and in all donors tested, ingenol reactivated proviruses mainly in TCM/TTM CD4+ cells, while the majority of bryostatin-reactivated cells had a TEM phenotype. This suggests that cellular subsets might respond differently to distinct LRAs [19]. Further studies are required to investigate whether combinations of multiple LRAs will be potent and broad enough in specificity to reactivate sufficient fraction of the latent HIV reservoir to make a cure achievable.
As the RNA flow FISH assay is highly adaptable to other viral RNAs, it provides a powerful tool to further study cure strategies at the single-cell level, in both SIV-infected non-human primate models and HIV-infected subjects. Notably, single-cell studies studies for viral reactivation thus far have focused on CD4+ T cells from peripheral blood. As the frequency of infected cells is thought to be higher in tissue [43], FAST and RNA flow FISH will be important tools for the evaluation of tissue reservoirs. Importantly, such studies will indicate if results obtained from peripheral blood CD4 T cells mirror latency reversal mechanisms in tissues. One limitation however is that for both of these approaches, samples need to be disrupted to obtain a single cell suspension for analysis. Although this allows high-throughput investigations, important spatial information related to the anatomic localization of the viral reservoir within the tissue is lost. Thus, in situ microscopic techniques could act as complementary tools to comprehensively characterize tissue reservoir.
Visualization of HIV at the tissue level
Given these points, classical in situ-hybridization (ISH) methods have been adapted to detect HIV or SIV in anatomically intact tissue samples from infected subjects or non-human primate models respectively, as the access to human tissue samples is limited [44–46]. vRNA sequences are targeted by specific probes that can be radiolabeled for visualization. Alternatively, probes can be labeled with biotin or digoxygenin, and detected with a fluorescent or enzymatic approach to avoid the requirement for radioactive material [45]. However, these chromogenic detection methods have shown a lower sensitivity than radiolabeled probes. Combining this method with immunofluorescence or immunohistochemistry staining for cellular markers has allowed the phenotypic and spatial analysis of HIV/SIV-infected cells at the single-cell level directly within the tissue. Such studies thus contributed to important findings related to viral tissue dissemination, such as the spread and establishment of productive infection from vaginal tissue to lymph node over time during early infection [47], and identified principal anatomic sites of high-level replication such as the lymphoid tissue and gut [44,48,49]. Crucially the majority of this work has been performed in the context of untreated viral infection.
During long-term ART the frequency of actively transcribing vRNA+ cells in the lymphoid tissue is dramatically reduced compared with untreated infection (for example by ~99% in lymphoid tissue after 6 months of ART) [46,50,51]. Nevertheless, rare vRNA+ cells can still be detected in tissue from ART-treated donors by ISH; such ongoing viral transcription has been correlated with low antiretroviral drug concentrations in these sites [51,52]. In addition to the classical CD4 T cell reservoir, in lymphoid tissues a proportion of these vRNA+ cells may represent virus persisting in different cell populations [52]. Indeed, during both untreated and ART-treated infection, trapped viral particles have been identified in the follicular dendritic cell (FDC) network [44,46]. However, it remained unclear whether the FDCs in these networks were infected, and therefore contained viral DNA, or were acting as stores of free virus. As standard ISH approaches were not sensitive enough for the detection of proviral HIV DNA, PCR amplification of the target DNA sequences directly on the tissue section was combined with hybridization of the amplified sequences using radiolabeled probes (PCR-in situ or PCR-ISH) [53]. Using this approach it was demonstrated that FDCs retain infectious virus but are not actually infected; they are positive for vRNA but negative for vDNA or spliced vRNA [44,54]. Such findings would have been missed with single-cell techniques analyzing disrupted tissue samples, due to the challenges of isolating FDCs from lymphoid tissue [55].
Due to the marked decrease in viral burden on ART, many tissue sections need to be screened in order to quantify rare infected cells. Conventional ISH assays, as described above, are time-intensive as the detection of low copy numbers requires long exposure times or prior sequence amplification, thus limiting the application of this approach to cure studies. Recently, the RNAScope assay was developed to overcome these limitations. This approach uses the same principle as the RNA flow FISH technique described previously based on the recognition of pairs of target probes by branched DNA amplifiers, which was indeed originally developed for microscopy [35]. Adapted for the detection of SIV vRNA, this assay showed a sensitivity and specificity comparable to the techniques described above, with a false-positive detection rate of 1 vRNA+ cell per million cells analyzed [50]. Importantly, the Scope technique enabled detection of single vRNA sequences in a short time and could also be used to reliably identify single vDNA sequences with improved spatial resolution and without prior sequence amplification. In addition, both vRNA and vDNA can be visualized concurrently [50]. It is important to note, however, that the detection of vDNA+ cells provides - similar to PCR methods described above - a maximal estimate of the HIV reservoir as defective sequences as well as unintegrated forms are included for detection. Regardless, this dual RNA/DNAscope technique confirmed the key role of the B cell follicles as a substantial tissue reservoir, due to the persistence of both HIV-infected mononuclear cells with or without active viral transcription and virus trapped in the FDC network [44,50,56]. These findings highlight the need for drugs that can efficiently penetrate tissue sites, but also demonstrates that alternative strategies may needed for the eradication of the virus trapped on FDCs in B cell follicles.
Although previous studies have focused on the detection of infected cells spontaneously transcribing vRNAs, Scope assays will be valuable tools to evaluate viral reactivation and eradication strategies in situ. Combined imaging of the viral reservoir together with virus-specific immune cells, for example using in situ tetramer staining techniques [57], could provide valuable information about the effectivity of both HIV reservoir targeting and immune system enhancing strategies in vivo. As confocal microscopy is limited by the number of parameters that can be analyzed concurrently, Scope techniques could be used in combination with histo-cytometry for multiparametric analysis without losing spatial information [58].
Visualization of HIV at the whole organ/body level
The RNA/DNA-Scope technique provides important spatial information regarding the intra-tissue localization of HIV/SIV reservoirs, but can also be used to infer total tissue viral burdens [51]. To achieve this, in a recent study, the frequency of vDNA+ or vRNA+ cells was determined per gram of analyzed tissue, then the number of total infected cells calculated from the weight of the analyzed organ [51]. This approach confirmed that lymphoid tissue as a main contributor to the whole body SIV reservoir during ART, but also identified transcription-competent reservoirs in other organs such as brain, lung and heart. Applying this strategy to HIV-infected donors on ART, this technique provided an estimate of ~105 vDNA+ cells/g lymphoid tissue and, based on the assumption that 5% of vDNA+ cells are replication-competent [9]: a total body burden of ~4×108 replication-competent HIV-infected cells [51]. However, with this type of analysis, preselection of specific tissues of interest and the potential for sampling error may lead to biases. Furthermore, the invasive nature of tissue analysis prevents longitudinal imaging of the same subject, for example during a course of treatment. With these caveats in mind, non-invasive full body imaging methods could provide an unbiased and more encompassing view of HIV reservoir sites in the body, despite a limited anatomical spatial resolution.
Initial iterations of this approach utilized animal models using infected with SIV- or HIV-constructs expressing reporter molecules detectable by body-scanning devices [59]. To apply this concept to HIV-infected humans and avoid the use of reporter viruses, the Villinger laboratory adapted a technique developed for the monitoring of cancer therapies [60] Whole-body Env expression was monitored in SIV-infected non-human primates, using radionucleotide-labeled SIV gp120-specific antibodies and detected by positron-emission-tomography (PET) [61]. This approach enabled viral dissemination to be followed in untreated SIV infection, and importantly identified previously unconsidered or underestimated sanctuary sites in ART treatment, such as the nasal lymphoid tissue and lung. While this approach has not yet been applied to HIV-infected humans, it’s potential application for the non-invasive monitoring of cure strategies is clear. However, further work is required to determine if the PET-based approach is sensitive enough to detect the lower frequencies of translation-competent viral reservoirs in the context of HIV and ART. Importantly, and as expected, the antibodies used were not able to breach the blood-brain-barrier; therefore this approach cannot be used to study an important HIV reservoir site [62].
Future directions
Recent developments in microscopic and flow cytometric assays have overcome technical limitations that previously prevented the sensitive and specific detection of HIV in cells and tissue. Further advancements will continue to build upon already existing techniques or develop new assays to study new or more detailed aspects of the viral reservoir. For example, BASEScope is a recent advancement based on the RNAScope technique, and may allow sensitive and specific detection of RNAs using only one probe pair (commercially available from ACD/Bio-Techne). Although this concept has not yet been tested in the context of HIV reservoir studies, this assay could potentially be used for the detection of multiply-spliced vRNAs in situ as a surrogate marker for replication-competence at baseline and after reactivation of the latent reservoir.
We have focussed here on the flow cytometric advances that have enabled multi-parametric, single-cell analysis of HIV-infected cells. However, small particle imaging, an emerging field, could visualize HIV at the single virus level. Previously, this approach required custom-made cytometers sensitive enough to capture small particles, however the newly developed flow virometry technique overcomes this limitation [63]. HIV particles are detected with Env-specific antibodies coupled to magnetic nanoparticles. These nanoparticles can be detected by commercial flow cytometers, thus enabling quantification and phenotypic characterization of single viral particles in supernatants, body fluids or tissues [63]. Importantly, this assay allows to distinguish between viral particles with non-functional or functional Envs [64]. This assay could provide additional complementary information to study viral reactivation after LRA stimulation at both the cellular, and viral, level.
Conclusion
Here, we highlight recently developed tools for the sensitive and specific visualization of the HIV reservoir at single-cell, tissue and body level. The majority of these techniques rely on the detection of vRNA and/or viral proteins, thereby providing a closer estimate for replication-competence compared to classical reservoir measurements based on vDNA detection only. By providing a detailed insight into the phenotype and anatomic localization of single HIV-infected cells, such techniques support the development, evaluation and monitoring of optimal HIV cure strategies in vitro and in vivo.
Acknowledgments
Funding
This work was supported by National Institutes of Health (NIH) grants RO1 HL-092565 (D.E.K); NIAID UM1AI100663 (CHAVI-ID) (D.E.K; Dennis Burton, principal investigator) and the Canadian Institutes for Health Research (project grant # 377124: D.E.K). AEB is the recipient of a CIHR Postdoctoral Fellowship (award no. 152536); DEK is supported by a FRQS Senior Research Scholar Award (award no. 31035).
Figure 1 has been adapted using images from Servier Medical Art (http://servier.com/Powerpoint-image-bank).
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
References
Recent papers of particular interest have been highlighted as:
* important
** very important
- 1.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–7. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 4.Harrigan PR, Whaley M, Montaner JS. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS. 1999;13:F59–62. doi: 10.1097/00002030-199905280-00001. [DOI] [PubMed] [Google Scholar]
- 5.Dahabieh MS, Battivelli E, Verdin E. Understanding HIV Latency: The Road to an HIV Cure. Annu Rev Med. 2015;66:407–21. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang H-C, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF. Latency reversal and viral clearance to cure HIV-1. Science. 2016;353:aaf6517. doi: 10.1126/science.aaf6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–51. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiener B, Horsburgh BA, Eden J-S, Barton K, Schlub TE, Lee E, et al. Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants. Cell Rep. 2017;21:813–22. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76:10942–50. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brady T, Kelly BJ, Male F, Roth S, Bailey A, Malani N, et al. Quantitation of HIV DNA integration: effects of differential integration site distributions on Alu-PCR assays. J Virol Methods. 2013;189:53–7. doi: 10.1016/j.jviromet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mexas AM, Graf EH, Pace MJ, Yu JJ, Papasavvas E, Azzoni L, et al. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26:2295–306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J Virol. 2014;88:12385–96. doi: 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morón-López S, Puertas MC, Gálvez C, Navarro J, Carrasco A, Esteve M, et al. Sensitive quantification of the HIV-1 reservoir in gut-associated lymphoid tissue. PLoS One. 2017;12:e0175899. doi: 10.1371/journal.pone.0175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, et al. Quantitation of Replication-Competent HIV-1 in Populations of Resting CD4 T Cells. J Virol. 2014;88:14070–7. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner KM, Hosmane NN, Siliciano RF. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol. 2015;23:192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Charlebois R, et al. Single-Cell Characterization of Viral Translation-Competent Reservoirs in HIV-Infected Individuals. Cell Host Microbe. 2016;20:368–80. doi: 10.1016/j.chom.2016.07.015. This study provided the initial demonstration of single-cell detection of translation-competent cellular HIV reservoirs by RNA flow cytometry, directly in samples from HIV-infected individuals. The RNA flow FISH approach used combines the detection of Gag protein with GagPol mRNA and enabled substantial advances in specificity. Importantly, the authors demonstrate that this technique enables the size and cellular distribution of the latent, translation-competent reservoir after latency reversal to be studied at the single-cell level. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graf EH, Pace MJ, Peterson BA, Lynch LJ, Chukwulebe SB, Mexas AM, et al. Gag-Positive Reservoir Cells Are Susceptible to HIV-Specific Cytotoxic T Lymphocyte Mediated Clearance In Vitro and Can Be Detected In Vivo. PLoS One. 2013;8:e71879. doi: 10.1371/journal.pone.0071879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.DeMaster LK, Liu X, VanBelzen DJ, Trinité B, Zheng L, Agosto LM, et al. A Subset of CD4/CD8 Double-Negative T Cells Expresses HIV Proteins in Patients on Antiretroviral Therapy. J Virol. 2015;90:2165–79. doi: 10.1128/JVI.01913-15. This study combined classical detection of HIV Gag protein with a cellular hallmark of HIV infection, CD4 downregulation, to reduce the false-positive rate associated with Gag staining alone. This enabled the identification of rare cells expressing HIV Gag directly ex vivo in samples from subjects on ART. The use of FAST (Fiber-optic Array Scanning Technology), enabled a relatively high-throughput for a microscopy-based technique. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Baxter AE, Niessl J, Fromentin R, Richard J, Porichis F, Massanella M, et al. Multiparametric characterization of rare HIV-infected cells using an RNA-flow FISH technique. Nat Protoc. 2017;12:2029–49. doi: 10.1038/nprot.2017.079. This paper provides a detailed protocol for the detection and characterization of translation-competent reservoir cells, identified by the concurrent expression of HIV Gag protein and vRNAs using the RNA flow FISH technique. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, et al. A rare-cell detector for cancer. Proc Natl Acad Sci U S A. 2004;101:10501–4. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das M, Riess JW, Frankel P, Schwartz E, Bennis R, Hsieh HB, et al. ERCC1 expression in circulating tumor cells (CTCs) using a novel detection platform correlates with progression-free survival (PFS) in patients with metastatic non-small-cell lung cancer (NSCLC) receiving platinum chemotherapy. Lung Cancer. 2012;77:421–6. doi: 10.1016/j.lungcan.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350:508–11. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 26.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–64. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 28.Geleziunas R, Bour S, Wainberg MA. Correlation between high level gp160 expression and reduced CD4 biosynthesis in clonal derivatives of human immunodeficiency virus type 1-infected U-937 cells. J Gen Virol. 1994;75(Pt 4):857–65. doi: 10.1099/0022-1317-75-4-857. [DOI] [PubMed] [Google Scholar]
- 29.Fujita K, Silver J, Omura S. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J Gen Virol. 1997;78:619–25. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- 30.Wildum S, Schindler M, Munch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 Down-Modulation and Resistance of Human Immunodeficiency Virus Type 1-Infected T Cells to Superinfection. J Virol. 2006;80:8047–59. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson BK, Till M, Otto P, Goolsby C, Furtado MR, McBride LJ, et al. Detection of HIV-1 DNA and messenger RNA in individual cells by PCR-driven in situ hybridization and flow cytometry. Science. 1993;260:976–9. doi: 10.1126/science.8493534. [DOI] [PubMed] [Google Scholar]
- 32.Patterson BK, Mosiman VL, Cantarero L, Furtado M, Bhattacharya M, Goolsby C. Detection of HIV-RNA-positive monocytes in peripheral blood of HIV-positive patients by simultaneous flow cytometric analysis of intracellular HIV RNA and cellular immunophenotype. Cytometry. 1998;31:265–74. doi: 10.1002/(sici)1097-0320(19980401)31:4<265::aid-cyto6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 33.Patterson BK, Czerniewski MA, Pottage J, Agnoli M, Kessler H, Landay A. Monitoring HIV-1 treatment in immune-cell subsets with ultrasensitive fluorescence-in-situ hybridisation. Lancet. 1999;353:211–2. doi: 10.1016/S0140-6736(05)77222-6. [DOI] [PubMed] [Google Scholar]
- 34.Chargin A, Yin F, Song M, Subramaniam S, Knutson G, Patterson BK. Identification and characterization of HIV-1 latent viral reservoirs in peripheral blood. J Clin Microbiol. 2015;53:60–6. doi: 10.1128/JCM.02539-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Flanagan J, Su N, Wang L-C, Bui S, Nielson A, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–9. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Porichis F, Hart MG, Griesbeck M, Everett HL, Hassan M, Baxter AE, et al. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat Commun. 2014;5:5641. doi: 10.1038/ncomms6641. This paper provides the initial description of the commerical RNA flow cytometry approach discussed in this review. The authors demonstrated the high-throughput detection of cellular mRNAs using the RNA flow FISH assay based on a branched DNA labeling technique originally developed for microscopy. This study provided a framework for many of the RNA flow studies discussed here. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Martrus G, Niehrs A, Cornelis R, Rechtien A, García-Beltran W, Lütgehetmann M, et al. Kinetics of HIV-1 Latency Reversal Quantified on the Single-Cell Level Using a Novel Flow-Based Technique. J Virol. 2016;90:9018–28. doi: 10.1128/JVI.01448-16. This study provided the demonstration that the RNA flow cytometry approach could be applied to in vitro models of HIV latency to detect and characterize cells expressing HIV mRNA and/or HIV protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Grau-Expósito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, et al. A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4 T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. MBio. 2017;8:e00876–17. doi: 10.1128/mBio.00876-17. This work describes a key advance for the RNA flow FISH technique; the identification of GagPol vRNA+ cells directly in samples from HIV-infected individuals, without the requirement for concurrent Gag protein detection, thus identifying the transcription-competent reservoir. The authors demonstrate that this approach enables the identification of the transcription-competent reservoir after latency reversal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagnarelli P, Valenza A, Menzo S, Sampaolesi R, Varaldo PE, Butini L, et al. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J Virol. 1996;70:7603–13. doi: 10.1128/jvi.70.11.7603-7613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang G, Mendes EA, Kaiser P, Sankaran-Walters S, Tang Y, Weber MG, et al. Reactivation of HIV latency by a newly modified Ingenol derivative via protein kinase Cδ-NF-κB signaling. AIDS. 2014;28:1555–66. doi: 10.1097/QAD.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeChristopher BA, Loy BA, Marsden MD, Schrier AJ, Zack JA, Wender PA. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat Chem. 2012;4:705–10. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton K, Winckelmann A, Palmer S. HIV-1 Reservoirs During Suppressive Therapy. Trends Microbiol. 2016;24:345–55. doi: 10.1016/j.tim.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Embretson J, Zupancic M, Ribas JL, Burke A, Racz P, Tenner-Racz K, et al. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature. 1993;362:359–62. doi: 10.1038/362359a0. [DOI] [PubMed] [Google Scholar]
- 45.Fox CH, Cottler-Fox M. In situ hybridization in HIV research. Microsc Res Tech. 1993;25:78–84. doi: 10.1002/jemt.1070250111. [DOI] [PubMed] [Google Scholar]
- 46.Cavert W, Notermans DW, Staskus K, Wietgrefe SW, Zupancic M, Gebhard K, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–4. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 47.Miller CJ, Li Q, Abel K, Kim E-Y, Ma Z-M, Wietgrefe S, et al. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005;79:9217–27. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Q, Duan L, Estes JD, Ma Z-M, Rourke T, Wang Y, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–52. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 49.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–70. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Jr, et al. Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathog Immun. 2016;1:68–106. doi: 10.20411/pai.v1i1.100. This study provided an initial demonstration of the in situ-hybridization Scope approach for the detection and localization of vRNA and vDNA in lymphoid tissues from SIV-infected non-human primates or HIV-infected subjects. Importantly, this assay shows greater speed compared to previous hybridization techniques and allows the concurrent detection of vRNA and vDNA on the same slide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51**.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med [Internet] 2017 doi: 10.1038/nm.4411. Available from: http://dx.doi.org/10.1038/nm.4411This study was the first to investigate how the in situ-hybridization DNA/RNAScope technique could be used to evaluate the contribution of various tissues to the SIV reservoir in ART-treated SIV-infected non-human primates. Based on these results, the authors use tissue biopsies from ART-treated HIV-infected subjects to estimate the total-body size of the replication-competent HIV reservoir. [DOI] [PMC free article] [PubMed]
- 52.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111:2307–12. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haase AT, Retzel EF, Staskus KA. Amplification and detection of lentiviral DNA inside cells. Proc Natl Acad Sci U S A. 1990;87:4971–5. doi: 10.1073/pnas.87.13.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reinhart TA, Rogan MJ, Viglianti GA, Rausch DM, Elden LE, Haase AT. A new approach to investigating the relationship between productive infection and cytopathicity in vivo. Nat Med. 1997;3:218–21. doi: 10.1038/nm0297-218. [DOI] [PubMed] [Google Scholar]
- 55.Usui K, Honda S-I, Yoshizawa Y, Nakahashi-Oda C, Tahara-Hanaoka S, Shibuya K, et al. Isolation and characterization of naïve follicular dendritic cells. Mol Immunol. 2012;50:172–6. doi: 10.1016/j.molimm.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 56.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–9. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Skinner PJ, Ha S-J, Duan L, Mattila TL, Hage A, et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323:1726–9. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–76. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song J, Cai Z, White AG, Jin T, Wang X, Kadayakkara D, et al. Visualization and quantification of simian immunodeficiency virus-infected cells using non-invasive molecular imaging. J Gen Virol. 2015;96:3131–42. doi: 10.1099/jgv.0.000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mestel R. Cancer: Imaging with antibodies. Nature. 2017;543:743–6. doi: 10.1038/543743a. [DOI] [PubMed] [Google Scholar]
- 61**.Santangelo PJ, Rogers KA, Zurla C, Blanchard EL, Gumber S, Strait K, et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat Methods. 2015;12:427–32. doi: 10.1038/nmeth.3320. This key method paper details the use of a non-invasive immuno-PET/CT approach to analyze the anatomic localization of viral protein-expressing cells throughout the whole body in untreated and ART-treated SIV-infected non-human primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marban C, Forouzanfar F, Ait-Ammar A, Fahmi F, El Mekdad H, Daouad F, et al. Targeting the Brain Reservoirs: Toward an HIV Cure. Front Immunol [Internet] 2016:7. doi: 10.3389/fimmu.2016.00397. Available from: http://dx.doi.org/10.3389/fimmu.2016.00397. [DOI] [PMC free article] [PubMed]
- 63.Arakelyan A, Fitzgerald W, Margolis L, Grivel J-C. Nanoparticle-based flow virometry for the analysis of individual virions. J Clin Invest. 2013;123:3716–27. doi: 10.1172/JCI67042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arakelyan A, Fitzgerald W, King DF, Rogers P, Cheeseman HM, Grivel J-C, et al. Flow virometry analysis of envelope glycoprotein conformations on individual HIV virions. Sci Rep. 2017;7:948. doi: 10.1038/s41598-017-00935-w. [DOI] [PMC free article] [PubMed] [Google Scholar]