Abstract
Aims/hypothesis
Metabolomic profiling offers the potential to reveal metabolic pathways relevant to the pathophysiology of diabetes and improve diabetes risk prediction.
Methods
We prospectively analysed known metabolites using an untargeted approach in serum specimens from baseline (1987–1989) and incident diabetes through to 31 December 2015 in a subset of 2939 Atherosclerosis Risk in Communities (ARIC) study participants with metabolomics data and without prevalent diabetes.
Results
Among the 245 named compounds identified, seven metabolites were significantly associated with incident diabetes after Bonferroni correction and covariate adjustment; these included a food additive (erythritol) and compounds involved in amino acid metabolism [isoleucine, leucine, valine, asparagine, 3-(4-hydoxyphenyl)lactate] and glucose metabolism (trehalose). Higher levels of metabolites were associated with increased risk of incident diabetes (HR per 1 SD increase in isoleucine 2.96, 95% CI 2.02, 4.35, p=3.18×10−8; HR per 1 SD increase in trehalose 1.16, 95% CI 1.09, 1.25, p=1.87×10−5), with the exception of asparagine, which was associated with a lower risk of diabetes (HR per 1 SD increase in asparagine 0.78, 95% CI 0.71, 0.85, p=4.19×10−8). The seven metabolites modestly improved prediction of incident diabetes beyond fasting glucose and established risk factors (C statistics 0.744 vs 0.735, p=0.001 for the difference in C statistics).
Conclusions/interpretation
Branched chain amino acids may play a role in diabetes development. Our study is the first to report asparagine as a protective biomarker of diabetes risk. The serum metabolome reflects known and novel metabolic disturbances that improve prediction of diabetes.
Keywords: Amino acids, Branched chain amino acids, Diabetes, Metabolic pathways, Metabolomics
Introduction
The overall burden of type 2 diabetes in the USA is high and is increasing [1,2]. It has been estimated that 21 million adults or approximately 10% of the US population had type 2 diabetes in 2010, and the prevalence of diabetes has nearly doubled in the past two decades [1]. Risk factors for diabetes, particularly obesity, are well characterised [3]. However, the metabolic disturbances leading to the development of diabetes are complex and not yet fully understood.
Recent advances in metabolomic profiling allow for the comprehensive characterisation of metabolism through the detection of many, small metabolites [4]. An untargeted and unbiased metabolomic approach maximises the potential for the discovery of novel markers and could provide new insights about the pathophysiology of diabetes [5]. Given the availability of metabolomic data and the ascertainment of diabetes incidence, the Atherosclerosis Risk in Communities (ARIC) study offers an opportunity to characterise the metabolomic fingerprint of diabetes.
In a systematic review and meta-analysis, 19 prospective studies were identified that investigated metabolites and risk of diabetes [6]. In a pooled analysis of 1940 individuals with diabetes from a total of 8000 participants, higher levels of branched chain amino acids (isoleucine, leucine and valine) and aromatic amino acids (tyrosine and phenylalanine) were significantly associated with a higher risk of incident diabetes. Glycine and glutamine were inversely associated with diabetes risk. The majority of these studies were conducted in European and US white study populations, with the exception of the Insulin Resistance Atherosclerosis Study (IRAS), which included European, Hispanic and African-American study participants and the Strong Heart Family Study of American Indians [7,8]. The individual studies adjusted for a limited number of covariates, and not all studies adjusted for fasting glucose.
The objective of the present study was to examine known and novel blood biomarkers identified through an untargeted metabolomic profile in association with incident diabetes in the community-based ARIC study population. The identification of novel diabetes biomarkers could advance our knowledge of the pathophysiological mechanisms underlying diabetes, and could improve the ability to predict the future development of diabetes.
Methods
Study design and study population
The ARIC study is a community-based cohort study in which 15,792 participants were randomly selected and recruited from four study centres: suburban Minneapolis, MN; Washington County, MD; Jackson, MS; and Forsyth County, NC. At the time of study enrolment in 1987–1989 (visit 1), participants were 45–64 years of age. Participants attended subsequent follow-up study visits in 1990–1992 (visit 2), 1993–1995 (visit 3), 1996–1998 (visit 4) and 2011–2013 (visit 5). For the present study, we conducted a prospective analysis of serum metabolites and diabetes incidence among ARIC study participants with available metabolomics data.
The study population for the present study consisted of black and white participants for whom metabolomic profiling was performed using fasting serum specimens that had been stored at −80°C since collection at baseline (visit 1, 1987–1989). Those participants without available metabolomics data, those with missing covariates, those who were not fasting at baseline and those with diabetes at baseline were excluded from the analysis. Prevalent diabetes at baseline was defined as fasting glucose ≥ 7.0 mmol/l, non-fasting glucose ≥ 11.1 mmol/l, self-reported diagnosis of diabetes or use of medication for diabetes within the previous 2 weeks. The analytic sample size for the present study was 2939. Study participants provided informed consent, the protocol was approved by the institutional review board, and procedures were followed in accordance with the Declaration of Helsinki.
Participants included in this analysis (n=2939) were generally similar to the overall ARIC study population (n=15,792) with respect to baseline characteristics (see electronic supplementary material [ESM] Table 1). By design, there was a larger proportion of African-Americans (56.7% vs 27.0%, respectively) and a lower mean blood level of fasting glucose (5.5 mmol/l vs 6.0 mmol/l, respectively) in the analytic study population compared with the ARIC study population; this was due to the exclusion of participants with diabetes at baseline in this analysis of incident diabetes.
Metabolomic profiling
Metabolites were measured from stored serum specimens by Metabolon (Durham, NC, USA) using an untargeted approach with a Waters ACQUITY ultra-performance liquid chromatography system and a ThermoFisher Scientific Q-Exactive high resolution mass spectrometer with a heated electrospray ionisation source and Orbitrap mass analyser [9]. Metabolomic profiling was conducted in two batches. The first batch was a random sample of ARIC study participants, and the second batch consisted of participants with sequencing data. In the present study, we included the metabolites that were detected in both black and white participants (corresponding with the two batches) and had a low rate of missing values (≤25%). For the remaining 285 metabolites that were detected and semi-quantified in the two batches, outliers were winsorised at the 99% level [10]. Missing values were imputed to the lowest detectable value for that metabolite within each batch. Metabolites were normalised to the median and then log-transformed.
In a subset of 97 specimens profiled in both batches, the Pearson correlation coefficient ranged from −0.09 to 0.99, with a mean of 0.63 and median of 0.71. Forty metabolites with a weak correlation (r<0.3) between batches were excluded from the analysis. After applying these inclusion criteria, 245 named metabolites were included in the analysis. Within this subset, there was a high correlation (r>0.9) between the glucose metabolite detected by this untargeted platform and glucose level measured using the standard clinical assay.
Ascertainment of incident diabetes
The incidence of diabetes was ascertained from baseline through to the end of follow-up on 31 December 2015. Incident diabetes was defined as elevated glucose at any of the four subsequent study visits (fasting glucose ≥7.0 mmol/l or non-fasting glucose ≥11.1 mmol/l), self-report of a diabetes diagnosis at a study visit or annual follow-up telephone interview, or self-report of diabetes medication use during a study visit or annual follow-up telephone interview [11]. Blood glucose levels were measured using the modified hexokinase/glucose-6-phosphate dehydrogenase method.
Medical history and medication use were assessed via an in-person questionnaire with a trained interviewer at each of the study visits. Participants were asked to fast for 12 h before the study visit. Fasting status was defined as at least 8 h since the last time food had been consumed. Annual follow-up telephone interviews were conducted to ascertain medication use and health status.
Measurement of covariates
Structured questionnaires were administered by trained study staff at the baseline study visit in order to collect information on demographics (age, sex, and race), socioeconomic status (education level), health behaviours (smoking status and physical activity) and health history (history of cardiovascular disease). Anthropometrics, including height and weight, were measured during the baseline study visit. BMI was calculated as weight in kilograms divided by the square of the height in meters. Blood pressure was measured three times using a random zero sphygmomanometer after resting for 5 min and after avoiding physical activity, smoking, food consumption and cold weather for 30 min. The mean of the second and third blood pressure measurements was used in the analysis. Blood specimens were collected in order to quantify biochemical indicators of health status.
HDL-cholesterol was determined by measuring cholesterol in the supernatant fraction after precipitation with magnesium chloride and dextran sulphate. Total cholesterol and triacylglycerol were measured using enzymatic methods. LDL-cholesterol was calculated using the Friedewald equation based on measured levels of total cholesterol, HDL-cholesterol and triacylglycerol [12]. Serum creatinine was measured by the modified kinetic Jaffé method. (eGFR was calculated using the 2009 Chronic Kidney Disease Epidemiology equation based on serum creatinine, age, sex and race [13].
Statistical analysis
We reported baseline characteristics using descriptive statistics for the overall study population and according to incident diabetes status. We used Cox proportional hazards regression to evaluate the prospective association between metabolites and incident diabetes. HRs and corresponding 95% CI were calculated per 1 SD increase in each log-transformed metabolite. We compared three multivariable regression models to account for potential confounding factors. Model 1 was minimally adjusted for demographic characteristics (age, sex and race) and study design features (centre and batch). To identify metabolites that were associated with incident diabetes independent of established diabetes risk factors, model 2 included all the variables in model 1 plus education level, systolic and diastolic blood pressures, BMI, HDL-cholesterol, LDL-cholesterol, smoking status, physical activity level, history of cardiovascular disease and eGFR. To examine whether any of the metabolites were associated with incident diabetes independent of the strongest biomarker of diabetes status, model 3 included all the variables in model 2 plus fasting glucose measured as per the ARIC study protocol as described above. We calculated Harrell’s C statistic for models with and without the significant metabolites, and tested for the difference between C statistics in order to evaluate the ability of the metabolites to improve the prediction of incident diabetes beyond established risk factors for diabetes (model 2) and fasting glucose (model 3) [14].
To reduce the likelihood of detecting false-positive findings, we adjusted the significance threshold by the Bonferroni method (0.05/245=2.04×10−4) to account for multiple comparisons. The strength of the association (HRs) is presented for all three models for the metabolites that were significantly associated with incident diabetes in model 1. Metabolites were plotted according to the size of the p value. We calculated Pearson’s correlation coefficients between all significant metabolites to describe their interrelationship. We stratified by race and tested for the interaction.
Results
In our study population of 2939 participants, the mean age at baseline was 53.3 years, mean BMI was 28.2 kg/m2, 59.7% were female and 56.7% were black (Table 1). A total of 1126 study participants developed diabetes over a median follow-up of 20 years. Those participants who developed diabetes during follow-up had higher systolic and diastolic blood pressures, BMI and eGFR; lower HDL-cholesterol and level of education; they were also more likely to be black than those who did not develop diabetes.
Table 1.
Baseline characteristics in the overall study population and according to incident diabetes status during 20 years of follow-up in a subset of ARIC study participants
| Variable | Overall (N=2939) | No incident diabetes (N=1813) | Incident diabetes (N=1126) |
|---|---|---|---|
| Age (years) | 53.3 (5.7) | 53.6 (5.8) | 52.8 (5.5) |
| Female sex | 1755 (59.7) | 1081 (59.6) | 674 (59.9) |
| African-American | 1665 (56.7) | 966 (53.3) | 699 (62.1) |
| Study centre | |||
| Forsyth County, NC | 485 (16.5) | 309 (17.0) | 176 (15.6) |
| Jackson, MI | 1566 (53.3) | 907 (50.0) | 659 (58.5) |
| Minneapolis, MN | 455 (15.5) | 316 (17.4) | 139 (12.3) |
| Washington County, MD | 433 (14.7) | 281 (15.5) | 152 (13.5) |
| Batch | |||
| Batch 1 | 1275 (43.4) | 735 (40.5) | 540 (48.0) |
| Batch 2 | 1664 (56.6) | 1078 (59.5) | 586 (52.0) |
| Education (years) | |||
| ≤11 | 835 (28.4) | 474 (26.1) | 361 (32.1) |
| 12–16 | 1029 (35.0) | 646 (35.6) | 383 (34.0) |
| 17–21 | 1075 (36.6) | 693 (38.2) | 382 (33.9) |
| Systolic blood pressure (mmHg) | 122.8 (19.8) | 121.4 (19.9) | 125.1 (19.4) |
| Diastolic blood pressure (mmHg) | 76.3 (12.3) | 75.3 (12.5) | 77.8 (11.8) |
| BMI (kg/m2) | 28.2 (5.6) | 27.2 (5.1) | 30.0 (6.0) |
| HDL-cholesterol (mmol/l) | 1.4 (0.5) | 1.5 (0.5) | 1.3 (0.4) |
| LDL-cholesterol (mmol/l) | 3.5 (1.0) | 3.5 (1.0) | 3.6 (1.0) |
| Physical activity index | 2.3 (0.8) | 2.3 (0.8) | 2.3 (0.7) |
| eGFR (ml min−1 [1.73 m]−2) | 106.4 (17.4) | 105.5 (17.5) | 107.7 (17.1) |
| Current smoking | 779 (26.5) | 500 (27.6) | 279 (24.8) |
| History of cardiovascular disease | 269 (9.2) | 154 (8.5) | 115 (10.2) |
| Fasting glucose (mmol/l) | 5.5 (0.6) | 5.3 (0.5) | 5.7 (0.6) |
Data are expressed as means (SD) and n (%)
In model 1, which included age, sex, race, centre and batch, a total of 73 metabolites were significantly associated with incident diabetes, representing eight classifications of compounds: amino acid (28), carbohydrate (4), cofactors and vitamins (1), energy (1), lipid (23), nucleotide (2), peptide (8), and xenobiotics (6) (ESM Table 2). For example, higher levels of glycine were strongly associated with a reduced risk of developing diabetes (HR per 1 SD increase 0.44; 95% CI 0.36, 0.55; p=1.3×10−13). For each SD increase in serum level of log-transformed glucose, the risk of incident diabetes was 14 times higher (HR 14.14; 95% CI 10.17, 19.66; p=6.9×10−56).
After adjustment for age, sex, race, centre, batch, education level, systolic blood pressure, diastolic blood pressure, BMI, HDL-cholesterol, LDL-cholesterol, smoking status, physical activity level, history of cardiovascular disease and eGFR (model 2), 47 serum metabolites remained significantly associated with incident diabetes after Bonferroni correction (ESM Table 2). The 47 serum metabolites were similarly representative of a wide variety of metabolic pathways, including amino acid (17), carbohydrate (5), energy (1), lipid (16), nucleotide (1), peptide (4) and xenobiotics (3). The majority of these metabolites (44/47, 94%) were significant in both model 1 and model 2. Three additional metabolites were statistically significant in model 2 [acisoga (N-[3-(2-oxopyrrolidin-1-yl)propyl]acetamide): HR 0.79; 95% CI 0.70, 0.89; p=1.59×10−4; erythronate: HR 1.53; 95% CI 1,23, 1.91; p=1.56×10−4; and eicosenoate: HR 1.35; 95% CI 1.16, 1.57; p=1.04×10−4] but not in model 1.
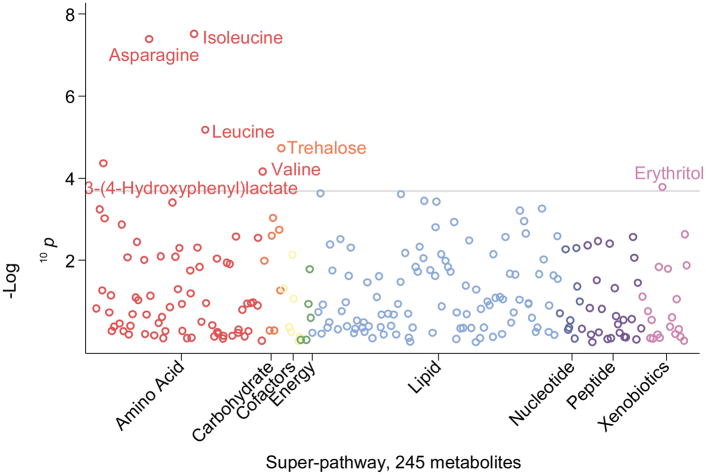
The magnitude of the associations of the majority of metabolites with future diabetes risk was substantially attenuated after additional adjustment for fasting glucose (model 3) (ESM Table 2). A total of seven metabolites remained significantly associated with incident diabetes, representing three classifications of metabolites: amino acid [isoleucine, asparagine, leucine, 3-(4-hydroxyphenyl)lactate and valine], carbohydrate (trehalose), and a xenobiotic or food additive (erythritol) (Table 2). The most robust associations between serum metabolites and incident diabetes in model 3 were observed for the branched chain amino acids: isoleucine (HR 2.96; 95% CI 2.02, 4.35), leucine (HR 2.37; 95% CI 1.63, 3.45) and valine (HR 2.41; 95% CI 1.56, 3.72). There was an inverse association between serum levels of asparagine and incident diabetes (HR 0.78; 95% CI 0.71, 0.85; p=4.19×10−8). The metabolites that had the smallest p values for their association with incident diabetes were involved in amino acid metabolism: asparagine, isoleucine and leucine (Fig. 1).
Table 2.
Serum metabolites significantly associated with incident diabetes according to metabolic pathway
| Pathway and sub-pathway | Metabolites | HR (95% CI)a | p value |
|---|---|---|---|
| Amino acid | |||
| Leucine, isoleucine and valine metabolism | Isoleucine | 2.96 (2.02, 4.35) | 3.18×10−8 |
| Alanine and aspartate metabolism | Asparagine | 0.78 (0.71, 0.85) | 4.19×10−8 |
| Leucine, isoleucine and valine metabolism | Leucine | 2.37 (1.63, 3.45) | 6.78×10−6 |
| Phenylalanine and tyrosine metabolism | 3-(4-Hydroxyphenyl)lactate | 1.55 (1.26, 1.92) | 4.11×10−5 |
| Leucine, isoleucine and valine metabolism | Valine | 2.41 (1.56, 3.72) | 6.95×10−5 |
|
| |||
| Carbohydrate | |||
| Disaccharides and oligosaccharides | Trehalose | 1.16 (1.09, 1.25) | 1.87×10−5 |
|
| |||
| Xenobiotic | |||
| Food component/plant | Erythritol | 1.66 (1.28, 2.16) | 1.63×10−4 |
HR per 1 SD increase in the log-transformed metabolite in multivariable regression models adjusted for covariates in model 3: age, sex, race, centre, batch, education level, systolic blood pressure, diastolic blood pressure, BMI, HDL-cholesterol, LDL-cholesterol, smoking status, physical activity level, history of cardiovascular disease, eGFR and fasting glucose
Fig. 1.
Plot of −log10 p values for the adjusted association between serum metabolites and incident diabetes mellitus (DM); adjusted for the covariates in model 3: age, sex, race, centre, batch, education level, systolic blood pressure, diastolic blood pressure, BMI, HDL-cholesterol, LDL-cholesterol, smoking status, physical activity level, history of cardiovascular disease, eGFR and fasting glucose. The width of each category of metabolites (super-pathway) reflects the number of metabolites within that category that were detected by the untargeted metabolomic approach in this study population
There was no statistically significant interaction for the association between the seven metabolites and incident diabetes by race (ESM Table 3). The direction of the associations were the same, and the strength of the associations were relatively similar, for the seven metabolites and for incident diabetes for the two race groups.
The branched chain amino acids (isoleucine, leucine and valine) were strongly correlated with each other (r >0.83; ESM Table 4). There was a moderate correlation between 3-(4-hydroxyphenyl)lactate and the branched chain amino acids (r=0.42–0.50). Erythritol was weakly correlated with the branched chain amino acids (r=0.23 to 0.28) and 3-(4-hydroxyphenyl)lactate (r=0.31). Asparagine and trehalose were not correlated or were weakly correlated with all other metabolites (r=−0.11–0.11).
The seven metabolites—isoleucine, asparagine, leucine, 3-(4-hydroxyphenyl)lactate, valine, trehalose and erythritol—improved prediction of incident diabetes when added to a model with established diabetes risk factors in model 2 (C statistic [95% CI] in the model without metabolites 0.669 [0.653, 0.684] vs the model including all seven metabolites 0.695 [0.680, 0.709]; p value for difference in C statistics <0.001; Table 3). The seven metabolites also improved the prediction of incident diabetes beyond fasting glucose and the other risk factors in model 3 (C statistic [95% CI] in the model without metabolites 0.735 [0.721, 0.749] vs the model including all seven metabolites 0.744 [0.730, 0.758]; p value for difference in C statistics=0.001).
Table 3.
Prediction of incident diabetes with seven significant metabolites beyond diabetes risk factors and fasting glucose
| Model | C statistic (95% CI) | Difference (95% CI) in C statistics | p value for difference in C statistics |
|---|---|---|---|
| Model 2a | 0.669 (0.653, 0.684) | 1 (Ref) | – |
| Model 2a + metabolitesb | 0.695 (0.680, 0.709) | 0.026 (0.016, 0.036) | <0.001 |
|
| |||
| Model 3c | 0.735 (0.721, 0.749) | 1 (Ref) | – |
| Model 3c + metabolitesb | 0.744 (0.730, 0.758) | 0.009 (0.004, 0.014) | 0.001 |
Model 2 was adjusted for age, sex, race, centre, batch, education level, systolic blood pressure, diastolic blood pressure, BMI, HDL-cholesterol, LDL-cholesterol, smoking status, physical activity level, history of cardiovascular disease and eGFR
Metabolites: isoleucine, asparagine, leucine, trehalose, 3-(4-hydroxyphenyl)lactate, valine and erythritol
Model 3 was adjusted for all the covariates in model 2 plus fasting glucose
Ref, reference
Discussion
In this study of 2939 middle-aged, black and white men and women, we identified seven named compounds that were independently associated with the development of diabetes over 20 years of follow-up after accounting for sociodemographics, diabetes risk factors and fasting glucose levels. These seven metabolites—isoleucine, leucine, valine, asparagine, 3-(4-hydoxyphenyl)lactate, trehalose and erythritol—improved the prediction of diabetes beyond established diabetes risk factors and fasting glucose. These metabolites represented three distinct categories of metabolic pathways, i.e. amino acids, carbohydrates and a xenobiotic (food additive). In models that were not adjusted for fasting glucose, 47 serum metabolites were significantly associated with diabetes, representing a wide variety of metabolic pathways and suggesting that diabetes is a state of substantial metabolic disruption. The compounds that were detected by our metabolomic platform and found to be associated with incident diabetes consisted of established markers of diabetes, including glucose, and compounds consumed by individuals with diabetes, including erythritol, thereby providing proof of concept for this untargeted metabolomic approach. Novel markers of diabetes were also identified, including branched chain amino acids, asparagine, trehalose and 3-(4-hydoxyphenyl)lactate, which point to potential mechanisms of diabetes development.
Our study findings are consistent with current knowledge about diabetes [6]. In models that were not adjusted for fasting glucose, the metabolite with the greatest magnitude of association with incident diabetes was, as expected, glucose. The concentration of glucose in the blood is the most widely used biomarker to screen and diagnose diabetes [15]. In models that adjusted for fasting glucose, trehalose was the only compound representative of carbohydrate metabolism that remained significantly associated with incident diabetes. Trehalose is a disaccharide of two glucose molecules, which is added to food and other manufactured products to prevent dehydration and protein denaturation [16,17]. In a prior analysis among black participants in the ARIC study, this serum metabolite was reported to be significantly associated with the TREH genetic variant as well as incident diabetes [18]. Individuals who were at risk of developing diabetes had elevated serum levels of the glucose metabolite and related compounds involved in carbohydrate metabolism, even after excluding participants with diabetes at baseline.
This untargeted metabolomic profile also included xenobiotics or exogenous substances, such as food components and drugs. Erythritol was significantly associated with incident diabetes in the fully adjusted model, which probably reflects a higher consumption of this compound among individuals with a higher risk of developing diabetes. Specifically, erythritol is a low-calorie sweetener that is added to food as a substitute for simple sugars since it has little to no impact on blood levels of insulin and glucose [19,20]. Erythritol was previously detected by a metabolomic profile and found to be associated with diabetes in a case–control study of 100 participants nested within the KORA (Cooperative Health Research in the Region of Augsburg) study and with elevated glucose in the TwinsUK cohort consisting of 2204 women [21,22].
The class of metabolites with the most significant hits for the association with diabetes was amino acids. It is noteworthy that higher serum levels of all of the branched chain amino acids (leucine, isoleucine and valine) were associated with an increased risk of diabetes. Even after adjustment for baseline glucose, the branched chain amino acids remained statistically significantly associated with incident diabetes. In a meta-analysis of eight prospective studies with metabolomic profiling, branched chain amino acids were consistently and significantly associated with diabetes and other measures of impaired glucose metabolism [6,7,23–29]. However, the aetiology of risk of diabetes mediated by branched chain amino acids has yet to be determined. One purported mechanism is that leucine activates mTORC-1 (mammalian target of rapamycin complex-1) and S6K1 (ribosomal protein S6 kinase), leading to serine phosphorylation of IRS-1 and IRS-2, which results in insulin resistance [30]. Another theory is that the metabolism of branched chain amino acids leads to an accumulation of toxic intermediates, beta cell mitochondrial dysfunction and insulin resistance [31,32].
In addition to the three branched chain amino acids, we identified two other amino acid-related metabolites that were significantly associated with incident diabetes, i.e. 3-(4-hydroxyphenyl)lactate and asparagine. The metabolite 3-(4-hydoxyphenyl)lactate is a byproduct of the degradation of tyrosine, an aromatic amino acid [33]. Whereas the aromatic amino acids tyrosine and phenylalanine have been consistently associated with diabetes risk in a meta-analysis of prospective studies with metabolomic profiling, 3-(4-hydoxyphenyl)lactate has not previously been identified as a compound of interest [6]. Tyrosine is considered to be both glucogenic and ketogenic in that the catabolism of tyrosine yields fumarate, which is an intermediate of the tricarboxylic acid (TCA) cycle, and acetoacetate, which can be used to synthesise ketone bodies. The process of converting amino acid degradation products to glucose is stimulated by a high blood glucagon to insulin ratio, such as in the setting of untreated diabetes. The metabolite 3-(4-hydoxyphenyl)lactate acts as an antioxidant by decreasing the production of reactive oxidative species, which are present during states of oxidative stress, for example among individuals at risk of developing diabetes [34,35].
Asparagine, an amino acid, was the sole metabolite in our study that had an inverse association with diabetes risk. Similar to tyrosine, asparagine is a glucogenic amino acid because oxaloacetate, a byproduct of asparagine catabolism, can be used in the TCA cycle to synthesise glucose. Asparagine is readily converted to aspartate and then undergoes transamination to form glutamate. Glutamate, along with glycine and cysteine, is a constituent of the tripeptide glutathione, which is a major antioxidant and thus protects against chronic diseases [36,37]. Higher blood levels of glutamine and glutamate have consistently been shown to be associated with a lower risk of diabetes in a meta-analysis of prospective metabolomic research studies [6]. Asparagine was reported as being significantly associated with insulin and HOMA, but not glucose, in the Framingham Heart Study [28]. No known metabolomics studies have previously identified asparagine as an independent predictor of incident diabetes.
Some study limitations should be considered in the interpretation of our results. Using a discovery approach to comprehensively detect a broad spectrum of diabetes biomarkers, we obtained relative measures of serum metabolites. Subsequent research using targeted assays will be needed to quantify absolute levels of promising new markers of diabetes risk. Metabolomic profiling was conducted using specimens in storage for over 20 years. Degradation of metabolites over time would be expected to be non-differential by incident diabetes case status. Furthermore, we found that the correlation between glucose measured with metabolomic profiling and glucose measured using the standard clinical chemistry method was high (>0.9). As with any observational study, the reported associations could, in part, be explained by residual confounding. However, we were able to account for multiple covariates that are established risk factors for diabetes in multivariable regression models. There was a small but statistically significant increase in the C statistic as a measure of diabetes risk prediction with the seven metabolites vs established risk factors. Nonetheless, these metabolites may represent metabolic pathways that would be worthwhile pursuing in future research.
There are several strengths of the present study that deserve mention. Compared with other metabolomics studies, the present study was conducted with a relatively large sample of 2939 study participants, with a substantial number of individuals with incident diabetes identified over an extended follow-up period of over 20 years. The prospective analysis allowed for the characterisation of metabolic disturbances apparent among those individuals at risk of subsequently developing diabetes. Our study included both black and white men and women from four communities in the USA, allowing for broad generalisability. Nonetheless, replication of these results will be necessary in similarly diverse study populations. In addition, we conducted a comprehensive and unbiased examination of the serum metabolomic profile using a leading metabolomics platform providing coverage of known pathways of carbohydrate metabolism and maximising the opportunity for the discovery of new diabetes biomarkers. Finally, we employed a conservative approach to account for multiple testing, i.e. Bonferroni correction, in order to reduce the likelihood of false-positive results. Given that some of the metabolites are correlated with each other, the use of the Bonferroni correction was probably an overly conservative approach and may have resulted in some false-negative results (true associations that we have not detected as statistically significant).
In conclusion, we identified seven serum metabolites that were independently associated with and improved the prediction of incident diabetes after accounting for sociodemographic factors, study design features, established risk factors for diabetes and fasting glucose: isoleucine, leucine, valine, asparagine, 3-(4-hydoxyphenyl)lactate, trehalose and erythritol. These metabolites may be useful as a panel of biomarkers to assess future risk of diabetes. This study provides clues to the early metabolic features associated with future development of diabetes in middle-aged adults, which may inform strategies for the prevention and individualised treatment of diabetes. Future research is warranted to precisely quantify these biomarkers and determine their role in diabetes pathophysiology.
Supplementary Material
Research in context.
What is already known about this subject?
The metabolic disturbances leading to diabetes development are complex and not yet fully understood
Untargeted metabolomics allows for the potential discovery of novel markers and could provide new insights into the pathophysiology of diabetes
In previous studies, higher levels of branched chain and aromatic amino acids were associated with a higher risk of incident diabetes, and glycine and glutamine were inversely associated with diabetes risk
What Is the key question?
Are blood biomarkers identified through untargeted metabolomics associated with incident diabetes?
What are the new findings?
Our study is the first to report asparagine as a protective biomarker of diabetes risk
Seven metabolites significantly improved prediction of incident diabetes beyond fasting glucose and established risk factors; these included a food additive (erythritol) and compounds involved in amino acid metabolism [isoleucine, leucine, valine, asparagine, 3-(4-hydoxyphenyl)lactate] and glucose metabolism (trehalose)
How might this Impact on clinical practice In the foreseeable future?
This study provides clues to the early metabolic features associated with future development of diabetes in middle-aged adults, which may inform strategies for the prevention and individualised treatment of diabetes
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions.
Funding The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). CMR is supported by a mentored research scientist development award from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (K01 DK107782). ES is supported by NIH/NIDDK grants K24 DK106414 and R01 DK089174. JC is partially supported by the Chronic Kidney Disease Biomarkers Consortium from the NIDDK (U01 DK085689). AK is supported by Deutsche Forschungsgemeinschaft (DFG 3598/3-1 and DFG 3598/4-1).
Abbreviations
- ARIC
Atherosclerosis Risk in Communities
- TCA
Tricarboxylic acid
Footnotes
Data availability Selected data elements of the ARIC study are available upon request through the National Health, Lung, and Blood Institute (NHLBI) Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC).
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement CMR proposed the study, planned the statistical analysis and wrote the manuscript. EB obtained the metabolomics data. BY, ZZ, PC and AT contributed to the statistical analysis. AK, LEW, JC, EB and ES provided methodological and content-related expertise. All authors provided substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; drafted the article or revised it critically for important intellectual content; and provided final approval of the version to be published. CMR is responsible for the integrity of the work as a whole.
References
- 1.Selvin E, Parrinello CM, Sacks DB, Coresh J. Trends in prevalence and control of diabetes in the United States, 1988–1994 and 1999–2010. Ann Intern Med. 2014;160:517–525. doi: 10.7326/M13-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988–2012. JAMA. 2015;314:1021–1029. doi: 10.1001/jama.2015.10029. [DOI] [PubMed] [Google Scholar]
- 3.DeFronzo RA, Ferrannini E, Groop L, et al. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 4.Tzoulaki I, Ebbels TM, Valdes A, Elliott P, Ioannidis JP. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol. 2014;180:129–139. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 5.Pallares-Mendez R, Aguilar-Salinas CA, Cruz-Bautista I, Del Bosque-Plata L. Metabolomics in diabetes, a review. Ann Med. 2016;48:89–102. doi: 10.3109/07853890.2015.1137630. [DOI] [PubMed] [Google Scholar]
- 6.Guasch-Ferre M, Hruby A, Toledo E, et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39:833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer ND, Stevens RD, Antinozzi PA, et al. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab. 2015;100:E463–E468. doi: 10.1210/jc.2014-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Zhu Y, Hyun N, et al. Novel metabolic markers for the risk of diabetes development in American Indians. Diabetes Care. 2015;38:220–227. doi: 10.2337/dc14-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 10.Dixon WJ. Simplified estimation from censored normal samples. Annals of Mathematical Statistics. 1960;31:385–391. [Google Scholar]
- 11.Selvin E, Steffes MW, Zhu H, et al. Glycated hemoglobin, diabetes, and cardiovascular risk in nondiabetic adults. N Engl J Med. 2010;362:800–811. doi: 10.1056/NEJMoa0908359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 13.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.American Diabetes Association. 2. Classification and diagnosis of diabetes. Diabetes Care. 2017;40(Suppl 1):S11–S24. doi: 10.2337/dc17-S005. [DOI] [PubMed] [Google Scholar]
- 16.Richards AB, Krakowka S, Dexter LB, et al. Trehalose: a review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem Toxicol. 2002;40:871–898. doi: 10.1016/s0278-6915(02)00011-x. [DOI] [PubMed] [Google Scholar]
- 17.Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18:24–36. doi: 10.1002/pro.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu B, Zheng Y, Alexander D, Morrison AC, Coresh J, Boerwinkle E. Genetic determinants influencing human serum metabolome among African Americans. PLoS Genet. 2014;10:e1004212. doi: 10.1371/journal.pgen.1004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda K, Nakayama K, Oku T. Serum glucose and insulin levels and erythritol balance after oral administration of erythritol in healthy subjects. Eur J Clin Nutr. 1994;48:286–292. [PubMed] [Google Scholar]
- 20.den Hartog GJ, Boots AW, Adam-Perrot A, et al. Erythritol is a sweet antioxidant. Nutrition. 2010;26:449–458. doi: 10.1016/j.nut.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Suhre K, Meisinger C, Doring A, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menni C, Fauman E, Erte I, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62:4270–4276. doi: 10.2337/db13-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stancakova A, Civelek M, Saleem NK, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 2012;61:1895–1902. doi: 10.2337/db11-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrannini E, Natali A, Camastra S, et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62:1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang-Sattler R, Yu Z, Herder C, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125:2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tillin T, Hughes AD, Wang Q, et al. Diabetes risk and amino acid profiles: cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall And Brent REvisited) Study. Diabetologia. 2015;58:968–979. doi: 10.1007/s00125-015-3517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson KC, Chen G, Xu Y, Hajnal A, Lynch CJ. Alloisoleucine differentiates the branched-chain aminoacidemia of Zucker and dietary obese rats. Obesity (Silver Spring) 2014;22:1212–1215. doi: 10.1002/oby.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mrochek JE, Dinsmore SR, Ohrt DW. Monitoring phenylalanine-tyrosine metabolism by high-resolution liquid chromatography of urine. Clin Chem. 1973;19:927–936. [PubMed] [Google Scholar]
- 34.Beloborodova N, Bairamov I, Olenin A, Shubina V, Teplova V, Fedotcheva N. Effect of phenolic acids of microbial origin on production of reactive oxygen species in mitochondria and neutrophils. J Biomed Sci. 2012;19:89. doi: 10.1186/1423-0127-19-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107:1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 37.Anderson ME. Glutathione: an overview of biosynthesis and modulation. Chem Biol Interact. 1998;111–112:1–14. doi: 10.1016/s0009-2797(97)00146-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.