Abstract
There continues to be a need to develop in vivo high-throughput screening (HTS) and computational methods to screen chemicals for interaction with the estrogen, androgen, and thyroid pathways and as complements to in vitro HTS assays. This study explored the utility of an embryonic zebrafish HTS approach to identify and classify endocrine bioactivity using phenotypically-anchored transcriptome profiling. Transcriptome analysis was conducted on zebrafish embryos exposed to 25 estrogen-, androgen-, or thyroid-active chemicals at concentrations that elicited adverse malformations or mortality at 120 hours post-fertilization in 80% of animals exposed. Analysis of the top 1000 significant differentially expressed transcripts and developmental toxicity profiles across all treatments identified a unique transcriptional and phenotypic signature for thyroid hormone receptor agonists. This unique signature has the potential to be used as a tiered in vivo HTS and may aid in identifying chemicals that interact with the thyroid hormone receptor.
Keywords: Transcriptomics, zebrafish, phenotypic anchoring, endocrine disruptors, high-throughput, ToxCast
1. Introduction
A large body of empirical and clinical evidence has shown that environmental chemicals can disrupt the endocrine systems of humans and wildlife, leading to developmental perturbations that produce in some cases reproductive malformations, teratogenicity, and neurocognitive deficits (e.g., reviewed in [1–5]). In response, the U.S. EPA’s Endocrine Disruptor Screening Program (EDSP) was established in 1998 and has employed a two-tiered strategy to screen and test chemicals for their potential to interact with the estrogen, androgen, and thyroid hormone (TH) pathways. The EDSP Tier 1 battery consists of 11 assays, both in vivo and in vitro, intended to be considered collectively in weight-of-evidence (WoE) evaluations, while EDSP Tier 2 testing includes more in-depth studies to establish dose-response relationships for chemicals with demonstrated bioactivity under Tier 1 [6–8].
Upwards of 10,000 chemicals need screening and testing by the EDSP for estrogen, androgen, and TH disruption potential, with hundreds of new chemicals being added every year [9]. Such a large chemical space presents challenges when relying on traditional in vivo studies for evaluation of potential endocrine effects; requiring extensive time, money, and animals to fully implement. This problem has been broadly recognized, and many regulatory agencies and other organizations have been seeking to develop high-throughput, high-content, and next-generation technologies to apply to chemical screens [10–18]. This has resulted in the establishment of several research programs such as the USEPA Toxicity Forecaster (ToxCast™) program and the inter-agency Toxicology in the 21st Century (Tox21) collaboration [19–23]. Likewise, the EDSP has adopted a similar framework, EDSP in the 21st Century, to begin to include high-throughput in vitro, in silico, and computational approaches with the goal of more efficiently screening chemicals for estrogen, androgen, or TH bioactivity [14, 24]. While progress continues to be made [12–18], challenges remain regarding the use of in vitro and in silico approaches, including limited metabolic competency, the homogeneity of cell cultures, in vivo extrapolation, and lack of exposure estimates [25–32]. Continuing advances to develop in vivo biosensor systems with higher throughput and content capabilities may be useful alongside in vitro HTS assays to help address some of the challenges and limitations presented by cell-based technologies.
One such model that meets the requirements for a higher throughput in vivo system is the embryonic zebrafish. There are several advantages to using embryonic zebrafish as a model organism for chemical screening, including prolific spawning and fecundity, rapid, external development, embryonic transparency for screening, and higher content functional endpoints. Translationally, the zebrafish genome shares 71% homology with humans [33], and developmental and biological pathways are well-conserved. Throughout early development, nearly the entire repertoire of the genome is expressed [34]; therefore, any molecular target that a chemical has the potential to interact with will be present. This allows for the screening of large numbers of chemicals with diverse bioactivities. Zebrafish have already been used to examine the developmental and neurobehavioral toxicity of the chemicals in ToxCast phase I and II libraries [25, 27, 35]. Similarly, other studies have screened flame retardants [36–38], polycyclic aromatic hydrocarbons (PAHs) [39], phytoestrogens [40], chemicals that disrupt cardiovascular function [41], nanomaterials [42, 43], inflammatory compounds [44], as well as high-throughput neurobehavioral screening to identify chemicals with therapeutic potential as psychoactive drugs [45, 46].
Although developmental toxicity screens like those mentioned above provide a wealth of information regarding possible toxic effects of chemicals, adverse phenotypes alone are limiting and often correlated, providing little mechanistic information regarding a chemical’s specific bioactivity [27]. For example, developmental exposure to prototypical estrogen, androgen, or TH-active chemicals cause overlapping phenotypes such as craniofacial abnormalities, pericardial and yolk sac edemas, axial defects, and delayed hatching. Therefore, for many chemicals, the observed adverse phenotypes in 120 hpf zebrafish cannot usually be mapped to a specific mode of action (MOA) [47–50]. However, coupling HTS with transcriptomics can offer a two-tiered approach to categorize chemicals based on developmental bioactivity profiles and transcriptional changes likely associated with a chemical’s MOA. Within the context of the zebrafish model, transcriptomics has been used to identify putative MOAs and classify biological activities for a variety of chemicals including PAHs [51, 52], antimicrobials [53], heavy metals, [39] endocrine disrupting compounds [50, 54, 55], and others [56].
Here, we explored the utility of using phenotypically-anchored transcriptomics in 48 hpf embryonic zebrafish to classify EDCs. We applied a systematic decision flow to identify 25 putative substances with estrogen, androgen, and TH bioactivity potential using our previous in vivo developmental toxicity assessment of the ToxCast phase I and II chemicals [27]. To understand the possible linkages between chemical-mediated changes in gene expression and adverse outcomes, we elected to use a concentration of each chemical that elicited adverse phenotypic effects, including mortality, in 80% of the exposed animals by 120 hpf (EC80) as sampling was from a pooled population. This concentration was independently identified for all 25 chemicals prior to transcriptomic microarray analysis. Clustered correlation analysis of the top 1000 differentially regulated transcripts identified a cluster of four substances, 3,3′,5-triiodothyronine (T3), 3,3′,5-triiodothyroacetic acid (Triac; a T3 analogue), 3,3′,5,5′-tetraiodothyroacetic acid (Tetrac; a thyroxine (T4) analogue), and the pharma compound, CP-634384, which had highly correlated transcriptional profiles that were unique compared to the remaining 21 chemicals. TH receptor (TR) is present and activated by maternal TH during early development preceding embryonic TH production [57, 58]; thyroid follicle development occurs in zebrafish starting at 36 hpf with endogenous TH production of T4 by 72 hpf [59]. Analysis of this subset of thyroid-active chemicals revealed 27 overlapping transcripts, consisting of several known thyroid-regulated genes, which may serve as a diagnostic signature of exposure to specific TR agonists in 48 hpf zebrafish. Furthermore, analysis of the developmental toxicity profiles of these four chemicals were highly similar and had a distinct phenotype in the gut, which may be indicative of overproduction of heme-containing proteins. Overall, we demonstrate the viability of using phenotypically-anchored transcriptomics in zebrafish as a tool to screen and classify the endocrine disrupting potential of chemicals.
2. Materials and Methods
2.1 EDC Chemical Selection
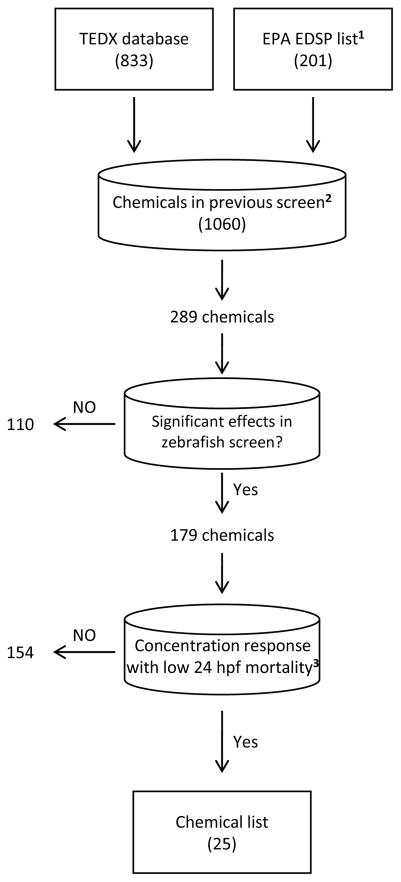
Candidate EDC selection went through a tiered decision tree as shown in Figure 1. We first began with a universe of known and expected EDCs that are available in The Endocrine Disruptor Exchange Database (TEDX; http://endocrinedisruption.org/endocrine-disruption/tedx-list-of-potential-endocrine-disruptors/overview; accessed Nov. 2013) and the first and draft second list of chemicals for tier 1 screening in the USEPA Endocrine Disruptor Screening Program [7, 60]. This consisted of a universe of 1034 chemicals. We then queried these chemicals for those that were included in our previous zebrafish developmental toxicity screen of ToxCast Phase I and II chemicals [27]. This resulted in 289 unique chemicals. From this list, we mined our larval zebrafish developmental screening database [27] to identify chemicals which displayed high quality concentration response curves in which there was either no significant lowest effect level (as calculated in [27]), or if a significant lowest effect level was observed at or above the second highest tested concentration (6.4 or 64 μM depending on chemical solubility in DMSO) for 24 hpf mortality as we will be sampling at 48 hpf. This filtering protocol resulted in the selection of 25 chemicals for further evaluation. Table 1 identifies the supplier, chemical stock purity, and concentration ranges tested. Test solutions were prepared in dimethyl sulfoxide (DMSO; Avantor Performance Materials, Center Valley, PA). A majority of compounds were gifted by the USEPA and were chemicals used in the ToxCast program. Information regarding the USEPA ToxCast chemicals (vendor, ToxCast bottle identification, chemical purity, QA/QC metrics and methods of determination) are provided in Supplemental Table 1. We did not perform analytical chemistry of the USEPA-procured chemicals and chemical purity was assumed to be at the same level provided by the USEPA. All other compounds were obtained from Sigma Aldrich (St. Louis, MO). Chemical stocks were allowed a maximum of 3 freeze/thaw cycles before new stocks were prepared.
Figure 1.
Chemical selection workflow based on the Endocrine Disruptor Exchange (TEDX)and USEPA EDC lists and our zebrafish developmental toxicity assessment of ToxCast Phase I and II chemicals. 1 Denotes the initial list and second list of chemicals for tier 1 screening in the USEPA EDSP. 2Chemicals from Truong et al [27]. 3A chemical with low 24 hpf mortality was defined as having a significant lowest effect level at or above the second highest tested concentration, or no significant lowest effect level, for 24 hpf mortality (as defined by [27]).
Table 1.
The 25 endocrine active compounds used in this study.
| Chemical name [CAS No.] | Abbreviation | Supplier | Purity | Concentration range (μM) |
|---|---|---|---|---|
| 17α-ethinylestradiol [57-63-6] | 17α-EE2 | USEPA | 99% | 0, 4, 8, 16, 32, 64 |
| 17β-estradiol [50-28-2] | 17β-E2 | Sigma Aldrich | 100% | 0, 4, 8, 12, 16, 24 |
| 17-methyltestosterone [58-18-4] | 17-MT | USEPA | 98% | 0, 15, 30, 40, 50, 60 |
| 3,3′,5,5′-tetraiodothyroacetic acid [67-30-1] | Tetrac | Sigma Aldrich | 100% | 0, 0.0016, 0.008, 0.04, 0.2, 1 |
| 3,3′,5-triiodothyronine [6893-02-3] | T3 | USEPA | 99% | 0, 0.00016, 0.0008, 0.004, 0.02, 0.1 |
| ({4-[(7-hydroxy-2,3-dihydro-1H-inden-4-yl)oxy]-3,5-dimethylphenyl}amino)(oxo)acetic acid [290352-28-2] | CP-634384 | USEPA | ≥90% | 0, 0.0016, 0.008, 0.04, 0.2, 1 |
| 5α-dihydrotestosterone [521-18-6] | 5α-DHT | USEPA | 98% | 0, 12.5, 25, 50, 75, 100 |
| Abamectin [71751-41-2] | Abamectin | Sigma Aldrich | 98.9% | 0, 0.25, 1, 4, 16, 64 |
| Bisphenol A [80-05-7] | BPA | Sigma Aldrich | 100% | 0, 15, 30, 40, 50, 60 |
| Bisphenol AF [1478-61-1] | BPAF | USEPA | 99.1% | 0, 5, 10, 15, 25, 35 |
| Butylparaben [94-26-8] | Butylparaben | USEPA | 99% | 0, 0.25, 1, 4, 16, 64 |
| Diisobutyl phthalate [84-69-5] | Diisobutyl phthalate | Sigma Aldrich | 99% | 0, 2.5, 5, 10, 12.5, 15 |
| Dimethipin [55290-64-7] | Dimethipin | USEPA | 99.5% | 0, 12.5, 25, 50, 75, 100 |
| Endrin [72-20-8] | Endrin | Sigma Aldrich | 99.3% | 0, 0.125, 0.25, 0.5, 2, 4 |
| Genistein [446-72-0] | Genistein | USEPA | 98% | 0, 5, 10, 15, 25, 30 |
| Haloperidol [52-86-8] | Haloperidol | USEPA | 99.4% | 0, 0.016, 0.08, 0.4, 2, 10 |
| Kepone [143-50-0] | Kepone | USEPA | 99.9% | 0, 0.016, 0.08, 0.4, 2, 10 |
| Pentachlorophenol [87-86-5] | PCP | USEPA | ≥90% | 0, 0.25, 0.5, 1, 1.5, 2 |
| Perfluorooctane sulfonic acid [1763-23-1] | PFOS | USEPA | 100% | 0, 0.5, 2, 4, 8, 10 |
| Propiconazole [60207-90-1] | Propiconazole | Sigma Aldrich | 99.2% | 0, 15, 30, 40, 50, 60 |
| Propylparaben [94-13-3] | Propylparaben | Sigma Aldrich | 100% | 0, 5, 10, 15, 25, 35 |
| Raloxifene hydrochloride [82640-04-8] | Raloxifene hydrochloride | USEPA | 98% | 0, 15, 30, 40, 50, 60 |
| 3,3′,5-triiodothyroacetic acid [51-24-1] | Triac | Sigma Aldrich | 99% | 0, 0.00016, 0.0008, 0.004, 0.02, 0.1 |
| Vinclozolin [50471-44-8] | Vinclozolin | Sigma Aldrich | 99.6% | 0, 6.25, 12.5, 25, 50, 100 |
| Ziram [137-30-4] | Ziram | USEPA | 99.2% | 0, 0.0016, 0.008, 0.04, 0.2, 1 |
2.2 Zebrafish
All zebrafish handling and use were conducted according to the Oregon State University Institutional Animal Care and Use Committee procedures. Wild-type Tropical 5D zebrafish were maintained at the Sinnhuber Aquatic Research Laboratory (Corvallis, OR) on a 28°C recirculating water system with a 14:10 hour light/dark photoperiod. Embryo collection was performed each morning from group spawns of adult zebrafish. In brief, group tanks of adults with a 1:1 male female ratio were set up the day prior to spawning. Embryos were collected from the tanks using an internal collection apparatus each morning, cleaned, developmentally staged in spans of no more than one hour, and kept in sterile petri dishes under the same conditions as adult zebrafish prior to exposures [61].
2.3 Chemical exposures
At 6 hpf, the chorions were removed enzymatically and the embryos were placed, in 96-well microplates pre-filled with 100 μL embryo medium as previously described [62]. For the EC80 range finding studies, zebrafish were exposed under static conditions to nominal graded concentrations of each chemical from 6–120 hpf (n = 32 embryos per exposure concentration; Table 1). Zebrafish were assessed for developmental toxicity across 22 endpoints at 24 and 120 hpf as previously described [63]. We conducted range finding studies based on the initial concentration response curves performed in the embryonic zebrafish high-throughput screen (HTS) of the ToxCast chemicals [27], selecting concentrations that were expected to elicit no response and up to 100% adverse effects, to allow for appropriate resolution for curve fitting. To calculate the EC80 for each chemical, the developmental toxicity data was consolidated to “no effect” and “effect” scores across all the endpoints. Each animal was assigned a 0 or a 1 if there were no adverse malformations or if the animal had any mortality or malformation, respectively. We then performed logistic regression analysis on these binomial data for each chemical using custom R scripts; EC50, EC60, EC70, and EC80 values were calculated for each regression curve using the dose.p function [64]. For the microarray studies, embryos were exposed to the EC80 of each chemical from 6–48 hpf and RNA was isolated as described below, with four biological replicates per chemical, in four separate grouped batches. Each group RNA isolation batch had its own vehicle (0.64% DMSO) control group to account for batch variability. Batch 1 consisted of vehicle control, 17β-E2, 3,3′,5,5′-tetraiodothyroacetic acid (Tetrac), abamectin, BPA, propiconazole, 3,5,3′-triiodothyroacetic acid (Triac), ziram, haloperidol, and kepone. Batch 2 consisted of vehicle control, T3, 17α-EE2, genistein, PFOS, 17-MT, and CP-634384. Batch 3 consisted of vehicle control, endrin, dimethipin, propylparaben, vinclozolin, BPAF, and butylparaben. Batch 4 consisted of vehicle control, diisobutyl phthalate, 5α-DHT, raloxifene hydrochloride, and pentachlorophenol (PCP). For the qRT-PCR studies, embryos were exposed to vehicle control, the EC50, or EC80 of T3, Triac, Tetrac, or CP-634384 from 6–48 hpf prior to RNA isolation, cDNA synthesis, and qRT-PCR analysis as described below.
2.4 RNA isolation
The Zymo Quick-RNA MiniPrep kit (Irvine, CA; Cat No. R1055) or the Zymo Direct-zol RNA MiniPrep kit (Irvine, CA; Cat No. R2052) were used to isolate RNA for the microarray or qRT-PCR validation studies, respectively. Each replicate consisted of total mRNA collected from pools of eight 48 hpf embryos. In brief, embryos were homogenized using a bullet blender (Next Advance, Averill Park, NY) for 3 minutes at speed 8 in either 300 μl lysis buffer for microarray samples or 500 μl RNAzol RT (Molecular Research Center, Cincinnati, OH) for the qRT-PCR samples with 0.5 mm zirconium oxide beads. RNA was extracted according to the manufacturer’s protocols. For the microarray samples, the optional DNase I digestion step was performed. The quality and concentration for each RNA sample was determined using the Gen5 Take3 and SynergyMX spectrophotometer (BioTek Instruments, Inc., Winooski, VT).
2.5 Microarray processing
Microarrays were hybridized and processed by OakLabs GmBH (Hennigsdorf, Germany) as described previously [53]. In brief, RNA was isolated from pooled embryos with four biological replicates per treatment group. RNA integrity was assessed to ensure RNA integrity scores (RIN) of seven or above (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA). Total RNA (600 ng/sample) was placed in RNAstable® tubes (Biomatrica, San Diego, CA) and dried according to the manufacturer’s instructions for shipment. ArrayXS Zebrafish microarrays were used, which contain oligonucleotides for 48,370 coding and 8,075 non-coding sequences developed using the Ensembl ZV9 release 75 assembly (http://feb2014.archive.ensembl.org/Danio_rerio/Info/Index). RNA samples were re-hydrated and the Low Input Quick Amp WT Labeling Kit (Agilent Technologies) was used to generate cyanine 3-CT labeled cRNA according to the manufacturer’s protocols. Samples were randomly hybridized to ArrayXS Zebrafish microarrays using the Gene Expression Hybridization Kit (Agilent Technologies) by OakLabs GmBH, and scanned using the SureScan Microarray Scanner (Agilent Technologies). Image files were processed with Agilent Feature Extraction software (version 11) and raw intensity data were provided for further processing and analysis.
2.6 Microarray analysis
The Linear Models for Microarray and RNA-seq Data (limma) R/Bioconductor package was used for microarray data filtering, background correction, normalization, and statistical analyses [65, 66]. Arrays were first background corrected and quantile normalized [67]. We filtered all control probes and any probe that was less than 10% brighter than the negative control probes on at least four arrays. Data preprocessing and QA/QC visualization identified nine arrays that had a similar aberrant intensity profile across many probes, and so were removed from the analysis. These deleted probe sets included replicates in two vehicle control batches, Abamectin, Haloperidol, Kepone, two T3 replicates, CP-634384, and Raloxifene hydrochloride. To account for potential batch effects, we implemented the ComBat algorithm [68] to control for scan date variability as well as including the RNA isolation batches in the limma statistical model. Raw and normalized microarray data were uploaded to the Gene Expression Omnibus and are publicly available (GSE89780). Differential expression was performed by fitting the data to a paired linear model in limma considering treatment and the grouped RNA batch effects, and calculating moderated t-statistics for each transcript using the empirical Bayes method of borrow information between genes. P-values were corrected at a 5% false discovery rate (FDR) using the Benjamini-Hochberg method. Transcripts with FDR adjusted P-value ≤ 0.05 and ≥ 1.5-fold increase or decrease in expression compared to the controls were considered significant (Supplemental Data 1). To examine the similarity of transcriptional profiles across all the treatment groups, we performed clustered correlation analysis using custom R scripts. The correlation matrix consisted of a dissimilarity matrix (1-Pearson correlation coefficient) of the probe-wise fold-changes for the top 1000 significantly differentially expressed transcripts, ranked by P-value, across all treatment groups and was clustered using the agglomerative complete-linkage hierarchical clustering algorithm.
2.7 Quantitative real-time PCR
We validated eight of the 18 unique significantly differentially expressed genes after T3, Triac, Tetrac, and CP-634384 exposure using quantitative real-time PCR. These genes were selected based on their overall expression changes (e.g. si:ch211-103n10.5 and gch2 were the most significantly increased and decreased genes, respectively) and their relevance to TH signaling (e.g. dio3a and plp1b). Primers for the target genes are listed in Supplemental Table 1. A total of three biological replicates per treatment group, independent of the original microarray study, were used for the validation study. cDNA was generated from total RNA using the ABI High-capacity cDNA Reverse Transcription kit (Thermo Fisher, Waltham, MA) according to the manufacturer’s protocol. We performed 12.5 μl qRT-PCR reactions using 6.25 μl ABI Power SYBR Green PCR Master Mix (Thermo Fisher), 3.25 μl H2O, 0.5 μl of forward or reverse primer, and 2 μl of cDNA (12.5 ng/μl) using the StepOnePlus Real-Time PCR system (Thermo Fisher). Manufacturer’s recommended cycling conditions were used. β-actin normalized fold-change measurements were calculated according to the procedure described by Pfaffl [69]. Data were analyzed in R using a one-way ANOVA with Tukey post-hoc test or the Kruskal-Wallis test with Dunn’s post-hoc test for data that passed or failed normality testing, respectively.
3. Results and Discussion
3.1 Selection of Dose and Sampling Time Point
The selection of chemical test concentration and developmental time point are key variables when designing studies to define phenotypically-anchored transcriptomic signatures. One difficulty with these types of transcriptomic studies centers on reliably detecting reproducible and strong expression signatures indicative of chemical bioactivity. For example, it has been demonstrated that the use of lower concentrations (EC10 and EC20 values) to profile estrogenic and anti-androgenic chemicals provide transcriptional signatures that have proven difficult to interpret as biomarkers and for application in larger chemical screens [55]. Therefore, we opted to use higher test concentrations at the EC80 with the purpose of eliciting robust transcriptional changes detectable above background expression. It is also important to note that mortality was also considered as an adverse effect in the calculation of the EC80. However, for all chemicals tested, mortality was never the most prevalent adverse effect contributing to the calculated EC80 value (Supplemental Data 2). Another challenge with these types of studies centers on estimating the time course of exposure to effects. For this study, sampling for transcriptome analysis was at 48 hpf as this has been demonstrated to provide mechanistic information for several chemical classes and adverse outcomes [51–53, 55]. However, it is recognized that some chemicals, particularly those that interact with the estrogen and androgen pathways, may elicit the most detectable bioactivity during later periods of reproductive differentiation, although it is also known that the brain is an important target of estrogen signaling early in development [70].
3.2 EC80 Determination of 25 Hormones and EDCs
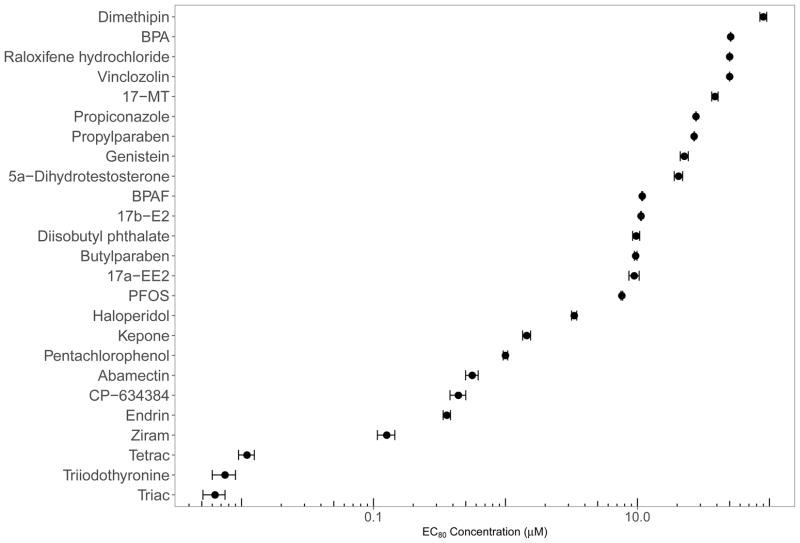
We calculated EC80 values for 23 of the 25 chemicals and observed a wide range of concentrations (Figure 2). We were unable to calculate an EC80 for Vinclozolin and Raloxifene hydrochloride due to no significant adverse effects at the highest concentration tested (100 μM) and solubility issues, respectively. As a result, these two chemicals were assigned an EC value of 50 μM for transcriptome analysis. Detailed developmental toxicity profiles across the 22 endpoints evaluated, the logistic regression plot, model output, and EC calculations for each chemical can be found in Supplemental Data 2. T3, Triac, and Tetrac, were the most potent chemicals tested with an EC80 range of 6.3–10.9 nM, supporting that embryonic zebrafish at this earliest developmental window are highly sensitive to TH or TH analogs. This is not an unexpected observation given the importance of TH in early stages of development. In contrast, model estrogens and androgens were less toxic, having EC80 values several orders of magnitude larger, ranging between 9.46–38.71 μM. For many of the chemicals tested, we observed adverse phenotypic profiles and calculated EC values similar to those reported by others [47–49, 71]. It should be noted that we did observe several chemicals where exposure to the EC80 elicited some adverse developmental progression effects at 24 and 48 hpf including 17β-E2, BPAF, diisobutyl phthalate, genistein, PCP, and ziram.
Figure 2.
Distribution of EC80 values for all 25 chemicals. EC80 concentrations for all 26 compounds ordered by potency. Vinclozolin and raloxifene hydrochloride elicited low toxicity in this assay, and so the EC value was arbitrarily set to 50 μM
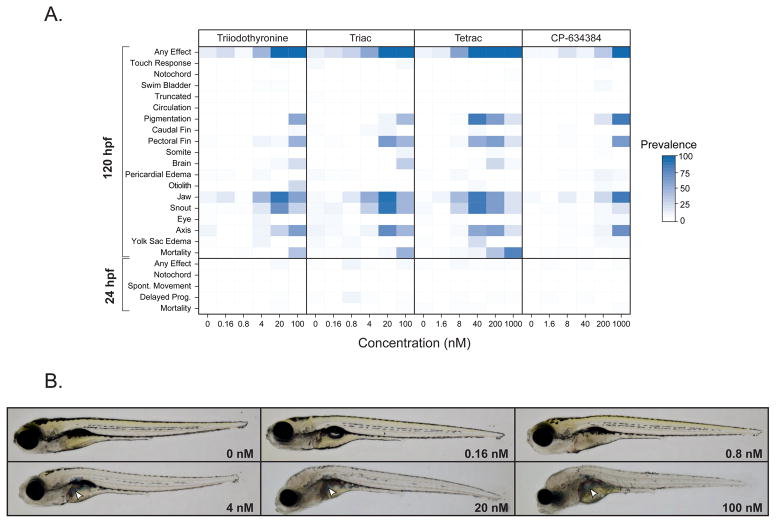
In general, we observed a diversity of phenotypic responses across all the chemicals tested, including common endpoints such as axial defects, pectoral and caudal fin malformations, craniofacial abnormalities, and yolk sac and pericardial edema. Clustered phenotypic profiling using the lowest effect level approach described previously [27] showed many chemicals elicited similar apical phenotypes across all endpoints, regardless of their predicted bioactivity via estrogen, androgen, or TH signaling pathways (Supplemental Figure 1). However, several compounds did elicit unique phenotypic responses. Of the 25 chemicals targeted, ziram, a thiocarbamate pesticide, was the only chemical that elicited notochord malformations at 24 and 120 hpf, a phenotype that has been well characterized previously for this chemical class [72–74]. The other chemicals that caused unique phenotypic responses after exposure were T3, Triac, Tetrac, and CP-634384. Triac and Tetrac are iodothyroacetic acid metabolites of iodothyronines that have been shown to be metabolized by similar mechanisms to TH; that is, deiodination and conjugation reactions but with less well-described formation pathways (reviewed in [75, 76]). Triac induces TR isoform- and response element-specific transcriptional bioactivity, having a higher affinity for the β isoform than T3 and sharing an equal affinity with T3 for α isoforms[77]. Similarly, Tetrac has similar transcriptional bioactivity as T4 [78] and was demonstrated to alleviate neuronal defects and inhibit thyroid stimulating hormone induction in a rodent model of Allan-Herndon-Dudley syndrome [79]. CP-634384 represents one of the failed pharmaceutical chemicals included in the ToxCast Phase II library. A search for the bioactivity of this chemical across the ToxCast/Tox21 or PubChem assays using the EPA Chemistry Dashboard (https://comptox.epa.gov/dashboard) showed that this chemical was highly active in TR receptor agonism assays [80]. Examination of the developmental toxicity profiles for these four compounds showed high similarity across the 22 phenotypic endpoints including developmental malformations (axial, craniofacial, and pectoral fin defects) observed among other compounds (Figure 3A).
Figure 3.
Developmental toxicity of T3, Triac, Tetrac, and CP-634384. A) Phenotypic profile for T3, Triac, Tetrac, and CP-634384 across the suite of 22 phenotypic endpoints evaluated at 24 and 120 hpf. B) Representative bright field images of 120 hpf embryos exposed to 0, 0.16, 0.8, 4, 20, or 100 nM Triac taken using the Keyence BZ-X710 fluorescent microscope (Keyence, Osaka, Japan). White arrows denote possible biliverdin accumulation in the liver of exposed embryos.
However, we did observe two phenotypes that were unique to these apparent TH-active compounds. Namely, the loss of pigmentation and the formation of blue-green pigment in the liver at higher exposure concentrations, as shown in representative images of embryos exposed to Triac (Figure 3B). A recent developmental toxicity screen of T3 as well as thyroid antagonists observed similar pigmentation in the liver but only among T3-exposed zebrafish [47]. Evidence has shown this altered liver pigmentation to be associated with accumulation of biliverdin, an intermediate product of heme catabolism. We also observed a notable decrease in the density of melanophores after exposure to these four chemicals (Figure 3B). This phenotype has been observed in other studies examining hyperthyroidism in adult and larval zebrafish and associated reductions in the proliferation and survival of melanophores [47, 81]. Another phenotype that has been observed in hypothyroid larval life-stages of fishes is underinflation of the anterior swim bladder [47, 82, 83]. Normal inflation of the swim bladder in zebrafish is reliant on a ‘swim-up’ behavior that includes zebrafish actively gulping air [84]. Detection of this phenotype can be confounded by administration of tricaine for 120 hpf evaluation. In our studies, we observed that the addition of tricaine resulted in a deflated swim bladder in some animals, including DMSO controls. Due to the challenge in interpreting this endpoint, this malformation was scored positive only in severe cases where the swim bladder was not visually detected, and did not contribute to the developmental toxicity of these four chemicals (Figure 3A).
3.3 Transcriptome profiling of chemicals that interact with the thyroid pathway
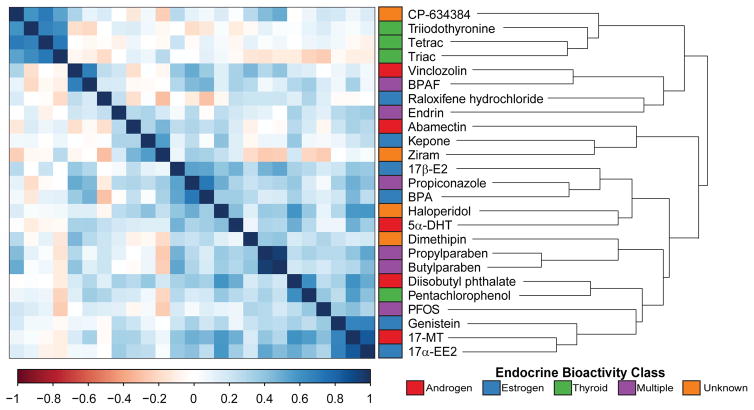
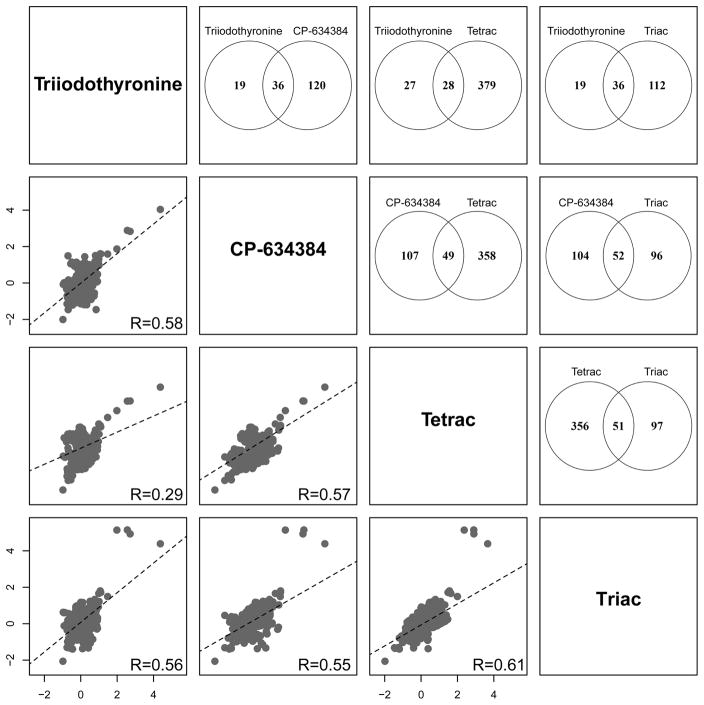
A clustered correlation analysis of the top 1000 significant differentially expressed transcripts across all tested chemicals resulted in a distinct separation of T3, Triac, Tetrac, and CP-634384 from the other compounds, with little to no correlation with all other chemicals (Figure 4). Exposure to the EC80 of T3, Triac, Tetrac, and CP-634384 resulted in 55, 148, 407, and 156 significant differentially expressed transcripts, respectively, for a total of 614 dysregulated transcripts across all four chemicals. As shown in Figure 5, we measured positive correlations for all pairwise comparisons of these four chemicals with the greatest transcriptional overlap between Triac and CP-634384 with a total of 52 transcripts. A total of 27 transcripts were significantly differentially expressed across all four treatments (Table 2), including several for the same gene target (akr, b2m, hbz, opn1lw2, and si:ch1073-459j12.1) representing a total of 18 unique gene targets. Interestingly, on average, we only observed 2–3 out of these 27 transcripts (<10%) significantly dysregulated in the other 21 chemicals, suggesting that these 27 transcripts represent a signature unique to T3, Triac, Tetrac, and CP-634384. This clustering pattern was consistent with the distinctive phenotypic observations for T3, Triac, Tetrac, and CP-634384, and indicative of highly correlated transcriptomic signatures related to thyroid pathway MOAs.
Figure 4.
Clustered correlation analysis of the top 1000 significantly expressed transcripts for the 25 EDCs. A dissimilarity matrix for the top 1000 significant transcripts (FDR adjusted P-value ≤ 0.05; fold change ≥ 1.5) for all pairwise comparisons was generated and hierarchically clustered using the complete linkage algorithm. Colored annotation bar denotes the suspected endocrine bioactivity class of the 25 chemicals.
Figure 5.
Scatter matrix and significant transcript overlap between T3, Triac, Tetrac, and CP-634384. Transcriptional profiles of the thyroid chemical group using pairwise scatter matrices of the 614 unique significant differentially expressed transcripts are shown in the bottom half of the plot. Dashed lines indicate the regression line for each comparison and the Pearson correlation coefficient is provided in the bottom right of each plot. Venn diagrams representing the pairwise overlap of significant transcripts for each treatment pair are shown in the top half of the plot.
Table 2.
Overlapping significant transcripts across T3, Triac, Tetrac, and CP-634384 (log2 fold change).
| Transcript ID | Gene Symbol | Triiodothyronine | Triac | Tetrac | CP-634384 |
|---|---|---|---|---|---|
| ENSDART00000027550 | ak4 | 0.82 | 0.72 | 0.59 | 1.01 |
| ENSDART00000145715 | ak4 | 0.71 | 0.73 | 0.67 | 0.90 |
| ENSDART00000134370 | ak4 | 0.67 | 0.72 | 0.96 | 0.81 |
| ENSDART00000150427 | b2m | 0.68 | 0.77 | 0.61 | 0.86 |
| ENSDART00000075127 | b2m | 0.93 | 0.91 | 0.64 | 1.17 |
| ENSDART00000076816 | cox6a2 | 0.73 | 1.32 | 0.75 | 0.98 |
| ENSDART00000122835 | dio3a | 0.77 | 1.02 | 1.27 | 1.49 |
| ENSDART00000131982 | dio3b | 0.95 | 1.65 | 1.60 | 1.30 |
| ENSDART00000132277 | opn1lw2 | 1.03 | 1.79 | 1.52 | 1.59 |
| ENSDART00000140547 | cox6a2 | 0.62 | 1.20 | 0.68 | 0.72 |
| ENSDART00000144302 | opn1lw2 | 0.74 | 1.19 | 0.75 | 1.17 |
| ENSDART00000150325 | b2m | 0.71 | 0.81 | 0.60 | 1.02 |
| ENSDART00000049895 | epd | 0.80 | 1.08 | 0.59 | 0.83 |
| ENSDART00000012265 | gch2 | −1.02 | −2.07 | −2.02 | −2.02 |
| GENSCAN00000005542 | GENSCAN00000005542 | −0.76 | −1.00 | −1.02 | −0.82 |
| ENSDART00000149920 | hbz | 2.52 | 5.13 | 2.86 | 2.87 |
| ENSDART00000066383 | hbz | 2.68 | 4.92 | 2.86 | 2.82 |
| ENSDART00000127604 | nfil3-6 | 1.44 | 1.48 | 1.96 | 1.57 |
| ENSDART00000065940 | opn1lw2 | 1.06 | 1.70 | 1.44 | 1.58 |
| ENSDART00000126540 | plp1b | 0.62 | 0.83 | 0.63 | 0.90 |
| ENSDART00000129033 | sb:cb827 | 0.97 | 1.21 | 1.04 | 0.96 |
| ENSDART00000025875 | si:ch1073-459j12.1 | 0.86 | 1.02 | 0.64 | 1.08 |
| ENSDART00000132115 | si:ch1073-459j12.1 | 0.88 | 0.91 | 0.59 | 1.03 |
| ENSDART00000077635 | si:ch211-103n10.5 | 4.34 | 4.38 | 3.62 | 4.02 |
| ENSDART00000147009 | si:dkey-251i10.2 | −0.79 | −1.32 | −1.27 | −1.16 |
| ENSDART00000109132 | tmem130 | −0.73 | −1.33 | −1.50 | −1.47 |
| ENSDART00000053077 | zgc:92880 | 1.95 | 5.13 | 2.33 | 1.85 |
Our data suggest that exposure to elevated exogenous TH or TH analogs produces pigmentation defects, and other morphological abnormalities, which may be a result of disruption in the normal cascade of developmental cues mediated by TR signaling. TH-mediated transcriptional regulation occurs through TR activity. In general, the unliganded TR acts as a constitutive transcriptional repressor when not bound by TH, and a transcriptional activator when bound by TH (as reviewed in [85]). Twenty-three of the 27 overlapping transcripts between T3, Triac, Tetrac, and CP-634384 were significantly increased (Table 2), suggesting that some of these genes may be transcriptionally repressed by unliganded TR at this life stage until later in development when endogenous TH is present. Notable TH-regulated transcripts that were shown to increase included dio3a, plp1b, hbz, zgc:92880, si:ch211-103n10.5, and b2m. Dio3 encoded by dio3a in zebrafish catalyzes the inactivation of T4 and T3 to 3,3′,5′-triiodothyronine (reverse-T3) and 3,3′-diiodothyronine (T2), respectively, and may be induced as a compensatory mechanism to inactivate the high levels of T3 and TH analogs due to exposure [86]. Developmental hypothyroidism causes delays and reductions in the production of the myelin sheath [87]. plp1b is a structural protein that is the predominant component of myelin and its expression is increased or decreased in response to a hyper- or hypothyroid state in the brain, respectively [87, 88]. Though not as straightforward as dio3a and plp1b, THs have also been shown to play a role in the switching of embryonic to adult hemoglobin, and the observed changes in expression for both hbz and zgc:92880, orthologous to the human fetal globin gene HBG1, may be involved in that process [54, 89]. The large increases in the expression of globin genes may contribute to the formation of the unique blue-green pigment detected in the livers of fish at 120 hpf. This altered liver pigmentation may be related to an adaptive response and merits additional study (Figure 3B). si:ch211-103n10.5, an orthologue of human HIST1H1C, codes for Histone H1.2, a histone complex protein that can bind to linker DNA between nucleosomes to form chromatin fibers, and is involved in the compaction of nucleosomes [90]. Histone proteins are normally regulated in a DNA replication-dependent manner and to our knowledge only one in vitro study has observed a similar effect of THs in increasing the abundance of histone proteins [91]. In that study, the effects of T3 on histone proteins were suggested to be due to increased translational efficiency of histones rather than increased transcription. However, this was never confirmed for histone H1, which may have an independent mechanism or may be regulated differently by TH. Two hypotheses for the si:ch211-103n10.5 induction may be a homeostatic response to the loss of TR repressive bioactivity in the presence of these four chemicals, or aberrant repression of genes through chromatin remodeling due to bioactivity of liganded TR in cells that participate in TH signaling [90]. Furthermore, the magnitude of differential expression of this gene coupled with the fact that it was measured in whole embryo homogenates suggests that this activity occurs in a large number of cells, possibly affecting many disparate cell types in 48 hpf embryos. b2m, a protein found on the surface of lymphocytes, is a component of class I histocompatibility antigens and has been shown to be elevated in hyperthyroid individuals [92–94]. The mechanism by which excessive TH levels increase b2m levels is unknown.
To demonstrate that the unique transcriptional signatures measured could be used as a possible biomarker for exposure to TH-active compounds, we selected eight of the 27 significant overlapping transcripts for qRT-PCR validation at both the calculated EC50 and EC80 exposure levels. As shown in Table 3, all the transcripts except for gch2 validated in one or more of these chemicals at the EC80 exposure level. Of the transcripts that validated at the EC80, the transcriptional responses between the EC50 and EC80 had an average concordance of 86% between these chemicals (57%, 100%, 100%, and 87.5% concordance for T3, Triac, Tetrac, and CP-634384 respectively), and appeared to change in a concentration-dependent manner. We observed significant expression changes at both the EC80 and EC50 exposure levels for dio3b, nfil3-6, sb:cb827, and si:ch211-103n10.5. plp1b was similarly significantly elevated for all tested chemicals at both the EC50 and EC80 except for the EC50 of T3 (FDR adjusted P-value = 0.23). We also observed mixed significant responses for both hbz and zgc:92880, which were not affected after exposure to Triac, suggesting these transcripts may not be fully reliable as biomarkers at 48 hpf. The concordance between the clustering of T3, Triac, Tetrac, and CP-634384 in the developmental toxicity (Supplemental Figure 1) and transcriptome profiling approach (Figure 4) demonstrates that developmental toxicity screening alone can identify potent TR agonists. In this case, full transcriptome profiling for chemicals that elicit a similar pigmentation phenotype after developmental exposure would not be required; however, confirmation of bioactivity using the biomarker gene set identified in this study would be useful.
Table 3.
qRT-PCR validation of target transcripts in the unique transcriptional signature of T3, Triac, Tetrac, and CP-634384.
| Gene | T3 | Triac | Tetrac | CP-634384 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Microarray | EC50 | EC80 | Microarray | EC50 | EC80 | Microarray | EC50 | EC80 | Microarray | EC50 | EC80 | |
| dio3a | 0.77 | 1.96 ± 0.16 | 2.04 ± 0.55 | 1.02 | 1.19 ± 0.13 | 1.42 ± 0.24 | 1.27 | 1.85 ± 0.10 | 2.04 ± 0.69 | 1.49 | 1.63 ± 0.1 | 1.75 ± 0.32 |
| gch2 | −1.02 | −1.02 ± 0.29 | −0.62 ± 0.56 | −2.07 | −0.37 ± 0.33 | −0.88 ± 0.47 | −2.02 | −0.04 ± 0.09 | −1.24 ± 1.75 | −2.02 | −0.38 ± 0.22 | −0.35 ± 0.47 |
| hbz | 2.68 | 0.89 ± 0.09 | 1.06 ± 0.63 | 4.92 | 0.11 ± 0.20 | −0.12 ± 0.29 | 2.86 | 0.77 ± 0.26 | 1.20 ± 0.44 | 2.82 | 0.63 ± 0.02 | 0.91 ± 0.16 |
| nfil3-6 | 1.44 | 0.59 ± 0.05 | 0.77 ± 0.36 | 1.48 | 0.89 ± 0.18 | 1.01 ± 0.29 | 1.96 | 1.48 ± 0.13 | 1.71 ± 0.34 | 1.57 | 1.24 ± 0.12 | 1.36 ± 0.06 |
| plp1b | 0.62 | 0.92 ± 0.68 | 2.58 ± 0.67 | 0.83 | 2.07 ± 0.14 | 1.51 ± 0.33 | 0.63 | 2.18 ± 0.28 | 2.09 ± 0.58 | 0.90 | 2.03 ± 0.36 | 2.24 ± 0.97 |
| sb:cb827 | 0.97 | 1.48 ± 0.45 | 1.53 ± 0.42 | 1.21 | 1.47 ± 0.23 | 1.58 ± 0.27 | 1.04 | 1.78 ± 0.21 | 1.83 ± 0.21 | 0.96 | 1.27 ± 0.33 | 1.86 ± 0.08 |
| si:ch211-103n10.5 | 4.34 | 1.33 ± 0.10 | 1.69 ± 0.49 | 4.38 | 1.58 ± 0.27 | 1.73 ± 0.59 | 3.62 | 1.65 ± 0.09 | 2.18 ± 0.47 | 4.02 | 1.29 ± 0.01 | 1.55 ± 0.15 |
| zgc:92880 | 1.95 | 0.84 ± 0.43 | 1.11 ± 0.26 | 5.13 | −0.25 ± 0.41 | −0.13 ± 0.13 | 2.33 | 0.98 ± 0.30 | 1.30 ± 0.45 | 1.85 | 0.27 ± 0.22 | 0.83 ± 0.25 |
Note: Shading denotes statistically significant log2 fold changes in expression compared to vehicle control (FDR adjusted P-value ≤ 0.05)
Although we did observe unique transcriptional profiles for T3, Triac, Tetrac, and CP-634384, we did not observe correlated transcriptional signatures with other test chemicals with known TH bioactivity. In vivo studies of PFOS exposure in rodents have measured decreased serum T4 and T3 after acute [95] and chronic exposures [96, 97]. The relationship of PFOS-induced thyroid bioactivity to subsequent developmental pathology in experimental mammalian models continues to need study. Mechanistic research has shown that increased hepatic glucuronidation and TH catabolism, with a potential compensatory response of increasing Dio1 (converting T4 to active T3) expression, play a role in the reductions in circulating TH [98–100]. Developmental exposure to PFOS for 15 days in zebrafish resulted in similar increases in expression of several TH-responsive and homeostatic genes, such as slc5a5 (sodium/iodide symporter) and dio1; however, PFOS exposure increased total T3 levels and had no effect on T4 levels, which may be due to compensatory activity related to increases in dio1 [101]. Based on these postulated mechanisms of PFOS interference with TH catabolism and possibly biosynthesis, it is not surprising that the transcriptional signatures for T3, Triac, Tetrac, and CP-634384 differed from that of PFOS. PCP is another chemical with suspected TH bioactivity that did not have a similar transcriptional profile as the four TR agonists. Developmental exposure to PCP in zebrafish results in a hyperthyroid phenotype with increased T3 levels and increased expression of several genes along the hypothalamus-pituitary-thyroid axis such as tshba, slc5a5, dio1, dio2, dio3a, thra, thrb, and ugt1ab [102, 103]. Although we observed significant increases in dio3a and thrb, with the latter observed in only T3, Triac, and CP-634384, these transcriptional responses were absent in our PCP samples. This difference may be due to concentration differences between studies, where our EC80 for PCP was 1.02 μM compared to the transcriptionally-active concentration of 38 nM (10 μg/L) in Cheng et al. [103]. Transcriptome studies examining the developmental toxicity of PCP at higher concentrations (190–760 nM) have demonstrated that PCP acts as an uncoupler of oxidative phosphorylation during early development [104, 105]. Transcriptional profiles at 24 hpf after exposure to 760 nM (200 μg/L) of PCP caused disruption in genes involved in somitogenesis and lens formation, consistent with non-thyroidal disruption of development [105]. Overall, these studies present a contrasting concentration-dependent mechanism of PCP toxicity during zebrafish development, where low concentrations (≤ 50 nM) appear to disrupt the TH pathway but higher concentrations elicit a toxic response involving general developmental processes. Because the EC80 exposure level in our analysis, 1.02 μM, is near the highest concentration used in Xu et al. [105], we hypothesize that the transcriptional profile we observed is likely similar to the non-thyroidal developmental effects of PCP. Additional studies of PCP, using the experimental approach presented here, are needed to determine whether PCP has TR agonist-like bioactivity at the concentrations used in Cheng et al. [103]. This demonstrates that not every chemical screened using the phenotypically-anchored transcriptome approach at an EC80 exposure level described here will be wholly sensitive to a chemical’s MOA, and may be complicated by transcriptional changes associated with non-specific toxicity observed at higher concentrations, thus highlighting the need for additional orthogonal assays to fully classify chemical bioactivity.
3.4 Transcriptome profiling of chemicals that interact with estrogen and androgen pathways
Estrogen receptor (ER) expression in zebrafish development first appears in the brain at 24 hpf followed by the liver, pancreas, neuromasts, and the heart at later developmental stages [106–108]. Similarly, androgen receptor (AR) expression can be identified in the brain and olfactory placodes, and pronephros at 24 hpf, with expression in specific brain regions and retina from 72–120 hpf [109]. The developmental expression of ER and AR, along with steroid hormone synthesis and aromatase enzymes, supports a functional role for estrogen and androgen signaling during early development outside of their primary role in reproductive differentiation (as reviewed in [110]). Furthermore, developmental exposure to estrogen/androgen agonist and antagonist chemicals induce neurobehavioral changes and adverse developmental effects, which suggests the possibility of conserved transcriptional responses from chemicals that modulate the activity of these receptors [40, 71, 107]. However, unlike T3, Triac, Tetrac, and CP-634384, we did not observe a strong transcriptional signature for estrogenic and androgenic chemicals tested. In a similar transcriptome study of developmental exposure to 1 μM 17β-estradiol, expression profiles of 24, 48, 72, and 96 hpf zebrafish were highly disparate and only one overlapping gene, vtg1, was found across all timepoints [107]. The lack of concordance in correlated transcriptional signatures in our study, and in Hao et al. [111], may be related to the biological importance of TR signaling in early development [112, 113] compared to estrogen and androgen pathways, although the role of steroidal signaling in brain development continues to evolve. TH levels are known to be involved in life-stage transitions in zebrafish as well as anuran metamorphosis, likely involving highly conserved transcriptional responses for many genes after exposure to TR agonists [114, 115]. The strong transcriptional signature for TH-related chemicals support this hypothesis and our data suggest 48 hpf is an appropriate timepoint to identify these types of chemicals. It is possible that examining transcriptional profiles beyond 48 hpf may identify sets of genes that are sensitive to estrogenic and androgenic chemicals. Including additional time points following chemical exposures would likely increase the sensitivity and reliability of any biomarkers of exposure for estrogenic and androgenic chemicals, and provide added information regarding the molecular events that precede the adverse phenotypic responses observed for these chemicals. However, this would need to be performed at lower concentrations to limit secondary effects associated with the progression of toxicity. These varied experimental paradigms will become more feasible as the cost of measuring the transcriptome decreases. Similarly, since we were interested in identifying discriminatory transcriptional signatures indicative of specific endocrine effects, we have focused our analysis at the transcript level and did not perform any downstream functional analyses, such as pathway or gene set analysis. Several transcriptome analyses have observed a lack of correlation between related chemicals [55, 107] or species comparisons of the same chemicals [116] at the transcript level, only to observe significant overlap in functional relationships between exposures at the biological pathway or gene ontology level.
4. Conclusion
Developing new models with the potential to more efficiently screen chemicals for endocrine bioactivity is paramount to be responsive to public health considerations underlying statutory requirements of the EDSP. Furthermore, additional work is needed to define in vivo models for these types of screens to supplement in vitro and in silico HTS assays. Here, we used phenotypically-anchored transcriptomics with a diverse set of 25 EDCs to identify unique transcriptional signatures that could predict a chemical’s potential to interact with the estrogen, androgen, and/or thyroid pathways. Although the transcriptome analysis presented here only consisted of one timepoint, and therefore represents only a snapshot of the transcriptome changes due to chemical exposure, it can model chemical-induced expression changes across all cell and tissue types present in 48 hpf zebrafish. Using this approach, we demonstrated that chemicals that act as specific TR agonists induce a unique transcriptional profile that is indicative of strong TR agonism, potentially mediated by loss of the basal repressive signaling of unliganded TRs. The identification and subsequent validation of many of the transcriptional changes observed between all four chemicals, alongside their involvement in TH signaling, suggests that these transcripts may serve as a suite of biomarkers of TR agonists in 48 hpf zebrafish. Moreover, the correlation between the unique 120 hpf phenotype, i.e. loss of pigmentation and biliverdin accumulation, suggest that developmental toxicity screening alone, followed by qRT-PCR confirmation using the identified biomarker gene set, is sufficient to identify potent TR agonists. Further study is needed to determine whether these transcripts continue to be disrupted or if any additional signatures become apparent later during development due to exposure to TR agonists. The lack of unique transcriptional signatures for estrogenic, androgenic, as well as chemicals that disrupt the TH pathway outside of specific TR agonism, suggests additional work is needed to better define the optimal developmental timepoint for the application of this approach for chemicals that interact with these endocrine pathways. However, this work demonstrates the promise of phenotypically-anchored whole genome transcriptomics in zebrafish to more rapidly screen and classify potential EDCs based on their global expression profiles.
Supplementary Material
Acknowledgments
We would like to thank Greg Gonnerman, Carrie Barton, and the staff at the Sinnhuber Aquatic Research Laboratory for help and support with fish husbandry, spawning, and embryo screening. We would also like to acknowledge Dr. Susan Tilton for statistical modeling advice, Dr. Amber Roegner for assistance during RNA collections, and Dr. Lisa Truong and Dr. Michael Simonich for manuscript editing and preparation. This research was supported by NIH P30 ES000210, T32 ES007060, P42 ES016465 and EPA #R835168. Pacific Northwest National Laboratory is a multi-program laboratory operated by Battelle for the U.S. Department of Energy under Contract DE-AC05-76RL01830.
Abbreviations
- EDC
Endocrine disrupting chemical
- TR
thyroid hormone receptor
- USEPA
United States Environmental Protection Agency
- EDSP
Endocrine Disruptor Screening Program
- hpf
hours post-fertilization
References
- 1.Bergman A, Heindel JJ, Kasten T, Kidd KA, Jobling S, Neira M, Zoeller RT, Becher G, Bjerregaard P, Bornman R, Brandt I, Kortenkamp A, Muir D, Drisse MN, Ochieng R, Skakkebaek NE, Bylehn AS, Iguchi T, Toppari J, Woodruff TJ. The impact of endocrine disruption: a consensus statement on the state of the science. Environ Health Perspect. 2013;121(4):A104–6. doi: 10.1289/ehp.1205448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colborn T, vom Saal FS, Soto AM. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ Health Perspect. 1993;101(5):378–84. doi: 10.1289/ehp.93101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC. Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev. 2009;30(4):293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gore AC, Chappell VA, Fenton SE, Flaws JA, Nadal A, Prins GS, Toppari J, Zoeller RT. EDC-2: The Endocrine Society’s Second Scientific Statement on Endocrine-Disrupting Chemicals. Endocr Rev. 2015;36(6):E1–E150. doi: 10.1210/er.2015-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavlock RJ, Daston GP, DeRosa C, Fenner-Crisp P, Gray LE, Kaattari S, Lucier G, Luster M, Mac MJ, Maczka C, Miller R, Moore J, Rolland R, Scott G, Sheehan DM, Sinks T, Tilson HA. Research needs for the risk assessment of health and environmental effects of endocrine disruptors: a report of the U.S. EPA-sponsored workshop. Environ Health Perspect. 1996;104(Suppl 4):715–40. doi: 10.1289/ehp.96104s4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.U.S. EPA. [Accessed October 2016];Endocrine Disruptor Screening Program Tier 1 Assays: Considerations for Use in Human Health and Ecological Risk Assessments. 2013 https://www.epa.gov/sites/production/files/2015-07/documents/use_of_tier_1_data_in_risk_assessment.pdf.
- 7.U.S. EPA. [Accessed October 2016];Endocrine Disruptor Screening Program (EDSP) 1998 Federal Register Notices. 2016 https://www.epa.gov/endocrine-disruption/endocrine-disruptor-screening-program-edsp-1998-federal-register-notices.
- 8.U.S. EPA; Endocrine Disruptor Screening Program, editor. Weight-of-Evidence: Evaluating Results of EDSP Tier 1 Screening to Identify the Need for Tier 2 Testing. 2011. pp. 1–47. [Google Scholar]
- 9.U.S. EPA. [Accessed September 2016];U.S. Environmental Protection Agency Endocrine Disruptor Screening Program Comprehensive Management Plan. 2012 https://www.epa.gov/sites/production/files/2015-08/documents/edsp-comprehensive-management-plan-2012.pdf.
- 10.National Research Council. Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press; Washington, DC: 2007. [Google Scholar]
- 11.NTP. A National Toxicology Program for the 21st Century: A Roadmap for the Future. 2004. pp. 1–14. [Google Scholar]
- 12.N. EPA. Integrated Bioactivity and Exposure Ranking: A Computational Approach for the Prioritization and Screening of Chemicals in the Endocrine Disruptor Screening Program. Presentation to the December 2014 meeting of the Scientific Advisory Panel for the EPA Federal Insecticide, Fungicide and Rodenticide Act [Internet]; Washington, DC: US EPA; 2014. [Google Scholar]
- 13.OECD. New Scoping Document on in vitro and ex vivo Assays for the Identification of Modulators of Thyroid Hormone Signalling. OECD Publishing; [Google Scholar]
- 14.N.R. Council. Endocrine Disruptor Screening and Testing Advisory Committee. 1999. [Google Scholar]
- 15.Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, Houck KA, Martin MT, Sipes N, Richard AM, Mansouri K, Setzer RW, Knudsen TB, Crofton KM, Thomas RS. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol Sci. 2015;148(1):137–54. doi: 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D, Sakamuru S, Xia M, Huang R, Judson R. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem Res Toxicol. 2017;30(4):946–964. doi: 10.1021/acs.chemrestox.6b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS. Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ Sci Technol. 2015;49(14):8804–14. doi: 10.1021/acs.est.5b02641. [DOI] [PubMed] [Google Scholar]
- 18.Murk AJ, Rijntjes E, Blaauboer BJ, Clewell R, Crofton KM, Dingemans MM, Furlow JD, Kavlock R, Kohrle J, Opitz R, Traas T, Visser TJ, Xia M, Gutleb AC. Mechanism-based testing strategy using in vitro approaches for identification of thyroid hormone disrupting chemicals. Toxicol in Vitro. 2013;27(4):1320–46. doi: 10.1016/j.tiv.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Persp. 2013;121(7):756. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319(5865):906–7. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol Sci. 2007;95(1):5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 22.Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, Dix DJ. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect. 2010;118(4):485–92. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavlock RJ, Austin CP, Tice RR. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009;29(4):485–7. doi: 10.1111/j.1539-6924.2008.01168.x. discussion 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S. EPA. [Accessed October 2016];Endocrine Disruptor Screening Program for the 21st Century: (EDSP21 Work Plan) The Incorporation of In Silico Models and In Vitro High Throughput Assays in the Endocrine Disruptor Screening Program (EDSP) for Prioritization and Screening. 2011 https://www.epa.gov/sites/production/files/2015-07/documents/edsp21_work_plan_summary_overview_final.pdf.
- 25.Padilla S, Corum D, Padnos B, Hunter DL, Beam A, Houck KA, Sipes N, Kleinstreuer N, Knudsen T, Dix DJ, Reif DM. Zebrafish developmental screening of the ToxCast Phase I chemical library. Reprod Toxicol. 2012;33(2):174–87. doi: 10.1016/j.reprotox.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Rotroff DM, Wetmore BA, Dix DJ, Ferguson SS, Clewell HJ, Houck KA, Lecluyse EL, Andersen ME, Judson RS, Smith CM, Sochaski MA, Kavlock RJ, Boellmann F, Martin MT, Reif DM, Wambaugh JF, Thomas RS. Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol Sci. 2010;117(2):348–58. doi: 10.1093/toxsci/kfq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL. Multidimensional in vivo hazard assessment using zebrafish. Toxicol Sci. 2014;137(1):212–33. doi: 10.1093/toxsci/kft235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reif DM, Martin MT, Tan SW, Houck KA, Judson RS, Richard AM, Knudsen TB, Dix DJ, Kavlock RJ. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ Health Perspect. 2010;118(12):1714–20. doi: 10.1289/ehp.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotroff DM, Dix DJ, Houck KA, Knudsen TB, Martin MT, McLaurin KW, Reif DM, Crofton KM, Singh AV, Xia M, Huang R, Judson RS. Using in vitro high throughput screening assays to identify potential endocrine-disrupting chemicals. Environ Health Perspect. 2013;121(1):7–14. doi: 10.1289/ehp.1205065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, Knudsen TB, Kancherla J, Mansouri K, Patlewicz G, Williams AJ, Little SB, Crofton KM, Thomas RS. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem Res Toxicol. 2016;29(8):1225–51. doi: 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- 31.Boyd WA, Smith MV, Co CA, Pirone JR, Rice JR, Shockley KR, Freedman JH. Developmental Effects of the ToxCast Phase I and Phase II Chemicals in Caenorhabditis elegans and Corresponding Responses in Zebrafish, Rats, and Rabbits. Environ Health Perspect. 2016;124(5):586–93. doi: 10.1289/ehp.1409645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, Joliet O, Frame A, Rabinowitz J, Knudsen TB, Judson RS, Egeghy P, Vallero D, Cohen Hubal EA. High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ Sci Technol. 2013;47(15):8479–88. doi: 10.1021/es400482g. [DOI] [PubMed] [Google Scholar]
- 33.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Threadgold G, Harden G, Ware D, Begum S, Mortimore B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Lloyd C, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Lanz C, Raddatz G, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Schuster SC, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vesterlund L, Jiao H, Unneberg P, Hovatta O, Kere J. The zebrafish transcriptome during early development. BMC Dev Biol. 2011;11:30. doi: 10.1186/1471-213X-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reif DM, Truong L, Mandrell D, Marvel S, Zhang G, Tanguay RL. High-throughput characterization of chemical-associated embryonic behavioral changes predicts teratogenic outcomes. Arch Toxicol. 2016;90(6):1459–70. doi: 10.1007/s00204-015-1554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dishaw LV, Hunter DL, Padnos B, Padilla S, Stapleton HM. Developmental exposure to organophosphate flame retardants elicits overt toxicity and alters behavior in early life stage zebrafish (Danio rerio) Toxicol Sci. 2014;142(2):445–54. doi: 10.1093/toxsci/kfu194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveri AN, Bailey JM, Levin ED. Developmental exposure to organophosphate flame retardants causes behavioral effects in larval and adult zebrafish. Neurotoxicol Teratol. 2015;52(Pt B):220–7. doi: 10.1016/j.ntt.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarema KA, Hunter DL, Shaffer RM, Behl M, Padilla S. Acute and developmental behavioral effects of flame retardants and related chemicals in zebrafish. Neurotoxicol Teratol. 2015;52(Pt B):194–209. doi: 10.1016/j.ntt.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barranco A, Escudero L, Sanz Landaluze J, Rainieri S. Detection of exposure effects of mixtures of heavy polycyclic aromatic hydrocarbons in zebrafish embryos. J Appl Toxicol. 2017;37(3):253–264. doi: 10.1002/jat.3353. [DOI] [PubMed] [Google Scholar]
- 40.Bugel SM, Bonventre JA, Tanguay RL. Comparative Developmental Toxicity of Flavonoids Using an Integrative Zebrafish System. Toxicol Sci. 2016;154(1):55–68. doi: 10.1093/toxsci/kfw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yozzo KL, Isales GM, Raftery TD, Volz DC. High-content screening assay for identification of chemicals impacting cardiovascular function in zebrafish embryos. Environ Sci Technol. 2013;47(19):11302–10. doi: 10.1021/es403360y. [DOI] [PubMed] [Google Scholar]
- 42.Harper SL, Carriere JL, Miller JM, Hutchison JE, Maddux BL, Tanguay RL. Systematic evaluation of nanomaterial toxicity: utility of standardized materials and rapid assays. ACS Nano. 2011;5(6):4688–97. doi: 10.1021/nn200546k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.George S, Xia T, Rallo R, Zhao Y, Ji Z, Lin S, Wang X, Zhang H, France B, Schoenfeld D, Damoiseaux R, Liu R, Bradley KA, Cohen Y, Nel AE. Use of a High-Throughput Screening Approach Coupled with In Vivo Zebrafish Embryo Screening to Develop Hazard Ranking for Engineered Nanomaterials. ACS Nano. 2011;5(3):1805–17. doi: 10.1021/nn102734s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.d’Alencon CA, Pena OA, Wittmann C, Gallardo VE, Jones RA, Loosli F, Liebel U, Grabher C, Allende ML. A high-throughput chemically induced inflammation assay in zebrafish. BMC biology. 2010;8:151. doi: 10.1186/1741-7007-8-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokel D, Bryan J, Laggner C, White R, Cheung CY, Mateus R, Healey D, Kim S, Werdich AA, Haggarty SJ, Macrae CA, Shoichet B, Peterson RT. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruni G, Lakhani P, Kokel D. Discovering novel neuroactive drugs through high-throughput behavior-based chemical screening in the zebrafish. Frontiers in pharmacology. 2014;5:153. doi: 10.3389/fphar.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jomaa B, Hermsen SA, Kessels MY, van den Berg JH, Peijnenburg AA, Aarts JM, Piersma AH, Rietjens IM. Developmental toxicity of thyroid-active compounds in a zebrafish embryotoxicity test. ALTEX. 2014;31(3):303–17. doi: 10.14573/altex.1402011. [DOI] [PubMed] [Google Scholar]
- 48.Kim DJ, Seok SH, Baek MW, Lee HY, Na YR, Park SH, Lee HK, Dutta NK, Kawakami K, Park JH. Developmental toxicity and brain aromatase induction by high genistein concentrations in zebrafish embryos. Toxicol Mech Methods. 2009;19(3):251–6. doi: 10.1080/15376510802563330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivero-Wendt CL, Oliveira R, Monteiro MS, Domingues I, Soares AM, Grisolia CK. Steroid androgen 17alpha-methyltestosterone induces malformations and biochemical alterations in zebrafish embryos. Environ Toxicol Pharmacol. 2016;44:107–13. doi: 10.1016/j.etap.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Saili KS, Tilton SC, Waters KM, Tanguay RL. Global gene expression analysis reveals pathway differences between teratogenic and non-teratogenic exposure concentrations of bisphenol A and 17beta-estradiol in embryonic zebrafish. Reprod Toxicol. 2013;38:89–101. doi: 10.1016/j.reprotox.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodale BC, La Du J, Tilton SC, Sullivan CM, Bisson WH, Waters KM, Tanguay RL. Ligand-Specific Transcriptional Mechanisms Underlie Aryl Hydrocarbon Receptor-Mediated Developmental Toxicity of Oxygenated PAHs. Toxicol Sci. 2015;147(2):397–411. doi: 10.1093/toxsci/kfv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodale BC, Tilton SC, Corvi MM, Wilson GR, Janszen DB, Anderson KA, Waters KM, Tanguay RL. Structurally distinct polycyclic aromatic hydrocarbons induce differential transcriptional responses in developing zebrafish. Toxicol Appl Pharmacol. 2013;272(3):656–70. doi: 10.1016/j.taap.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haggard DE, Noyes PD, Waters KM, Tanguay RL. Phenotypically anchored transcriptome profiling of developmental exposure to the antimicrobial agent, triclosan,reveals hepatotoxicity in embryonic zebrafish. Toxicol Appl Pharmacol. 2016;308:32–45. doi: 10.1016/j.taap.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pelayo S, Oliveira E, Thienpont B, Babin PJ, Raldua D, Andre M, Pina B. Triiodothyronine-induced changes in the zebrafish transcriptome during the eleutheroembryonic stage: implications for bisphenol A developmental toxicity. Aquat Toxicol. 2012;110–111:114–22. doi: 10.1016/j.aquatox.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 55.Schiller V, Wichmann A, Kriehuber R, Schafers C, Fischer R, Fenske M. Transcriptome alterations in zebrafish embryos after exposure to environmental estrogens and anti-androgens can reveal endocrine disruption. Reprod Toxicol. 2013;42:210–23. doi: 10.1016/j.reprotox.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Schuttler A, Reiche K, Altenburger R, Busch W. The Transcriptome of the Zebrafish Embryo After Chemical Exposure: A Meta-Analysis. Toxicol Sci. 2017;157(2):291–304. doi: 10.1093/toxsci/kfx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walpita CN, Van der Geyten S, Rurangwa E, Darras VM. The effect of 3,5,3′-triiodothyronine supplementation on zebrafish (Danio rerio) embryonic development and expression of iodothyronine deiodinases and thyroid hormone receptors. Gen Comp Endocrinol. 2007;152(2–3):206–14. doi: 10.1016/j.ygcen.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 58.Chang J, Wang M, Gui W, Zhao Y, Yu L, Zhu G. Changes in thyroid hormone levels during zebrafish development. Zoolog Sci. 2012;29(3):181–4. doi: 10.2108/zsj.29.181. [DOI] [PubMed] [Google Scholar]
- 59.Porazzi P, Calebiro D, Benato F, Tiso N, Persani L. Thyroid gland development and function in the zebrafish model. Mol Cell Endocrinol. 2009;312(1–2):14–23. doi: 10.1016/j.mce.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 60.U.S. EPA. [Accessed December 2016];Overview of the First List of Chemicals for Tier 1 Screening under the Endocrine Disruptor Screening Program. 2016 https://www.epa.gov/endocrine-disruption/overview-first-list-chemicals-tier-1-screening-under-endocrine-disruptor.
- 61.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 62.Mandrell D, Truong L, Jephson C, Sarker MR, Moore A, Lang C, Simonich MT, Tanguay RL. Automated zebrafish chorion removal and single embryo placement: optimizing throughput of zebrafish developmental toxicity screens. J Lab Autom. 2012;17(1):66–74. doi: 10.1177/2211068211432197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Truong L, Harper SL, Tanguay RL. Evaluation of Embryotoxicity Using the Zebrafish Model. In: Gautier J-C, editor. Drug Safety Evaluation: Methods and Protocols. Humana Press; Totowa, NJ: 2011. pp. 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.RC. Team. R language definition. Vienna, Austria: R foundation for statistical computing; 2000. [Google Scholar]
- 65.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 66.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silver JD, Ritchie ME, Smyth GK. Microarray background correction: maximum likelihood estimation for the normal-exponential convolution. Biostatistics. 2009;10(2):352–63. doi: 10.1093/biostatistics/kxn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 69.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9) doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kishida M, Callard GV. Distinct Cytochrome P450 Aromatase Isoforms in Zebrafish (Danio rerio) Brain and Ovary Are Differentially Programmed and Estrogen Regulated during Early Development**This research was supported by grants from the National Science Foundation (IBN-96-05053) and the NIH (P42 ES-07381). The nucleotide sequences reported in this paper have been submitted to the GenBank/EMBL Data Bank with accession numbers AF226619 and AF226620. Endocrinology. 2001;142(2):740–750. doi: 10.1210/endo.142.2.7928. [DOI] [PubMed] [Google Scholar]
- 71.Saili KS, Corvi MM, Weber DN, Patel AU, Das SR, Przybyla J, Anderson KA, Tanguay RL. Neurodevelopmental low-dose bisphenol A exposure leads to early life-stage hyperactivity and learning deficits in adult zebrafish. Toxicology. 2012;291(1–3):83–92. doi: 10.1016/j.tox.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haendel MA, Tilton F, Bailey GS, Tanguay RL. Developmental toxicity of the dithiocarbamate pesticide sodium metam in zebrafish. Toxicol Sci. 2004;81(2):390–400. doi: 10.1093/toxsci/kfh202. [DOI] [PubMed] [Google Scholar]
- 73.Teraoka H, Urakawa S, Nanba S, Nagai Y, Dong W, Imagawa T, Tanguay RL, Svoboda K, Handley-Goldstone HM, Stegeman JJ, Hiraga T. Muscular contractions in the zebrafish embryo are necessary to reveal thiuram-induced notochord distortions. Toxicol Appl Pharmacol. 2006;212(1):24–34. doi: 10.1016/j.taap.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 74.Tilton F, La Du JK, Vue M, Alzarban N, Tanguay RL. Dithiocarbamates have a common toxic effect on zebrafish body axis formation. Toxicol Appl Pharmacol. 2006;216(1):55–68. doi: 10.1016/j.taap.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 75.Groeneweg S, Peeters RP, Visser TJ, Visser WE. Triiodothyroacetic acid in health and disease. Journal of Endocrinology. 2017 doi: 10.1530/JOE-17-0113. [DOI] [PubMed] [Google Scholar]
- 76.Hoefig CS, Zucchi R, Köhrle J. Thyronamines and derivatives: physiological relevance, pharmacological actions, and future research directions. Thyroid. 2016;26(12):1656–1673. doi: 10.1089/thy.2016.0178. [DOI] [PubMed] [Google Scholar]
- 77.Takeda T, Suzuki S, Liu RT, DeGroot LJ. Triiodothyroacetic acid has unique potential for therapy of resistance to thyroid hormone. J Clin Endocrinol Metab. 1995;80(7):2033–40. doi: 10.1210/jcem.80.7.7608251. [DOI] [PubMed] [Google Scholar]
- 78.Freitas J, Cano P, Craig-Veit C, Goodson ML, Furlow JD, Murk AJ. Detection of thyroid hormone receptor disruptors by a novel stable in vitro reporter gene assay. Toxicol in Vitro. 2011;25(1):257–66. doi: 10.1016/j.tiv.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 79.Horn S, Kersseboom S, Mayerl S, Muller J, Groba C, Trajkovic-Arsic M, Ackermann T, Visser TJ, Heuer H. Tetrac can replace thyroid hormone during brain development in mouse mutants deficient in the thyroid hormone transporter mct8. Endocrinology. 2013;154(2):968–79. doi: 10.1210/en.2012-1628. [DOI] [PubMed] [Google Scholar]
- 80.Li JJ, Mitchell LH, Dow RL. Thyroid receptor agonists for the treatment of androgenetic alopecia. Bioorg Med Chem Lett. 2010;20(1):306–8. doi: 10.1016/j.bmcl.2009.10.109. [DOI] [PubMed] [Google Scholar]
- 81.McMenamin SK, Bain EJ, McCann AE, Patterson LB, Eom DS, Waller ZP, Hamill JC, Kuhlman JA, Eisen JS, Parichy DM. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345(6202):1358–61. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bagci E, Heijlen M, Vergauwen L, Hagenaars A, Houbrechts AM, Esguerra CV, Blust R, Darras VM, Knapen D. Deiodinase knockdown during early zebrafish development affects growth, development, energy metabolism, motility and phototransduction. PLoS One. 2015;10(4):e0123285. doi: 10.1371/journal.pone.0123285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stinckens E, Vergauwen L, Schroeder AL, Maho W, Blackwell BR, Witters H, Blust R, Ankley GT, Covaci A, Villeneuve DL, Knapen D. Impaired anterior swim bladder inflation following exposure to the thyroid peroxidase inhibitor 2-mercaptobenzothiazole part II: Zebrafish. Aquat Toxicol. 2016;173:204–17. doi: 10.1016/j.aquatox.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 84.Lindsey BW, Smith FM, Croll RP. From inflation to flotation: contribution of the swimbladder to whole-body density and swimming depth during development of the zebrafish (Danio rerio) Zebrafish. 2010;7(1):85–96. doi: 10.1089/zeb.2009.0616. [DOI] [PubMed] [Google Scholar]
- 85.Stewart MD, Wong J. Transcriptional repression by the thyroid hormone receptor: function of corepressor complexes, Current Opinion in Endocrinology. Diabetes and Obesity. 2004;11(4):218–225. [Google Scholar]
- 86.Guo C, Chen X, Song H, Maynard MA, Zhou Y, Lobanov AV, Gladyshev VN, Ganis JJ, Wiley D, Jugo RH, Lee NY, Castroneves LA, Zon LI, Scanlan TS, Feldman HA, Huang SA. Intrinsic expression of a multiexon type 3 deiodinase gene controls zebrafish embryo size. Endocrinology. 2014;155(10):4069–80. doi: 10.1210/en.2013-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernal J. Thyroid Hormones in Brain Development and Function. In: De Groot LJ, Beck-Peccoz P, Chrousos G, Dungan K, Grossman A, Hershman JM, Koch C, McLachlan R, New M, Rebar R, Singer F, Vinik A, Weickert MO, editors. Endotext. South Dartmouth (MA): 2000. [Google Scholar]
- 88.Ferreira AA, Pereira MJ, Manhaes AC, Barradas PC. Ultrastructural identification of oligodendrocyte/myelin proteins in corpus callosum of hypothyroid animals. Int J Dev Neurosci. 2007;25(2):87–94. doi: 10.1016/j.ijdevneu.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 89.Flavin M, Duprat AM, Rosa J. Effect of thyroid hormones on the switch from larval to adult hemoglobin synthesis in the salamander Pleurodeles waltlii. Cell Differ. 1982;11(1):27–33. doi: 10.1016/0045-6039(82)90013-6. [DOI] [PubMed] [Google Scholar]
- 90.Kim JM, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Scientific reports. 2015;5:16714. doi: 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zambrano A, Garcia-Carpizo V, Villamuera R, Aranda A. Thyroid hormone increases bulk histones expression by enhancing translational efficiency. Mol Endocrinol. 2015;29(1):68–75. doi: 10.1210/me.2014-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Escobar-Morreale HF, Serrano-Gotarredona J, Villar LM, Garcia-Robles R, Gonzalez-Porque P, Sancho JM, Varela C. Methimazole has no dose-related effect on the serum concentrations of soluble class I major histocompatibility complex antigens, soluble interleukin-2 receptor, and beta 2-microglobulin in patients with Graves’ disease. Thyroid. 1996;6(1):29–36. doi: 10.1089/thy.1996.6.29. [DOI] [PubMed] [Google Scholar]
- 93.Lervang HH, Moller-Petersen J, Ditzel J. Serum beta 2-microglobulin levels in thyroid diseases. J Intern Med. 1989;226(4):261–4. doi: 10.1111/j.1365-2796.1989.tb01391.x. [DOI] [PubMed] [Google Scholar]
- 94.Roiter I, Da Rin G, De Menis E, Foscolo GC, Legovini P, Conte N. Increased serum beta 2-microglobulin concentrations in hyperthyroid states. J Clin Pathol. 1991;44(1):73–4. doi: 10.1136/jcp.44.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang S-C, Thibodeaux JR, Eastvold ML, Ehresman DJ, Bjork JA, Froehlich JW, Lau C, Singh RJ, Wallace KB, Butenhoff JL. Thyroid hormone status and pituitary function in adult rats given oral doses of perfluorooctanesulfonate (PFOS) Toxicology. 2008;243(3):330–339. doi: 10.1016/j.tox.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 96.Yu YX, Tao S, Liu WX, Lu XX, Wang XJ, Wong MH. Dietary Intake and Human Milk Residues of Hexachlorocyclohexane Isomers in Two Chinese Cities. Environmental Science & Technology. 2009;43(13):4830–4835. doi: 10.1021/es900082v. [DOI] [PubMed] [Google Scholar]
- 97.Lau C, Thibodeaux JR, Hanson RG, Rogers JM, Grey BE, Stanton ME, Butenhoff JL, Stevenson LA. Exposure to perfluorooctane sulfonate during pregnancy in rat and mouse. II: postnatal evaluation. Toxicol Sci. 2003;74(2):382–92. doi: 10.1093/toxsci/kfg122. [DOI] [PubMed] [Google Scholar]
- 98.Coperchini F, Awwad O, Rotondi M, Santini F, Imbriani M, Chiovato L. Thyroid disruption by perfluorooctane sulfonate (PFOS) and perfluorooctanoate (PFOA) Journal of Endocrinological Investigation. 2017;40(2):105–121. doi: 10.1007/s40618-016-0572-z. [DOI] [PubMed] [Google Scholar]
- 99.Yu WG, Liu W, Jin YH. Effects of perfluorooctane sulfonate on rat thyroid hormone biosynthesis and metabolism. Environ Toxicol Chem. 2009;28(5):990–6. doi: 10.1897/08-345.1. [DOI] [PubMed] [Google Scholar]
- 100.Yu WG, Liu W, Liu L, Jin YH. Perfluorooctane sulfonate increased hepatic expression of OAPT2 and MRP2 in rats. Arch Toxicol. 2011;85(6):613–21. doi: 10.1007/s00204-010-0613-x. [DOI] [PubMed] [Google Scholar]
- 101.Shi X, Liu C, Wu G, Zhou B. Waterborne exposure to PFOS causes disruption of the hypothalamus–pituitary–thyroid axis in zebrafish larvae. Chemosphere. 2009;77(7):1010–1018. doi: 10.1016/j.chemosphere.2009.07.074. [DOI] [PubMed] [Google Scholar]
- 102.Guo Y, Zhou B. Thyroid endocrine system disruption by pentachlorophenol: an in vitro and in vivo assay. Aquat Toxicol. 2013;142–143:138–45. doi: 10.1016/j.aquatox.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 103.Cheng Y, Ekker M, Chan HM. Relative developmental toxicities of pentachloroanisole and pentachlorophenol in a zebrafish model (Danio rerio) Ecotox Environ Safe. 2015;112(Supplement C):7–14. doi: 10.1016/j.ecoenv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 104.Xu T, Zhao J, Hu P, Dong Z, Li J, Zhang H, Yin D, Zhao Q. Pentachlorophenol exposure causes Warburg-like effects in zebrafish embryos at gastrulation stage. Toxicol Appl Pharmacol. 2014;277(2):183–91. doi: 10.1016/j.taap.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 105.Xu T, Zhao J, Xu Z, Pan R, Yin D. The developmental effects of pentachlorophenol on zebrafish embryos during segmentation: A systematic view. Scientific reports. 2016;6:25929. doi: 10.1038/srep25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Froehlicher M, Liedtke A, Groh K, Lopez-Schier H, Neuhauss SC, Segner H, Eggen RI. Estrogen receptor subtype beta2 is involved in neuromast development in zebrafish (Danio rerio) larvae. Dev Biol. 2009;330(1):32–43. doi: 10.1016/j.ydbio.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 107.Hao R, Bondesson M, Singh AV, Riu A, McCollum CW, Knudsen TB, Gorelick DA, Gustafsson JA. Identification of estrogen target genes during zebrafish embryonic development through transcriptomic analysis. PLoS One. 2013;8(11):e79020. doi: 10.1371/journal.pone.0079020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bardet PL, Horard B, Robinson-Rechavi M, Laudet V, Vanacker JM. Characterization of oestrogen receptors in zebrafish (Danio rerio) J Mol Endocrinol. 2002;28(3):153–63. doi: 10.1677/jme.0.0280153. [DOI] [PubMed] [Google Scholar]
- 109.Gorelick DA, Watson W, Halpern ME. Androgen receptor gene expression in the developing and adult zebrafish brain. Dev Dyn. 2008;237(10):2987–95. doi: 10.1002/dvdy.21700. [DOI] [PubMed] [Google Scholar]
- 110.McCarthy MM. Estradiol and the Developing Brain. Physiological reviews. 2008;88(1):91–124. doi: 10.1152/physrev.00010.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hao N, Whitelaw ML. The emerging roles of AhR in physiology and immunity. Biochem Pharmacol. 2013;86(5):561–70. doi: 10.1016/j.bcp.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 112.Patel J, Landers K, Li H, Mortimer RH, Richard K. Thyroid hormones and fetal neurological development. Journal of Endocrinology. 2011;209(1):1–8. doi: 10.1530/JOE-10-0444. [DOI] [PubMed] [Google Scholar]
- 113.Zoeller RT, Rovet J. Timing of thyroid hormone action in the developing brain: clinical observations and experimental findings. J Neuroendocrinol. 2004;16(10):809–18. doi: 10.1111/j.1365-2826.2004.01243.x. [DOI] [PubMed] [Google Scholar]
- 114.Morvan-Dubois G, Demeneix BA, Sachs LM. Xenopus laevis as a model for studying thyroid hormone signalling: from development to metamorphosis. Mol Cell Endocrinol. 2008;293(1–2):71–9. doi: 10.1016/j.mce.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 115.Liu YW, Chan WK. Thyroid hormones are important for embryonic to larval transitory phase in zebrafish. Differentiation. 2002;70(1):36–45. doi: 10.1046/j.1432-0436.2002.700104.x. [DOI] [PubMed] [Google Scholar]
- 116.Driessen M, Vitins AP, Pennings JL, Kienhuis AS, Water B, van der Ven LT. A transcriptomics-based hepatotoxicity comparison between the zebrafish embryo and established human and rodent in vitro and in vivo models using cyclosporine A, amiodarone and acetaminophen. Toxicol Lett. 2015;232(2):403–12. doi: 10.1016/j.toxlet.2014.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.