Abstract
Mutations in the MeCP2 gene are responsible for the neurodevelopmental disorder Rett syndrome (RTT). MeCP2 is a DNA-binding protein whose abundance and ability to complex with histone deacetylase 3 is linked to the regulation of chromatin structure. Consequently, loss-of-function mutations in MeCP2 are predicted to have broad effects on gene expression. However, to date, studies in mouse models of RTT have identified a limited number of gene or pathway-level disruptions, and even fewer genes have been identified that could be considered amenable to classic drug discovery approaches. Here, we performed RNA sequencing (RNA-seq) on nine motor cortex and six cerebellar autopsy samples from RTT patients and controls. This approach identified 1887 significantly affected genes in the motor cortex and 2110 genes in the cerebellum, with a global trend toward increased expression. Pathway-level analysis identified enrichment in genes associated with mitogen-activated protein kinase signaling, long-term potentiation, and axon guidance. A survey of our RNA-seq results also identified a significant decrease in expression of the CHRM4 gene, which encodes a receptor [muscarinic acetylcholine receptor 4 (M4)] that is the subject of multiple large drug discovery efforts for schizophrenia and Alzheimer’s disease. We confirmed that CHRM4 expression was decreased in RTT patients, and, excitingly, we demonstrated that M4 potentiation normalizes social and cognitive phenotypes in Mecp2+/− mice. This work provides an experimental paradigm in which translationally relevant targets can be identified using transcriptomics in RTT autopsy samples, back-modeled in Mecp2+/− mice, and assessed for preclinical efficacy using existing pharmacological tool compounds.
Introduction
Rett syndrome (RTT) is a neurodevelopmental disorder that affects 1 in 20,000 live births (Rett, 1966). RTT is clinically diagnosed based on the presence of developmental regression, acquired microcephaly, stereotyped hand movements, and apneas, among other symptoms (Hagberg, 2002). Loss-of-function mutations in the MeCP2 gene underlie 90% of RTT cases (Amir et al., 1999). Specifically, pathogenic mutations are believed to either disrupt MeCP2 binding to methylated DNA and/or prevent association with transcriptional repression complexes (Lyst et al., 2013). Promise for the discovery of a treatment for RTT was bolstered when it was shown that re-expression of Mecp2 protein in symptomatic RTT model mice corrected phenotypic deficits (Luikenhuis et al., 2004; Giacometti et al., 2007; Guy et al., 2007), indicating that interventions given after the disease has developed may still be efficacious. However, existing data sets have only characterized a limited number of candidate genes that show dramatic changes in expression induced by loss of MeCP2 function, and even fewer have been identified that can be considered amenable to classic drug discovery approaches.
MeCP2 acts as a transcriptional repressor when associated with the nuclear receptor corepressor/silencing mediator for retinoid and thyroid receptors/histone deacetylase 3 complex, and disruption of this complex would be predicted to result in a loss of heterochromatin and an increase in global transcription. Beyond its role as a repressor, MeCP2 has additional functions as a transcriptional activator, splicing factor, and regulator of chromatin looping (Lyst and Bird, 2015). These diverse cellular functions complicate RTT transcriptional profiling efforts, which can produce variable and often discordant results depending on the brain region, cell type, species, and point in disease progression being assessed. For example, 7.6-fold more genes showed increased expression as opposed to decreased expression in peripheral blood lymphomonocytes from RTT patients (Pecorelli et al., 2013). Conversely, induced pluripotent stem cell–derived neurons carrying pathogenic RTT mutations exhibited a 10-fold decrease in gene expression (Li et al., 2013). Several studies have also surveyed gene expression in the brains of RTT model mice; however, these efforts have yet to identify disrupted genes or pathways with therapeutic utility (Tudor et al., 2002; Nuber et al., 2005; Chahrour et al., 2008; Ben-Shachar et al., 2009; Zhao et al., 2013).
To date, very few expression profiling efforts have been performed in the most relevant species and tissue type: brain samples from RTT patients. Several transcriptomics-based approaches have quantified a global decrease in gene expression in pooled cortical samples from RTT patients (Colantuoni et al., 2001; Lin et al., 2016). Additionally, a number of more targeted approaches have been employed to measure transcript levels of preclinical target genes such as brain-derived neurotrophic factor (BDNF) (Abuhatzira et al., 2007) and potassium-chloride transporter member 5 (KCC2) (Duarte et al., 2013; Tang et al., 2016). These studies have yielded mixed results that are presently not validated in the brains of RTT patients at the protein level.
Recently, we obtained a set of RTT samples from the motor cortex of RTT patients in sufficient quantity to conduct RNA and protein analysis. These samples were previously used to profile the translational relevance of the preclinical target genes GRM5 (mGlu5) (Gogliotti et al., 2016) and GRM7 (mGlu7) (Gogliotti et al., 2017). Building on these data sets, we next asked the question of whether novel genes or pathways could be identified using transcriptomics. We performed RNA sequencing (RNA-seq) analysis on motor cortex and cerebellar RTT autopsy samples and characterized a significant disruption in genes that were previously associated with RTT, as well as several novel targets with inherent therapeutic potential. Among these was the CHRM4 gene, which has been linked to RTT-related diseases like fragile X syndrome and idiopathic autism (Yonan et al., 2003; Veeraragavan et al., 2012). CHRM4 encodes a receptor [muscarinic acetylcholine receptor 4 (M4)] actively being targeted in drug discovery efforts for Alzheimer’s disease and schizophrenia (Foster et al., 2012), and the loss of Chrm4 (Chrm4−/−) results in RTT-like phenotypes in mice (Tzavara et al., 2003; Koshimizu et al., 2012). In addition to patients, our experiments also demonstrated that Chrm4 expression is decreased in Mecp2+/− mice, and excitingly, we show that an M4 positive allosteric modulator (PAM) normalizes social and cognitive phenotypes in this model. These data constitute an investigative paradigm by which changes in expression of a druggable target were first identified in human samples and then back-modeled to mice for further preclinical efficacy studies.
Materials and Methods
Study Design.
The selection of mouse model (Mecp2+/tm1.1 bird), sex, and sample size was based on the standards established by the National Institute of Mental Health and RTT research community (Katz et al., 2012). For phenotypic assays, mice were assigned randomly to dosing groups and quantitation was performed by a researcher that was blinded to genotype and treatment group or by automated software. Statistics were carried out using Prism 6.0 (GraphPad) and Excel (Microsoft). All data shown represent mean ± S.E.M. Statistical significance between groups was determined using two-tailed unpaired or paired Student’s t tests and one- or two-way analysis of variance, with Bonferroni’s or individual Student’s t test post-hoc analysis. Sample size, number of replicates, and statistical test are specified in Figures 1-4.
Fig. 1.
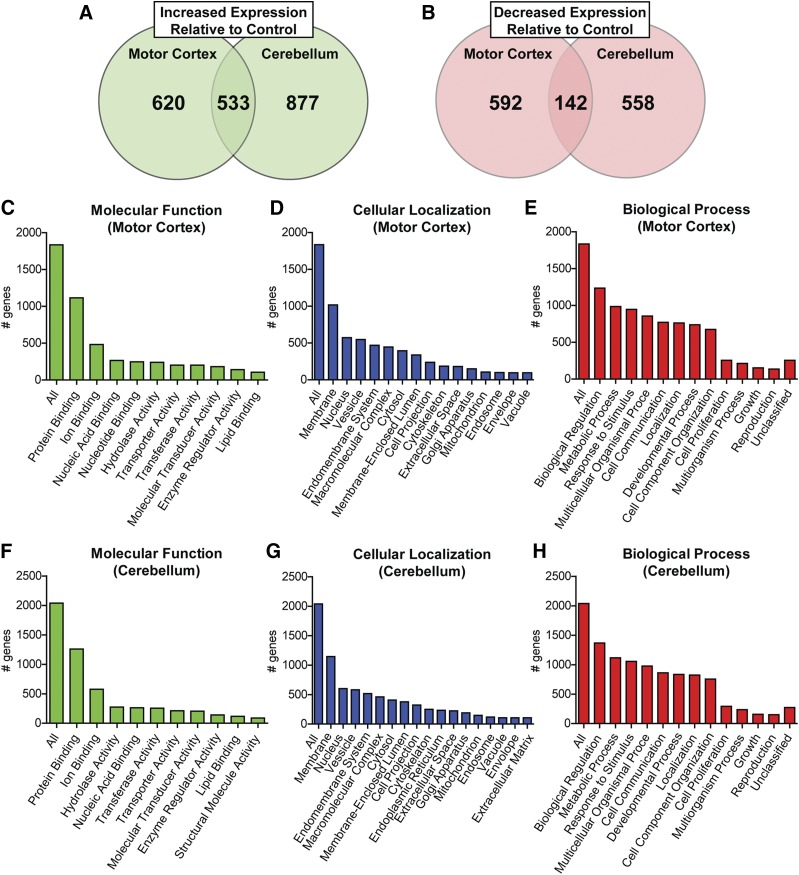
RNA-seq analysis of control and RTT autopsy samples from motor cortex and cerebellum. BA4 (motor cortex) N = 8 control, N = 9 RTT; cerebellum N = 8 control, N = 6 RTT. (A) Significantly increased expression was exhibited in 1153 genes in the motor cortex and 1410 genes in the cerebellum (Cuffdiff, P < 0.05) relative to age, sex, and PMI matched controls. Of these, 533 genes were increased in both brain regions. (B) Relative to controls, 734 genes in the motor cortex and 700 genes in the cerebellum were significantly decreased (Cuffdiff, P < 0.05). Of these, 142 genes were decreased in both brain regions. (C–H) Gene Ontology-Slim analysis of disrupted genes binned by molecular function, cellular localization, and biologic process: (C–E) motor cortex; (F–H) cerebellum.
Fig. 4.
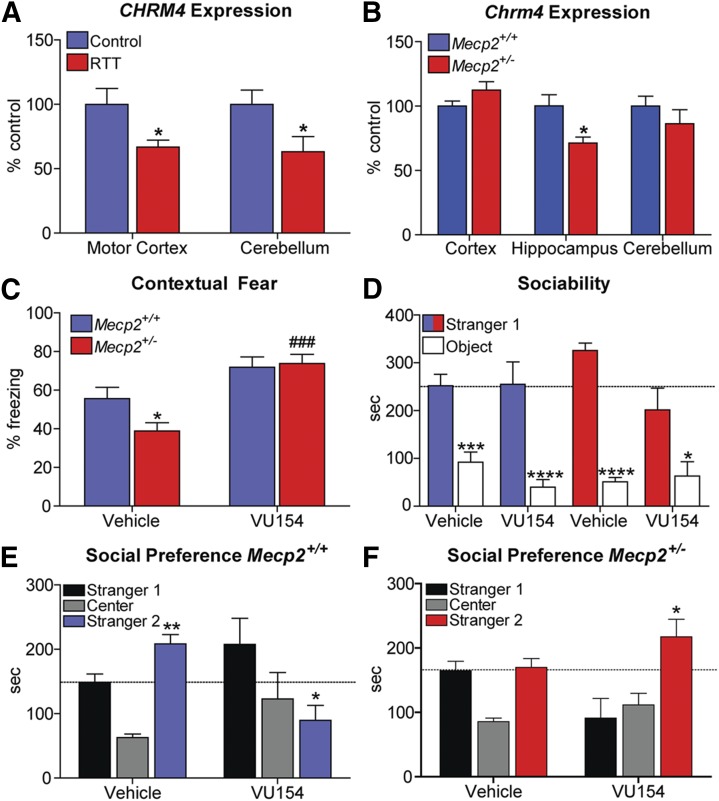
M4 receptor (CHRM4) expression is decreased in RTT patients and M4 potentiation rescues RTT phenotypes in mice. (A) Relative to controls (100%), CHRM4 mRNA expression is significantly decreased in the motor cortex (100.0% ± 12.4% control vs. 66.9% ± 5.3% RTT, *P < 0.05) and cerebellum (100.0% ± 11.1% control vs. 66.2% ± 11.8% RTT, *P < 0.05) of RTT autopsy samples. Student’s t test (B) Chrm4 expression is significantly decreased in the hippocampus of Mecp2+/− mice (20w) (100.0% ± 8.7% Mecp2+/+ vs. 71.3% ± 4.7% Mecp2+/−, *P < 0.05), but not the cerebellum (100.0% ± 7.5% Mecp2+/+ vs. 86.3% ± 6.4% RTT, *P < 0.05) or the cortex (100.0% ± 4.0% Mecp2+/+ vs. 112.4% ± 6.5% Mecp2+/−). N = 7–10 mice/brain region/genotype. (C) Contextual fear freezing response is attenuated in Mecp2+/− mice (Mecp2+/+ 55.6% ± 5.8% vs. Mecp2+/− 38.9% ± 4.2%, *P < 0.05). Administration of the M4 positive allosteric modulator VU0467154 (VU154, 3.3 mg/kg, i.p.) significantly normalizes conditioned fear responses in Mecp2+/− mice (73.8 ± 4.8, ###P < 0.001) relative to vehicle-treated Mecp2+/− mice. N = 10/treatment/genotype. Two-way analysis of variance (ANOVA) with Bonferroni post-hoc analysis. (D) Three-chamber social preference assay. Sociability was unaffected in phase 1 of the social preference assay, regardless of treatment or genotype. (E and F) Phase 2, three-chamber social preference assay. Unlike Mecp2+/+ mice, preference for the novel stranger (Stranger 2) is not observed in vehicle-treated Mecp2+/− mice (Mecp2+/+: Stranger 1: 148.7 ± 13.1s vs. Stranger 2: 208.5 ± 14.5s, **P < 0.01; Mecp2+/−: Stranger 1: 164.6 ± 14.7s vs. Stranger 2: 169.8 ± 14.0s). 3.3 mg/kg VU154 treatment restored social preference in Mecp2+/− mice (Stranger 1: 91.0 ± 30.6s vs. Stranger 2: 217.3 ± 27.5s, *P < 0.05), but disrupted social preference in Mecp2+/+ mice (Stranger 1: 207.6 ± 40.5s vs. Stranger 2: 89.6 ± 23.4s, *P < 0.05), indicative of genotype-specific effects. N = 10/treatment/genotype. Two-way ANOVA with Bonferroni post-hoc analysis. Data are expressed as mean ± S.E.M.
Ethics.
Human samples were obtained from the National Institutes of Health NeuroBioBank (neurobank.nih.gov) under Public Health Service contract HHSN-271-2013-00030. The tissues were post mortem and fully de-identified, and as such are classified as exempt from human subject research regulations. Animal work was conducted under the oversight of the Vanderbilt Institutional Animal Care and Use Committee under protocol M/14/263. Mice sacrificed for gene expression were euthanized using CO2 inhalation at a flow rate of 2 l/min, as recommended by the American Veterinary Medical Association (Schaumburg, IL, USA).
RNA-cDNA Preparation.
Autopsy samples were obtained from the National Institutes of Health NeuroBioBank. For N = 9, the RTT samples were of an age of 16.0 ± 2.3 years and a post-mortem interval (PMI) of 10.5 ± 2.5 hours; N = 8 controls were of an age of 18.8 ± 2.5 years and a PMI of 17.0 ± 2.8 hours. All samples were derived from female patients. Patient mutations assessed were as follows: R168X (one), R255X (four), R270X (two), and MeCP2-mutation negative (two). Approximately 1 g of motor cortex and cerebellum were impact dissociated under dry ice and then pulverized using mortar and pestle under liquid nitrogen. A homogenous mixture of pulverized sample was then allocated for downstream applications. Total RNA was prepared from 100 to 200 mg of tissue using standard Trizol-chloroform methodology. Total RNA was purified and DNase treated using the RNeasy kit (Qiagen, Hilden, Germany). RNA quality was confirmed using BioCube and BioAnalyzer instrumentation by the Vanderbilt Technologies for Advanced Genomics core (at Vanderbilt University Medical Center). cDNA was synthesized from 2 μg total RNA with the SuperScript VILO kit (Thermo Fisher, Waltham, MA). Cortex, cerebellum, and hippocampal samples from 20 week old Mecp2+/− mice were prepared using identical methodology.
RNA Sequencing.
RNA-seq was conducted by Vanderbilt Technologies for Advanced Genomics on total RNA samples that passed BioCube and BioAnalysis quality control. A cDNA library was prepared using the Illumina Access kit (San Diego, CA) and RNA-seq was performed on a HiSEq. 2500 Next Generation Sequencing instrument. Sequencing used 75 base pair, paired-end reads to assist with downstream alignment, and an average of 44.8 ± 1.7 million reads were obtained per sample. Sequencing analysis was done using RNA-seq for Eukaryotes Analysis Kit 3.0 by Maverix Biomics (San Mateo, CA). Raw sequencing reads produced by the Illumina sequencer were quality checked for potential sequencing issues and contaminants using FastQC (Babraham Institute, Babraham, UK). Adapter sequences and primers were trimmed from the sequencing reads using Trimmomatic (Bolger et al., 2014), then followed by removing polyA tail, polyN, and read portions with quality score below 28 using PRINSEQ (Schmieder and Edwards, 2011). Reads with a remaining length of fewer than 20 base pairs after trimming were discarded. A second round of quality check with FastQC was made to compare read quality before and after trimming. Trimmed reads were mapped to the human genome (GRCh37/hg19) using TopHat2 (Kim et al., 2013) with NCBI RefSeq (O’Leary et al., 2016) annotated genes as transcriptome index data. Read alignment coverage and summary statistics for visualization were computed using SAMtools (Li et al., 2009), BEDtools (Quinlan and Hall, 2010), and University of California Santa Cruz Genome Browser utilities (Meyer et al., 2013). Cufflinks 2.2.0 workflow (Trapnell et al., 2012) was adopted for abundance estimation and differential expression analysis. In brief, the sequencing reads aligned to RefSeq (O’Leary et al., 2016) annotated genes were quantified using Cuffquant. Cuffnorm was then used to normalize the gene expression levels across the studied samples with fragments per kilobase of transcript per million computed for sample correlation assessment. Differential expression analysis between samples was performed using Cuffdiff with the computed results from Cuffquant. Significant differentially expressed genes were determined by a false discovery rate of 0.05.
mRNA and Protein Analysis.
Quantitative real-time polymerase chain reaction (QRT-PCR) was performed on BioRad CFX96 instrumentation (Hercules, CA) using Taqman Fast Advanced Master Mix (Thermo Fisher) and Thermo Fisher Assay on Demand primer-probe kits. The assay identifications used were: BDNF (total: Hs02718934_s1, BDNF-I HS00538277_m1, BDNF II Hs00538278_m1, and BDNF III Hs04188535_m1), IGF1 (Hs01547656_m1), SLC12A5 (KCC2, Hs00221168_m1), SLC12A2 (NKCC1, Hs00169032_m1), HSPA6 (Hs04187232_g1), SNORD116 (Hs03309547_s1), CHRM4 (Hs00265219_s1), and Chrm4 (Mm00432514_s1). Human and mouse samples were normalized to the internal control G6PD (Hs00166169_m1; Mm00656735_g1). QRT-PCR data were analyzed using the ΔΔCt method.
Human and mouse protein from 200 mg of tissue was isolated using radioimmunoprecipitation assay buffer. Western blots were run using standard methodology (50 µg total protein per well). Primary antibodies were used at the following concentrations: KCC2 (1:1000, ab49917; Abcam), NKCC1 (1:1000, ab59791; Abcam, Cambridge, UK), HSPA6 (1:1000, ab69408; Abcam), and Tubulin (1:2000, ab6046; Abcam). The fluorescent secondary antibodies used were: goat anti-mouse 680 (1:5000; LiCor, Lincoln, NE) and goat anti-rabbit 800 (1:5000; LiCor). Images were acquired and fluorescence was quantified on a LiCor Odyssey Infrared Imaging System. The enzyme-linked immunosorbent assay kits used were: BDNF (ab99978; Abcam) and IGF1 (ab100545; Abcam). Total protein (150 μg) was loaded per well and samples were run in triplicate in accordance with the manufacturer’s instructions.
Contextual Fear Conditioning.
Mecp2+/− and Mecp2+/+ female mice (18–20 weeks old) were used for these studies. Animals were fear conditioned on day 1 of the task and the percentage of time spent freezing was assessed 24 hours later. On training day of the task, mice were injected 30 minutes prior to the task with either vehicle (10% Tween 80) or 3.3 mg/kg of VU0467154 (10 ml/kg, i.p.). Mice were then placed into an operant chamber with a shock grid (Med Associates Inc., Fairfax, VT) in the presence of a 10% vanilla odor cue. Following a 3-minute habituation period, mice were exposed to two 1 second, 0.7 mA foot shocks spaced 30 seconds apart. Mice remained in the context for an additional 15 seconds after the second foot shock. Twenty-four hours later, mice were placed back into the same shock chamber with a 10% vanilla odor cue and the percentage of time spent freezing during a 3-minute testing period was assessed by video software (Video Freeze; Med Associates Inc.)
Three-Chamber Social Preference Assay.
Thirty minutes prior to testing, 18–20 week old Mecp2+/− and Mecp2+/+ female mice were injected intraperitoneally with either vehicle (10% Tween 80) or 3.3 mg/kg of VU0467154 and returned to their home cage. Mice were then placed in a standard three-chamber apparatus and allowed to habituate for a period of 7 minutes. A novel juvenile mouse (stranger 1) that was restrained in a wire cage was then placed in one of the end chambers, and a toy rubber duck was placed in the opposing end chamber, with the center chamber empty. The test mouse was then allowed to explore all three chambers for 7 minutes before being returned to its home cage. After 3 hours, the test mouse was returned to the three-chamber apparatus and the ability to distinguish between stranger 1 and a novel stranger (stranger 2) was quantified for 7 minutes. Animal tracking was performed using AnyMaze software (Stoelting Co., Wood Dale, IL).
Results
Motor Cortex and Cerebellum RNA Sequencing.
We recently obtained six cerebellum samples from RTT patient autopsies to serve as a complement to our existing work in the motor cortex, as well as eight age, sex, and PMI matched controls [described further in Gogliotti et al. (2016, 2017) and Supplemental Table 1]. As a baseline characterization of these new samples, we quantified MeCP2, mGlu5, and mGlu7 protein expression and observed it to be significantly reduced in a manner similar to what we previously reported in the motor cortex (Supplemental Fig. 1) (Gogliotti et al., 2016, 2017).
We next sought to characterize gene disruption in an unbiased manner by performing differential RNA-seq to compare RTT motor cortex (N = 9) and cerebellum (N = 6) autopsy samples relative to controls (N = 8). A summary of patient demographics and RNA-seq quality control is provided in Supplemental Table 1. RNA-seq experiments were conducted by the Vanderbilt Technologies for Advanced Genomics staff and Maverix Biomics was contracted for bioinformatics analysis. RNA extraction, mRNA enrichment, and cDNA library preparation were performed on individual samples, and sequencing of each sample was conducted with paired-end 75 base pair high throughput sequencing using the Illumina HiSEq. 3000. Following trimming, an average Phred quality score of 41 was obtained in both brain regions, and no individual sample score of under 28 was quantified, indicating >99% accuracy in base assignment (Supplemental Fig. 2). Over 41 million high-quality reads were obtained per sample, of which 76.0% ± 1.2% were nonduplicate reads and 99.7% ± 0.02% were mapped to the University of California, San Francisco (UCSF) human reference genome (Supplemental Table 1). Principal component analysis showed that the RTT and control groups could be differentiated by genotype in both brain regions (Supplemental Fig. 3), and a heat map correlating expression of individual samples is provided in Supplemental Fig. 4.
Differential sequencing using Cuffdiff revealed that 1887 total genes were significantly altered in expression in the motor cortex and 2110 genes were significantly affected in the cerebellum (P < 0.05) (Supplemental Tables 2 and 3). Of these, 1153 genes were increased in the RTT motor cortex, as opposed to 734 decreased (Fig. 1A; Supplemental Table 2). The cerebellum was similarly affected, with 1410 genes increased and 700 decreased (Fig. 1B; Supplemental Table 3). A total of 675 genes showed parallel disruption in both brain regions, as highlighted in Supplemental Table 4. A Kyoto Encyclopedia of Genes and Genomes analysis of affected genes, independent of magnitude or direction, is presented in Supplemental Tables 5 and 6 and Table 1. A Gene Ontology/Slim analysis (webgestalt.org) of both brain regions showed enrichment (∼2-fold) of genes encoding proteins that are located at the membrane and involved in protein binding, as well as proteins whose cellular functions are associated with biologic regulation, metabolic processes, and response to cellular stimuli (Fig. 1, C–H).
TABLE 1.
Kyoto Encyclopedia of Genes and Genomes analysis of RTT RNA-seq gene lists
| Pathway | Number of Genes | FDR |
|---|---|---|
| Motor cortex | ||
| Synaptic vesicle cycle | 26 | 4.30 × 10−7 |
| ECM-receptor interaction | 29 | 1.70 × 10−6 |
| Calcium signaling | 47 | 4.58 × 10−6 |
| Regulation of actin cytoskeleton | 50 | 5.14 × 10−5 |
| MAPK signaling | 55 | 1.25 × 10−4 |
| Long-term potentiation | 23 | 1.42 × 10−4 |
| Focal adhesion | 46 | 1.54 × 10−4 |
| Advanced glycation endproducts signaling pathway | 28 | 2.16 × 10−4 |
| Oxytocin signaling pathway | 38 | 2.29 × 10−4 |
| Aldosterone synthesis and secretion | 24 | 2.92 × 10−4 |
| Cerebellum | ||
| ECM-receptor interaction | 28 | 2.99 × 10−4 |
| Malaria infection immune response | 18 | 3.49 × 10−3 |
| MAPK signaling pathway | 57 | 3.49 × 10−3 |
| Focal adhesion | 47 | 5.13 × 10−3 |
| Axon guidance | 42 | 5.51 × 10−3 |
| Gastric acid secretion | 22 | 8.20 × 10−3 |
| Staphylococcus aureus infection immune response | 18 | 8.20 × 10−3 |
| Cell adhesion molecules | 35 | 8.20 × 10−3 |
| Mineral absorption | 17 | 8.20 × 10−3 |
| Aldosterone synthesis and secretion | 23 | 8.69 × 10−3 |
ECM, extracellular matrix; FDR, false discovery rate; MAPK, mitogen-activated protein kinase.
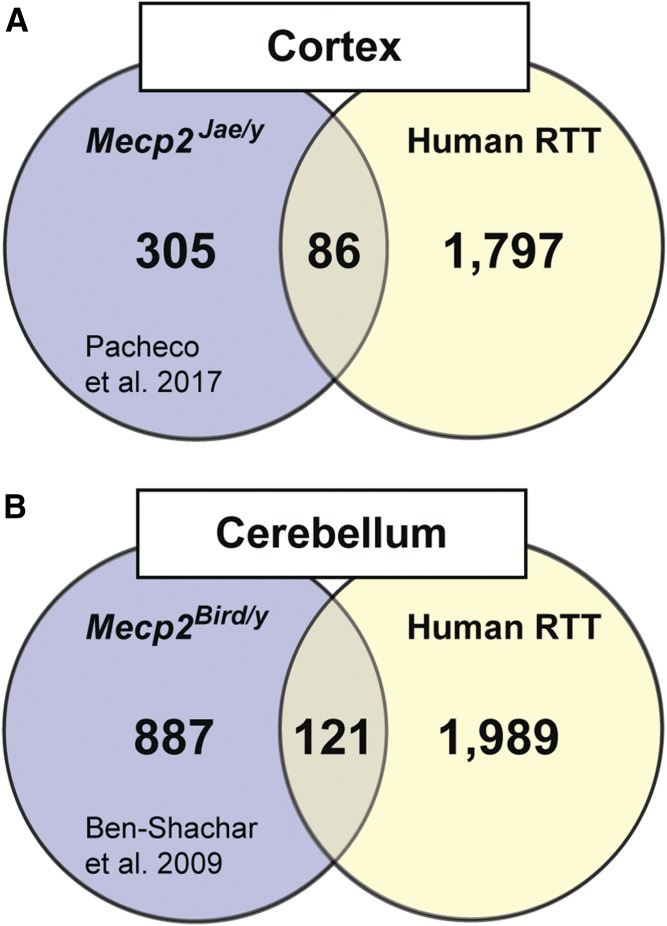
Given that transcriptional profiling has also been conducted in the cortex and cerebellum of RTT mouse models, we next compared the newly generated human data set to what was observed in Mecp2−/y mice. Our RNA-seq analysis identified 1887 genes with significantly disrupted expression in the motor cortex, and a previously study by Pacheco et al. (2017) quantified 391 genes disrupted in total cortex from Mecp2tm1.1Jae/y mice using a similar RNA-seq approach (Fig. 2A; Supplemental Table 7). An alignment of these gene lists indicated that 86 of the genes (21.9%) identified in the mouse were also affected in human patients. Similarly, here we report 2110 genes with significantly disrupted expression in the cerebellum, and a previous study by Ben-Shachar et al. (2009) reported 1008 affected genes in the Mecp2tm1.1bird/y cerebellum. An alignment of each gene list revealed that 121 genes, or 12.0% of the genes identified in the mouse, were also disrupted in human RTT patients (Fig. 2B; Supplemental Table 7). A Kyoto Encyclopedia of Genes and Genomes analysis of overlapping genes from each brain region failed to identify any significant pathway enrichments. While these results must be interpreted in the context of age and sex-related differences, they do highlight the potential challenges associated with translating expression profiling data sets from rodent models to clinical populations.
Fig. 2.
Comparison of transcriptional profiling in RTT patients relative to RTT mouse models reveals minimal overlap. (A) A separate RNA-seq analysis of total cortex in Mecp2Jae/y mice found 391 genes that were significantly disrupted (Pacheco et al., 2017), irrespective of direction, while our analysis of the motor cortex identified 1883 genes. An alignment of these two gene lists identified 86 genes that were similarly affected in both species, with no significant pathway enrichment (>5 genes) as assessed by Kyoto Encyclopedia of Genes and Genomes/Gene Ontology (KEGG-GO) analysis. (B) A microarray analysis of cerebellum in Mecp2Bird/y mice found 1008 genes that were significantly disrupted (Ben-Shachar et al., 2009), irrespective of direction, while our analysis of the cerebellum identified 2110 genes. An alignment of these two gene lists identified 121 genes that were similarly affected in both species, with no significant enrichment (>5 genes) as assessed by KEGG-GO analysis.
Disruption of RTT-Relevant Genes.
It is standard practice to validate a subset of differentially expressed genes using at least one orthogonal technique to assess the accuracy of transcriptomic approaches. To validate our RNA-seq results, we selected several genes that have previously been associated with RTT and/or idiopathic autism and quantified gene expression using QRT-PCR and western blot or enzyme-linked immunosorbent assay. The genes chosen for validation include two targets currently under clinical development for RTT, KCC2 (Duarte et al., 2013; Tang et al., 2016) and IGF1 (Tropea et al., 2009; Castro et al., 2014); a noncoding RNA within the Prader-Willi syndrome locus known to be epigenetically regulated by MeCP2, SNORD116-18 (Makedonski et al., 2005); and a gene implicated in the pathophysiology of idiopathic autism, HSPA6 (Garbett et al., 2008).
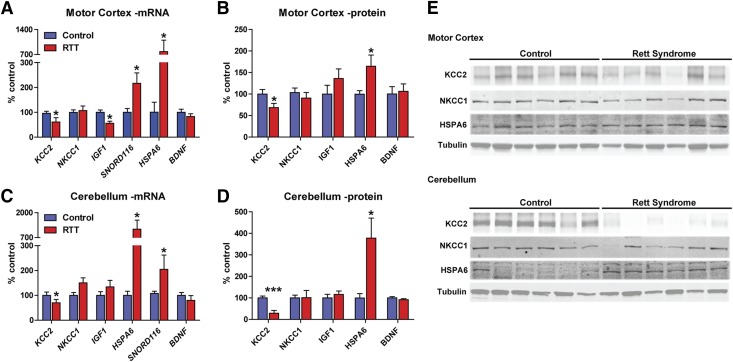
Consistent with our RNA-seq data, KCC2 mRNA and protein expression was significantly decreased in both brain regions, while expression of its co-receptor, NKCC1, was unchanged (Fig. 3). Likewise, IGF1 mRNA was significantly decreased in the motor cortex, but not in the cerebellum, as predicted by RNA-seq (Fig. 3). Interestingly, IGF1 protein was unchanged in both brain regions, suggesting the observed reduction in mRNA did not alter total protein levels for this gene. Consistent with the RNA-seq data, SNORD116-18 and HSPA6 levels were quantified as significantly increased (Fig. 3). HSPA6 is an especially intriguing example of the utility of transcriptional profiling in human patients because there is no murine Hspa6 homolog (Parsian et al., 2000); consequently, HSPA6 may not have been associated with RTT without directly profiling human samples.
Fig. 3.
Expression analysis of genes previously associated with RTT validates RNA-seq results. As orthogonal validation of our RNA-seq results, we selected several genes that had previously been associated with RTT in patients and model mice and quantified their expression using QRT-PCR, fluorescent western blotting, and enzyme-linked immunosorbent assay (ELISA). N = 8 control, N = 9 RTT in motor cortex and N = 6 RTT in cerebellum. (A and B) In the motor cortex, KCC2 expression was significantly reduced at the mRNA (100.0% ± 8.3% control vs. 61.0% ± 17.4% RTT, *P < 0.05) and protein levels (100.0% ± 10.6% control vs. 68.4% ± 9.8% RTT, *P < 0.05) relative to controls, while its co-receptor NKCC1 was unaffected. IGF1 mRNA expression was decreased in RTT samples (100.0 ± 9.3 control vs. 56.0% ± 8.4% RTT, *P < 0.05); however, this failed to translate to the protein level. SNORD116 and HSPA6 expression were significantly increased (SNORD116: 100.0% ± 15.3% control vs. 216.4% ± 42.3% RTT, *P < 0.05; HSPA6: 100.0% ± 40.3% control vs. 781.8% ± 40.3% RTT, *P < 0.05; HSPA6: 100.0% ± 8.3% control vs. 164.7% ± 25.9% RTT, *P < 0.05), as observed using RNA-seq, while the negative control BDNF was unaffected. Two-tailed Student’s t test comparing control vs. RTT for each gene/protein. (C and D) In the cerebellum, KCC2 expression was significantly reduced at the mRNA (100.0% ± 13.6% control vs. 70.2% ± 13.3% RTT, *P < 0.05) and protein levels (100.0% ± 8.5% control vs. 29.0% ± 12.7% RTT, ***P < 0.001) relative to controls, while its co-receptor NKCC1 was unaffected. Similar to motor cortex, SNORD116 and HSPA6 expression was also significantly increased in the cerebellum (SNORD116: 100.0% ± 8.8% control vs. 204.2% ± 57.9%, *P < 0.05; HSPA6: 100.0% ± 16.7% control vs. 1138.4% ± 464.3% RTT, *P < 0.05; HSPA6: 100.0% ± 20.0% control vs. 378.0% ± 93.0% RTT, *P < 0.05), while the negative control BDNF was still unaffected. Paired Student’s t test comparing control vs. RTT for each gene/protein. (E) Representative western blots for KCC2, NKCC1, and HSPA6 expression in human control and RTT patients. Note that IGF1 and BDNF were quantified by ELISA and SNORD116 is a noncoding RNA with no protein correlate.
As a final validation measure, we quantified mRNA and protein levels of the BDNF gene, whose expression would have been predicted to be affected based on extensive mouse and in vitro data (Chang et al., 2006; Wang et al., 2006; Ogier et al., 2007; Schmid et al., 2012), but was not identified in our RNA-seq data set. BDNF expression was measured by QRT-PCR and enzyme-linked immunosorbent assay analysis in both brain regions and no significant changes were observed, which is in agreement with our RNA-seq data (Fig. 3). Individual BDNF isoforms were also binned by shared promoter usage as previously described (Abuhatzira et al., 2007) and quantified by QRT-PCR. In these experiments, only transcripts driven by the BDNF II and III promoters exhibited decreased expression relative to controls, and only in the motor cortex (Supplemental Fig. 5). Since this finding represents a departure from what has been reported in the mouse, we also correlated BDNF protein expression with individual MeCP2 protein levels [presented in Gogliotti et al. (2016) and Supplemental Fig. 1] to determine whether this result could be attributed to preferential X inactivation of the mutant MeCP2 allele. We observed no significant correlation in the motor cortex, but did quantify an R2 value of 0.84 (**P = 0.01) in the cerebellum (Supplemental Fig. 5). It should be noted, however, that total MeCP2 protein was still significantly decreased in the cerebellum (Supplemental Fig. 1), suggesting that favorable X inactivation is likely not linked to BDNF expression in this sample set.
M4 as a Novel Target for RTT.
The overarching goal of performing RNA-seq experiments in autopsy samples was to identify novel genes that are disrupted in RTT patients that could be exploited therapeutically. In this regard, one salient finding of our experiments was that four of the five muscarinic acetylcholine receptors (mAChRs) exhibited significantly disrupted expression in at least one brain region (CHRM1, CHRM2, CHRM3, and CHRM4). Previously, cholinergic dysfunction was associated with altered brainstem activity in Mecp2−/y mice (Oginsky et al., 2014), and more recently has been linked to cognitive and social phenotypes in choline acetyltransferase-Mecp2 knockout mice (Zhang et al., 2016). Importantly, administration of the nonselective mAChR agonist xanomeline improved these same symptom domains in clinical trials for both Alzheimer’s disease and schizophrenia (Bodick et al., 1997; Shekhar et al., 2008). Of the four mAChR genes identified, we considered the CHRM4 (M4) gene to be the most important finding because 1) Chrm4−/− mice have phenotypic overlap in cognitive and social domains with both global and choline acetyltransferase-Mecp2 knockout mice (Tzavara et al., 2003; Koshimizu et al., 2012; Lyst and Bird, 2015; Zhang et al., 2016); 2) changes in Chrm4/CHRM4 expression have been reported in related autism-associated diseases such as fragile X syndrome, Pitt-Hopkins syndrome, and idiopathic autism (Yonan et al., 2003; Veeraragavan et al., 2012; Kennedy et al., 2016); and, 3) there are multiple clinical development campaigns for M4 modulators already underway, which could be expanded to include RTT as an indication (Chan et al., 2008; Bubser et al., 2014; Byun et al., 2014).
To determine whether M4 might be a viable target for RTT, we first confirmed that CHRM4 expression was affected in our RTT samples as quantified during transcriptional profiling. Using QRT-PCR, we confirmed that CHRM4 expression is decreased in the motor cortex (66% of controls, *P < 0.05) (Fig. 4A) and in the cerebellum (63% of controls, *P < 0.05). Note that the decrease cerebellar CHRM4 expression failed to reach significance with RNA-seq analysis due to high variability. However, a significant disruption was quantified using the more targeted QRT-PCR methodology; we were unable to validate any commercially available M4 antibody as selective and thus only transcripts were quantified. To determine whether these findings could be recapitulated in a mouse model of RTT, we harvested cortex, hippocampus, and cerebellum from 20-week-old Mecp2+/− mice and again assessed Chrm4 expression using QRT-PCR. In accordance with the recommendations of the National Institute of Mental Health and the RTT community (Katz et al., 2012), we profiled female Mecp2+/− mice at 20 weeks because this represents a symptomatic time point in a model that replicates the molecular etiology of the disease. We observed a significant decrease in Chrm4 mRNA in the hippocampus (71% of controls, *P < 0.05), but not in the cortex or cerebellum. Hippocampal tissue was not available from patient autopsies; however, these results demonstrate that decreased CHRM4 expression is a conserved finding between RTT patients and mouse models, but not in all brain regions (Fig. 4B).
Given that we quantified decreased Chrm4 expression in the hippocampus, we next sought to determine whether hippocampal-dependent phenotypes in RTT model mice would be responsive to M4 potentiation. During the course of optimization of M4 tool compounds, we have characterized several highly selective PAMs with properties suitable for in vivo studies (Chan et al., 2008; Bubser et al., 2014; Byun et al., 2014). One such compound, VU0467154 (VU154), has previously shown procognitive efficacy in a genetic model of Huntington’s disease and in pharmacological models of schizophrenia (Bubser et al., 2014; Pancani et al., 2015). To assess VU154’s efficacy on cognitive phenotypes in RTT model mice, we performed contextual fear conditioning in 20-week-old Mecp2+/− and Mecp2+/+ female mice. The contextual fear assay is an assessment of learning and memory that consists of placing a mouse in an activity chamber and applying a series of foot shocks. The mouse is then returned to its home cage and placed back in the activity chamber 24 hours later to assess fear expression. The time spent freezing is then used to quantify associative learning with regard to the aversive environment. Either 3.3 mg/kg VU154 or vehicle (10% Tween 80) was administered via intraperitoneal injection 30 minutes prior to training. As demonstrated in Fig. 4C, vehicle-treated Mecp2+/− mice show significantly less freezing in this assay compared with Mecp2+/+ controls. In support of our hypothesis, treatment with VU154 significantly increased freezing in Mecp2+/− mice such that they were indistinguishable from controls (Fig. 4C).
Both Chrm4−/− and cholinergic-Mecp2 knockout mice exhibit social deficits (Koshimizu et al., 2012; Zhang et al., 2016). To determine whether social behavior would be responsive to M4 potentiation, we performed a three-chamber social preference assay. In this assay, VU154- or vehicle-treated mice are first exposed to a stranger mouse (stranger 1) and an inanimate object. The mouse is then returned to its home cage for 3 hours, and placed back into the chamber and allowed to distinguish between the stranger 1 mouse and a novel stranger (stranger 2) as a measure of social recognition and/or preference. In phase 1 of the assay, both vehicle- and VU154-treated mice preferred interactions with the mouse over the toy (Fig. 4D). In phase 2 of the assay, vehicle-treated Mecp2+/+ mice showed preference for the novel stranger 2 mouse, while vehicle-treated Mecp2+/− mice failed to distinguish (Fig. 4E). Similar to contextual fear conditioning, VU154 administration normalized social recognition and/or preference for the novel stranger in Mecp2+/− mice to wild-type levels (Fig. 4F). It is of note that VU154 treatment disrupted social behavior in wild-type mice, which may suggest that precise M4 signaling is required for normal sociability (Koshimizu et al., 2012).
Discussion
Modest expression changes in a broad swath of genes have been reported in the brains of RTT patients and mouse models, some of which are tolerable, while others additively contribute to manifest the RTT phenotype (Colantuoni et al., 2001; Tudor et al., 2002; Nuber et al., 2005; Chahrour et al., 2008; Ben-Shachar et al., 2009; Pecorelli et al., 2013; Zhao et al., 2013; Lin et al., 2016). As such, identifying the proteins and pathways that mediate specific RTT symptoms is a key step toward identifying druggable access points that might be exploited therapeutically. Here, we performed an unbiased assessment of gene expression in motor cortex and cerebellar samples from RTT autopsy samples. The goal of these studies was to identify previously uncharacterized points of intervention in RTT patients that would allow subsequent preclinical discovery efforts to begin from a place of translational relevance. These studies identified over 2000 genes whose expression was significantly affected in the brains of RTT patients by mutations in MeCP2. As proof of concept, this list was then counter-screened for receptor classes believed to be amenable to classic drug discovery approaches and for those where clinical efficacy in neurologic diseases has already been observed. This secondary analysis led us to focus on the M4 receptor, which we quantified as significantly decreased in the motor cortex and cerebellum of RTT patients. We then validated M4 disruption in Mecp2+/− mice, and established that M4 positive modulation has efficacy in preclinical assays of cognition and sociability.
The two main mechanisms by which mutations in MeCP2 become pathogenic are by either preventing binding to methylated DNA or by preventing MeCP2 association with transcriptional repression complexes (Lyst et al., 2013). Based on these data, a loosening of chromatin structure and an overall increase in gene expression would be predicted in RTT. In general, a diminished ability to cluster heterochromatin is observed in vitro; however, gene expression changes in vivo are not always consistent with this model. Similar to what was reported in peripheral blood lymphomonocytes from RTT patients (Pecorelli et al., 2013), we quantified a global increase in gene expression within the motor cortex and cerebellum. However, other studies in induced pluripotent stem cells and in the frontal and temporal cortex of RTT autopsies have reported a global decrease in expression (Li et al., 2013; Lin et al., 2016). While brain region-specific effects and diverse MeCP2 functions seem likely contributors to this discrepancy, other factors like age, sex, PMI, ethnicity, and mutation type may also play a role. Another variable that is difficult to control for is sample size, which is often suboptimal due to the paucity of available autopsy samples. We believe that the current use of nine samples represents the largest transcriptional profiling effort conducted in brain samples from RTT patients to date, and the control cohort was matched for age, sex, and PMI. We focused on patients with three known MeCP2 mutations (R168X, R255X, and R270X), and two patients with MeCP2 mutation-negative RTT; however, the low sample size for each mutation enables the possibility that mutation type also represents a potential source of variability in our study. Such disparity across RTT expression profiling experiments advocates that they be analyzed with the caveat that the aforementioned variables significantly contribute to gene expression patterns, which if not taken into account will preclude comparison across multiple data sets.
Another important finding from our studies was that both targeted quantification of BDNF and the transcriptional profiling efforts of others in mice had limited overlap with the human patient samples examined here (Ben-Shachar et al., 2009; Pacheco et al., 2017). The latter discrepancy may simply be due to the use of male Mecp2−/y mice as opposed to our female RTT patient samples (Ben-Shachar et al., 2009); however, the BDNF data are more difficult to reconcile. The activity-dependent BDNF promoter has been reported to be preferentially affected by loss of Mecp2 in mice (Abuhatzira et al., 2007), and we also observed this in the motor cortex of our human samples. However, total BDNF protein levels have been reported to be decreased in multiple mouse models of RTT (Chang et al., 2006; Wang et al., 2006; Ogier et al., 2007; Schmid et al., 2012), which was not observed in either of the brain regions tested here. In RTT-model mice, decreased BDNF expression is only observed at post-symptomatic ages (Wang et al., 2006). Given that our samples represent two brain regions from advanced symptomatic/terminal patients, they offer a valuable comparator to the preclinical mouse work. When analyzed in concert, these data either suggest that only select BDNF isoforms are affected in clinical RTT populations, which exist in quantities too small to alter total-BDNF quantitation, and/or that a fundamental difference exists between mice and humans with regard to how loss of Mecp2 disrupts BDNF expression.
The main goal of our RNA-seq studies was to identify novel, translationally relevant intervention strategies for RTT. In this regard, we believe that our experiments demonstrating that CHRM4 expression is decreased in RTT patients and model mice represent a significant finding. mAChRs are a family of five G-protein-coupled receptors that can be subdivided into those that are Gq-coupled (M1, M3, and M5), and those that are Gi/o-coupled (M2 and M4). mAChRs garnered intense therapeutic interest following the encouraging results of two clinical trials, where the nonselective agonist xanomeline demonstrated procognitive efficacy in patients with Alzheimer’s disease and schizophrenia (Bodick et al., 1997; Shekhar et al., 2008). Unfortunately, peripherally mediated adverse effects, which are believed to be due to M2 and M3 activation in the gastrointestinal tract, derailed many of these efforts (Ehlert, 2003). However, hope has recently been renewed since it has now been shown that the selective targeting of M4 receptors with PAMs provides a mechanism to conserve efficacy and avoid adverse effects (Chan et al., 2008; Bubser et al., 2014; Byun et al., 2014; Foster et al., 2014; Pancani et al., 2015). This renewed interest has resulted in the development of highly selective tool compounds with properties suitable for in vivo studies (Chan et al., 2008; Bubser et al., 2014; Melancon et al., 2017; Wood et al., 2017). Using one such tool compound, VU0467154, we established that both conditioned fear and social preference phenotypes can be rescued by M4 potentiation in Mecp2+/− mice. These data align closely with experiments demonstrating that cell-specific deletion of Mecp2 from cholinergic neurons results in conditioned fear and social preference deficits (Zhang et al., 2016). Excitingly, this may indicate that M4 receptor modulation is a viable therapeutic approach for RTT that originated from direct expression profiling in patients, and for which much of the early stage drug discovery burden has already been completed. A formalized assessment of M4 PAM efficacy and adverse effect liability will now be required in multiple models of RTT, across a range of concentrations, and in chronic dosing paradigms to further validate this approach. Taken together, we believe that the data presented here justify exploration of M4 potentiation in the context of RTT. Furthermore, Anavex (New York, NY) has recently reported that the sigma 1/M1 agonist ANAVEX 2–73, which is currently undergoing phase 2 clinical trials for Alzheimer’s disease and received orphan drug status for RTT, also has a benefit in RTT model mice. These results may point to the global repression of muscarinic acetylcholine signaling in RTT, and more broadly a responsiveness of RTT model mice to positive modulation of this form of neurotransmission.
Acknowledgments
We acknowledge the patient families, Rettsyndrome.org, Harvard Brain Tissue Resource Center, and University of Maryland Brain Bank for the generous gift of Rett syndrome autopsy samples. We thank Sabina Berretta and Jose Bartolome for assistance in obtaining these samples.
Abbreviations
- BDNF
brain-derived neurotrophic factor
- KCC2
potassium-chloride transporter member 5
- mAChR
muscarinic acetylcholine receptor
- M4
muscarinic acetylcholine receptor 4
- PAM
positive allosteric modulator
- PMI
post-mortem interval
- QRT-PCR
quantitative real-time polymerase chain reaction
- RNA-seq
RNA sequencing
- RTT
Rett syndrome
Authorship Contributions
Participated in research design: Gogliotti, Niswender.
Conducted experiments: Gogliotti, Fisher, Stansley.
Contributed new reagents or analytic tools: Jones, Lindsley, Conn, Niswender.
Performed data analysis: Gogliotti, Niswender.
Wrote or contributed to the writing of the manuscript: Gogliotti, Fisher, Stansley, Niswender.
Footnotes
C.K.J., C.W.L., P.J.C., and C.M.N. are inventors of patents protecting M4 positive allosteric modulators and have received royalties and research funding from AstraZeneca and Lundbeck for the development of M4 positive allosteric modulators for schizophrenia and other central nervous system disorders. This funding was not used to support the studies in the current paper.
The Harvard Brain Tissue Resource Center is supported by a Public Health Service contract [Contract HHSN-271-2013-00030] and the University of Maryland Brain Bank is a Tissue Repository of the National Institutes of Health NeuroBioBank. R.G.G. received support by a Mentored Training Fellowship from Rettsyndrome.org, a Young Investigator Award from Brain and Behavior Research Foundation, and National Institutes of Health National Institute of Mental Health K01 [Grant MH112983]. N.M.F. was supported by National Institutes of Health T32 training grant [Grant GM007628-36], the Vanderbilt Program in Molecular Medicine through Vanderbilt University, and National Institutes of Health F31 [Grant MH113259]. B.J.S. was supported by a National Institutes of Health F32 training grant [Grant MH111124-01]. The authors would also like to acknowledge National Institute of Mental Health U54 [Grant MH084659] (C.W.L.), National Institute of Mental Health R01 [Grant MH087965-01] (P.J.C.), as well as a Basic Research grant from Rettsyndrome.org (C.M.N.), a Treatment Grant from Autism Speaks (C.M.N), and National Institute of Mental Health R21 [Grant MH102548] (C.M.N.).
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. (2007) MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics 2:214–222. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188. [DOI] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. (2009) Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet 18:2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodick NC, Offen WW, Levey AI, Cutler NR, Gauthier SG, Satlin A, Shannon HE, Tollefson GD, Rasmussen K, Bymaster FP, et al. (1997) Effects of xanomeline, a selective muscarinic receptor agonist, on cognitive function and behavioral symptoms in Alzheimer disease. Arch Neurol 54:465–473. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubser M, Bridges TM, Dencker D, Gould RW, Grannan M, Noetzel MJ, Lamsal A, Niswender CM, Daniels JS, Poslusney MS, et al. (2014) Selective activation of M4 muscarinic acetylcholine receptors reverses MK-801-induced behavioral impairments and enhances associative learning in rodents. ACS Chem Neurosci 5:920–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun NE, Grannan M, Bubser M, Barry RL, Thompson A, Rosanelli J, Gowrishankar R, Kelm ND, Damon S, Bridges TM, et al. (2014) Antipsychotic drug-like effects of the selective M4 muscarinic acetylcholine receptor positive allosteric modulator VU0152100. Neuropsychopharmacology 39:1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro J, Garcia RI, Kwok S, Banerjee A, Petravicz J, Woodson J, Mellios N, Tropea D, Sur M. (2014) Functional recovery with recombinant human IGF1 treatment in a mouse model of Rett Syndrome. Proc Natl Acad Sci USA 111:9941–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. (2008) MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WY, McKinzie DL, Bose S, Mitchell SN, Witkin JM, Thompson RC, Christopoulos A, Lazareno S, Birdsall NJ, Bymaster FP, et al. (2008) Allosteric modulation of the muscarinic M4 receptor as an approach to treating schizophrenia. Proc Natl Acad Sci USA 105:10978–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. (2006) The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron 49:341–348. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Jeon OH, Hyder K, Chenchik A, Khimani AH, Narayanan V, Hoffman EP, Kaufmann WE, Naidu S, Pevsner J. (2001) Gene expression profiling in postmortem Rett Syndrome brain: differential gene expression and patient classification. Neurobiol Dis 8:847–865. [DOI] [PubMed] [Google Scholar]
- Duarte ST, Armstrong J, Roche A, Ortez C, Pérez A, O’Callaghan MdelM, Pereira A, Sanmartí F, Ormazábal A, Artuch R, et al. (2013) Abnormal expression of cerebrospinal fluid cation chloride cotransporters in patients with Rett syndrome. PLoS One 8:e68851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert FJ. (2003) Contractile role of M2 and M3 muscarinic receptors in gastrointestinal, airway and urinary bladder smooth muscle. Life Sci 74:355–366. [DOI] [PubMed] [Google Scholar]
- Foster DJ, Choi DL, Conn PJ, Rook JM. (2014) Activation of M1 and M4 muscarinic receptors as potential treatments for Alzheimer’s disease and schizophrenia. Neuropsychiatr Dis Treat 10:183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DJ, Jones CK, Conn PJ. (2012) Emerging approaches for treatment of schizophrenia: modulation of cholinergic signaling. Discov Med 14:413–420. [PMC free article] [PubMed] [Google Scholar]
- Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, Mirnics K, Persico AM. (2008) Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis 30:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. (2007) Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA 104:1931–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Senter RK, Fisher NM, Adams J, Zamorano R, Walker AG, Blobaum AL, Engers DW, Hopkins CR, Daniels JS, et al. (2017) mGlu7 potentiation rescues cognitive, social, and respiratory phenotypes in a mouse model of Rett syndrome. Sci Transl Med 9:eaai7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Senter RK, Rook JM, Ghoshal A, Zamorano R, Malosh C, Stauffer SR, Bridges TM, Bartolome JM, Daniels JS, et al. (2016) mGlu5 positive allosteric modulation normalizes synaptic plasticity defects and motor phenotypes in a mouse model of Rett syndrome. Hum Mol Genet 25:1990–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. (2007) Reversal of neurological defects in a mouse model of Rett syndrome. Science 315:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B. (2002) Clinical manifestations and stages of Rett syndrome. Ment Retard Dev Disabil Res Rev 8:61–65. [DOI] [PubMed] [Google Scholar]
- Katz DM, Berger-Sweeney JE, Eubanks JH, Justice MJ, Neul JL, Pozzo-Miller L, Blue ME, Christian D, Crawley JN, Giustetto M, et al. (2012) Preclinical research in Rett syndrome: setting the foundation for translational success. Dis Model Mech 5:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, Lewis JW, Posey J, Strange SK, Guzman-Karlsson MC, et al. (2016) Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Reports 16:2666–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshimizu H, Leiter LM, Miyakawa T. (2012) M4 muscarinic receptor knockout mice display abnormal social behavior and decreased prepulse inhibition. Mol Brain 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Lovén J, Kwok SM, Feldman DA, Bateup HS, Gao Q, et al. (2013) Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell 13:446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Nicholls L, Assareh H, Fang Z, Amos TG, Edwards RJ, Assareh AA, Voineagu I. (2016) Transcriptome analysis of human brain tissue identifies reduced expression of complement complex C1Q genes in Rett syndrome. BMC Genomics 17:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S, Giacometti E, Beard CF, Jaenisch R. (2004) Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc Natl Acad Sci USA 101:6033–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Bird A. (2015) Rett syndrome: a complex disorder with simple roots. Nat Rev Genet 16:261–275. [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima Alves F, et al. (2013) Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci 16:898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonski K, Abuhatzira L, Kaufman Y, Razin A, Shemer R. (2005) MeCP2 deficiency in Rett syndrome causes epigenetic aberrations at the PWS/AS imprinting center that affects UBE3A expression. Hum Mol Genet 14:1049–1058. [DOI] [PubMed] [Google Scholar]
- Melancon BJ, Wood MR, Noetzel MJ, Nance KD, Engelberg EM, Han C, Lamsal A, Chang S, Cho HP, Byers FW, et al. (2017) Optimization of M4 positive allosteric modulators (PAMs): the discovery of VU0476406, a non-human primate in vivo tool compound for translational pharmacology. Bioorg Med Chem Lett 27:2296–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. (2013) The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41:D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al. (2005) Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet 14:2247–2256. [DOI] [PubMed] [Google Scholar]
- Ogier M, Wang H, Hong E, Wang Q, Greenberg ME, Katz DM. (2007) Brain-derived neurotrophic factor expression and respiratory function improve after ampakine treatment in a mouse model of Rett syndrome. J Neurosci 27:10912–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oginsky MF, Cui N, Zhong W, Johnson CM, Jiang C. (2014) Alterations in the cholinergic system of brain stem neurons in a mouse model of Rett syndrome. Am J Physiol Cell Physiol 307:C508–C520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco NL, Heaven MR, Holt LM, Crossman DK, Boggio KJ, Shaffer SA, Flint DL, Olsen ML. (2017) RNA sequencing and proteomics approaches reveal novel deficits in the cortex of Mecp2-deficient mice, a model for Rett syndrome. Mol Autism 8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancani T, Foster DJ, Moehle MS, Bichell TJ, Bradley E, Bridges TM, Klar R, Poslusney M, Rook JM, Daniels JS, et al. (2015) Allosteric activation of M4 muscarinic receptors improve behavioral and physiological alterations in early symptomatic YAC128 mice. Proc Natl Acad Sci USA 112:14078–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsian AJ, Sheren JE, Tao TY, Goswami PC, Malyapa R, Van Rheeden R, Watson MS, Hunt CR. (2000) The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim Biophys Acta 1494:201–205. [DOI] [PubMed] [Google Scholar]
- Pecorelli A, Leoni G, Cervellati F, Canali R, Signorini C, Leoncini S, Cortelazzo A, De Felice C, Ciccoli L, Hayek J, et al. (2013) Genes related to mitochondrial functions, protein degradation, and chromatin folding are differentially expressed in lymphomonocytes of Rett syndrome patients. Mediators Inflamm 2013:137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rett A. (1966) [On a unusual brain atrophy syndrome in hyperammonemia in childhood]. Wien Med Wochenschr 116:723–726. [PubMed] [Google Scholar]
- Schmid DA, Yang T, Ogier M, Adams I, Mirakhur Y, Wang Q, Massa SM, Longo FM, Katz DM. (2012) A TrkB small molecule partial agonist rescues TrkB phosphorylation deficits and improves respiratory function in a mouse model of Rett syndrome. J Neurosci 32:1803–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar A, Potter WZ, Lightfoot J, Lienemann J, Dubé S, Mallinckrodt C, Bymaster FP, McKinzie DL, Felder CC. (2008) Selective muscarinic receptor agonist xanomeline as a novel treatment approach for schizophrenia. Am J Psychiatry 165:1033–1039. [DOI] [PubMed] [Google Scholar]
- Tang X, Kim J, Zhou L, Wengert E, Zhang L, Wu Z, Carromeu C, Muotri AR, Marchetto MC, Gage FH, et al. (2016) KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci USA 113:751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. (2009) Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci USA 106:2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. (2002) Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci USA 99:15536–15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzavara ET, Bymaster FP, Felder CC, Wade M, Gomeza J, Wess J, McKinzie DL, Nomikos GG. (2003) Dysregulated hippocampal acetylcholine neurotransmission and impaired cognition in M2, M4 and M2/M4 muscarinic receptor knockout mice. Mol Psychiatry 8:673–679. [DOI] [PubMed] [Google Scholar]
- Veeraragavan S, Graham D, Bui N, Yuva-Paylor LA, Wess J, Paylor R. (2012) Genetic reduction of muscarinic M4 receptor modulates analgesic response and acoustic startle response in a mouse model of fragile X syndrome (FXS). Behav Brain Res 228:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chan SA, Ogier M, Hellard D, Wang Q, Smith C, Katz DM. (2006) Dysregulation of brain-derived neurotrophic factor expression and neurosecretory function in Mecp2 null mice. J Neurosci 26:10911–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MR, Noetzel MJ, Poslusney MS, Melancon BJ, Tarr JC, Lamsal A, Chang S, Luscombe VB, Weiner RL, Cho HP, et al. (2017) Challenges in the development of an M4 PAM in vivo tool compound: the discovery of VU0467154 and unexpected DMPK profiles of close analogs. Bioorg Med Chem Lett 27:171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonan AL, Palmer AA, Smith KC, Feldman I, Lee HK, Yonan JM, Fischer SG, Pavlidis P, Gilliam TC. (2003) Bioinformatic analysis of autism positional candidate genes using biological databases and computational gene network prediction. Genes Brain Behav 2:303–320. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cao SX, Sun P, He HY, Yang CH, Chen XJ, Shen CJ, Wang XD, Chen Z, Berg DK, et al. (2016) Loss of MeCP2 in cholinergic neurons causes part of RTT-like phenotypes via α7 receptor in hippocampus. Cell Res 26:728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YT, Goffin D, Johnson BS, Zhou Z. (2013) Loss of MeCP2 function is associated with distinct gene expression changes in the striatum. Neurobiol Dis 59:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]