Abstract
N-Methyl-d-aspartate (NMDA)–type glutamate receptors mediate excitatory synaptic transmission in the central nervous system and play critical roles in many neuronal processes. The physiologic roles of NMDA receptors are shaped by their functional properties, which are highly dependent on subunit composition. Most NMDA receptors are assembled from two GluN1 and two GluN2 subunits, but diversity in subunit composition is made possible by eight GluN1 splice variants (i.e., isoforms) and four distinct GluN2 subunits (GluN2A-D). We demonstrate using Förster resonance energy transfer and fluorescence lifetime imaging that GluN1-1a and GluN1-1b isoforms, which include or lack residues encoded by exon 5, form triheteromeric GluN1-1a/GluN1-1b/GluN2A (1a/1b/2A) and GluN1-1a/GluN1-1b/GluN2B (1a/1b/2B) receptors. We describe the selective expression of NMDA receptors containing two different GluN1 isoforms, and show that triheteromeric 1a/1b/2A and 1a/1b/2B receptors exhibit intermediate deactivation kinetics and pharmacological properties compared with the respective diheteromeric GluN1-1a/GluN1-1a/GluN2 and GluN1-1b/GluN1-1b/GluN2 receptors. These results highlight the intriguing possibility that neurons can finely tune NMDA receptor signaling by shifting the ratio of expressed GluN1-1a and GluN1-1b isoforms. Furthermore, we evaluate the contribution of channel pore residues to magnesium block and calcium permeability. These data point to the asymmetric contribution of pore residues in GluN1 and GluN2 to magnesium block, and reveal that a single copy of pore residues from GluN3 subunits strongly attenuates magnesium block and calcium permeability of NMDA receptors. Thus, the selective expression of NMDA receptors containing two distinct GluN1 isoforms provides new opportunities to study functional properties relevant to neuronal receptors.
Introduction
N-Methyl-d-aspartate (NMDA) receptors are ionotropic glutamate receptors that play critical roles in synaptic plasticity and neuronal development, but aberrant NMDA receptor signaling is implicated in a plethora of neurological and psychological diseases (Traynelis et al., 2010; Paoletti et al., 2013). The majority of NMDA receptors in the central nervous system (CNS) are tetrameric assemblies of two glycine/D-serine-binding GluN1 and two glutamate-binding GluN2 subunits (Traynelis et al., 2010; Paoletti et al., 2013). Four different GluN2 subunits have been identified (GluN2A-D) that display pronounced variation in regional and developmental expression levels, and each GluN2 subunit endows NMDA receptors with distinct functional and pharmacological properties (Monyer et al., 1992, 1994; Ishii et al., 1993; Akazawa et al., 1994; Vicini et al., 1998; Gielen et al., 2009; Yuan et al., 2009). The GluN2 subunits are therefore principle mediators of functional diversity among NMDA receptor subtypes and dictate their psychological roles in the CNS.
The GluN1 subunit, which is obligatory in all NMDA receptor subtypes, is expressed from a single gene, but has eight different isoforms that arise from alternative splicing of three exons (Durand et al., 1992; Nakanishi et al., 1992; Sugihara et al., 1992; Hollmann et al., 1993). Exon 5 encodes the N1 cassette of 21 highly charged amino acids in the extracellular GluN1 amino-terminal domain, while exons 21 and 22 encode C1 and C2 cassettes in the intracellular C-terminal domain (CTD). Alternative splicing of exons 21 and 22 has no discernible effects on the functional and pharmacological properties of NMDA receptors, but rather alter interactions with intracellular proteins that can influence the subcellular distribution of NMDA receptors (Scott et al., 2001; Mu et al., 2003). By contrast, GluN1 subunits that contain the N1 cassette (e.g., GluN1-1b), encoded by exon 5, reduce agonist potency of NMDA receptors and decrease sensitivity to inhibition by extracellular zinc and protons compared with GluN1 subunits lacking the N1 cassette (e.g., GluN1-1a) (Traynelis et al., 1995, 1998). The presence of the N1 cassette also accelerates deactivation of glutamate-activated NMDA receptor responses and shortens the time course of excitatory postsynaptic currents (Prybylowski et al., 2000; Rumbaugh et al., 2000; Vance et al., 2012; Swanger et al., 2015). Alternative splicing of exon 5 in the GluN1 subunit can therefore have psychological implications by modulating NMDA receptor properties relevant to synaptic transmission. Furthermore, the presence of the N1 cassette in GluN1-1b attenuates inhibition by GluN2B-selective antagonists, such as ifenprodil and CP-101,606 (Pahk and Williams, 1997; Mott et al., 1998), and reduces GluN2B-selective potentiation by extracellular polyamines, such as spermine (Durand et al., 1992, 1993; Zhang et al., 1994; Traynelis et al., 1995; Rumbaugh et al., 2000).
The GluN1 isoforms have different regional and developmental expression patterns (Standaert et al., 1993; Laurie and Seeburg, 1994; Zhong et al., 1995; Paupard et al., 1997; Prybylowski et al., 2000), and it has been shown using single-cell RT-PCR that mRNA encoding GluN1 splice variants with and without the N1 cassette can coexist in neuronal cells (Karlsson et al., 2002; Paarmann et al., 2005). This overlap in expression at the cellular level creates the intriguing possibility that neuronal NMDA receptors can contain two different GluN1 isoforms with and without the N1 cassette, although the existence of such receptor subtypes has not been demonstrated in vivo. Diheteromeric GluN1-1a/GluN1-1a/GluN2 (hereinafter denoted as 1a/1a/2) and GluN1-1b/GluN1-1b/GluN2 (hereinafter denoted as 1b/1b/2) receptors have been intensively studied in heterologous expressions systems (e.g., HEK293 cells and Xenopus oocytes), but properties of triheteromeric GluN1-1a/GluN1-1b/GluN2 (hereinafter denoted as 1a/1b/2) receptors are unresolved. We show that GluN1-1a and GluN1-1b can coassemble in functional NMDA receptors and describe a method for expression of triheteromeric 1a/1b/2 NMDA receptors in heterologous expressions systems without accompanying coexpression of diheteromeric 1a/1a/2 and 1b/1b/2 receptors.
Materials and Methods
DNA Constructs and Ligands.
Rat cDNAs for GluN1-1a (GenBank accession number U08261), GluN1-1b (GenBank accession number U08263), GluN2A (GenBank accession number D13211), and GluN2B (GenBank accession number U11419) were provided by Dr. S. Heinemann (Salk Institute, La Jolla, CA) and Dr. S. Nakanishi (Osaka Bioscience Institute, Osaka, Japan). Rat GluN2B cDNA was modified to remove a T7 RNA polymerase termination site located in the CTD as previously described (Hansen et al., 2014). DNA constructs encoding GluN1 isoforms for Förster resonance energy transfer (FRET) and fluorescence lifetime imaging (FLIM) experiments were generated by replacing residues 24–25, downstream from the signal peptide, with the cyan fluorescent protein (CFP) mCerulean3 (Markwardt et al., 2011) or the yellow fluorescent protein (YFP) mVenus (Nagai et al., 2002). CFP and YFP were flanked by N-terminal (RTGGSGGGSGGG) and C-terminal (GGGGSGGGSGRT) peptide linkers. The DNA construct for YFP fused to the amino-terminal domain of the GluA2 AMPA receptor subunit (YFPGluA2) has been previously described (Zachariassen et al., 2016). The cDNAs for peptide tags C1 and C2 were custom synthesized (Genscript, Piscataway, NJ) as previously described (Hansen et al., 2014) and inserted in place of the stop codon in the open reading frame (ORF) of GluN1-1a and GluN1-1b to generate 1aC1, 1aC2, 1bC1, and 1bC2. The constructs for expression of triheteromeric NMDA receptors containing two differentGluN2 subunits have been previously described (Hansen et al., 2014).
We developed a specialized DNA construct for expression of GluN1 in HEK293 cells, in which cDNA encoding enhanced green fluorescent protein (EGFP) was inserted between the CMV promoter in pCI-neo and the ORF of GluN1 (i.e., EGFP and GluN1 were not expressed as a fusion protein). This DNA construct resulted in high expression of EGFP and low expression of GluN1, while maintaining a linear relationship between EGFP and GluN1 expression. The ribosome scans the 5′-untranslated region of the mRNA for AUG initiation codons, and the first AUG in this DNA construct will initiate translation of the ORF for EGFP. However, the ribosome will frequently skip this first AUG and instead initiate translation from downstream AUG codons, which in this construct encodes GluN1. The placement of 0–3 AUGs between EGFP and GluN1, which initiate short upstream ORFs (Meijer and Thomas, 2002; Barbosa et al., 2013) encoding 42–58 amino acids, results in a gradual reduction of GluN1 expression, while maintaining high expression of EGFP.
Amino acids are numbered according to the full-length protein, including the signal peptide. Site-directed mutagenesis was performed using the QuikChange method (Agilent Technologies, Santa Clara, CA) and verified by DNA sequencing. For expression in Xenopus oocytes, DNA constructs were linearized by restriction enzymes to produce the template for in vitro cRNA synthesis (mMessage mMachine; Thermo Fisher Scientific, Waltham, MA). Ifenprodil and spermine were purchased from Hello Bio (Princeton, NJ), and all other ligands were purchased from Sigma-Aldrich (St. Louis, MO).
Confocal Imaging and Fluorescence Lifetime Measurements.
HEK293 cells were seeded in 12-well culture plates approximately 24 hours before transfection and cultured in Dulbecco’s modified Eagle’s medium with GlutaMax-I and sodium pyruvate (GIBCO; Thermo Fisher Scientific) supplemented with 10% dialyzed fetal bovine serum (GIBCO; Thermo Fisher Scientific), 10 U/ml penicillin, and 10 µg/ml streptomycin (GIBCO; Thermo Fisher Scientific). Cells were transfected using TransIT Transfection Reagent (Mirus Bio, Madison, WI) with plasmid cDNAs encoding CFP-GluN1, YFP-GluN1, and GluN2A at a ratio of 1:1:1 or CFP-GluN1 and GluN2A at a ratio of 1:1. To prevent NMDA receptor-mediated cytotoxicity, the antagonists D,L-2-amino-5-phosphonovalerate (200 µM), 7-chlorokynurenic acid (200 µM), and MK-801 (10 µM) were added to the culture medium.
Fluorescence lifetime measurements of CFP were conducted on live HEK293 cells approximately 48 hours following transfection. The cells were replated to 33 mm poly-D-lysine-coated (0.1 mg/ml) coverslips in culture medium 1 to 2 hours before measurements. Hanks’ balanced salt solution without divalent cations (GIBCO; Thermo Fisher Scientific) was used to detach cells (i.e., trypsin was not used) and the cells were allowed to recover in culture medium. Immediately before imaging, the cells were transferred to Hanks’ balanced salt solution with CaCl2 and MgCl2 (GIBCO; Thermo Fisher Scientific). Confocal images were acquired using a Zeiss 880 confocal microscope (Carl Zeiss, Oberkochen, Germany) with a 63× 1.4 Oil DIC M27 objective and an argon laser. CFP/YFP-tagged receptors were visualized using a 458-nm laser at 20% input power for CFP excitation and 514-nm laser at 15% input power for YFP excitation. CFP emission was collected between 463 and 492 nm and YFP emission was collected between 519 and 620 nm. For FRET-FLIM measurements, the cells were excited by an LDH-P-C-440 diode laser at 440 nm controlled by a PDL 828 Sepia II Driver (PicoQuant, West Springfield, MA). Excitation light was pulsed at a 20 MHz repetition rate through a 2T442 dichroic beam combiner (PicoQuant). Emission light was passed through BP482/35 and BP550/49 filters, detected by a 2PMA-Hydrid-40 Detector, and photon counts were recorded using a PicoHarp300 TCSPC Module (PicoQuant). Cells were scanned for 120–240 seconds to obtain counts of at least 1000 photons in the region of interest, selected as the cell surface membrane. The acquired FLIM images were analyzed using SymPhoTime 64 (PicoQuant) to determine the fluorescence lifetime (τ) of membrane-localized CFP by fitting the average fluorescence decay of all pixels in the region of interest to a monoexponential function for cells expressing CFP only and a two-exponential function for cells expressing both CFP and YFP.
Two-Electrode Voltage-Clamp Recordings.
Injection of cRNA and maintenance of Xenopus oocytes (purchased from Rob Weymouth at Xenopus 1, Dexter, MI) was performed as previously described (Hansen et al., 2013). For experiments with triheteromeric NMDA receptors that contain distinct GluN1 isoforms, the cRNAs encoding C1- and C2-tagged GluN1 as well as GluN2 were injected at a 1:1:1 ratio at a total volume of 50 nl, and the cRNA was diluted with RNase-free water to yield 2.5–3 ng total injected cRNA. For experiments with triheteromeric NMDA receptors that contain distinct GluN2 subunits, the cRNAs encoding GluN1 as well as C1- and C2-tagged GluN2 were injected at a 1:1:1 ratio at a total volume of 50 nl, and the cRNA was diluted with RNase-free water to yield 6 to 7 ng total injected cRNA. The injected Xenopus oocytes were incubated at 17°C for expression of diheteromeric NMDA receptors, but required incubation at 19°C for expression of triheteromeric NMDA receptors (Yi et al., 2017). Two-electrode voltage-clamp recordings (OC-725; Warner Instruments, Hamden, CT) were performed 2–4 days after injection at room temperature (20°C). Except for experiments involving extracellular Zn2+, the extracellular recording solution contained 90 mM NaCl, 1 mM KCl, 10 mM HEPES, 0.5 mM BaCl2, and 0.01 mM EDTA (pH 7.4 with NaOH). For the Zn2+ experiments, the pH of the extracellular solution was 7.4 and EDTA was replaced with 10 mM tricine to buffer extracellular Zn2+ as previously described (Traynelis et al., 1998). Recording electrodes were filled with 3.0 M KCl, and recordings were performed using a holding potential at −40 mV, unless otherwise stated and except for experiments with Zn2+ and spermine, in which the holding potential was −20 mV to minimize voltage-dependent channel block. For experiments with triheteromeric receptors, the fraction of current response from escaped receptors compared with the total current response was always determined on the day of the experiment, and experiments were only performed if the escape current response was <10% of the total current response.
Whole-Cell Voltage-Clamp Recordings.
HEK293 cells were plated onto poly-D-lysine-coated (0.1 mg/ml) glass coverslips approximately 48 hours before experiments and cultured in Dulbecco’s modified Eagle’s medium with GlutaMax-I and sodium pyruvate (GIBCO; Thermo Fisher Scientific) supplemented with 10% dialyzed fetal bovine serum (GIBCO; Thermo Fisher Scientific), 10 U/ml penicillin, and 10 µg/ml streptomycin (GIBCO; Thermo Fisher Scientific). Cells were transfected using the calcium phosphate precipitation method with plasmid cDNAs encoding GluN1 (together with EGFP in the pCI-neo vector) and GluN2 subunits at a ratio of 1:1. For experiments with triheteromeric NMDA receptors, plasmid cDNAs encoding the two distinct GluN1 isoforms and the GluN2 subunit were transfected at 1:1:5 (1aC2/1bC1/2A) or 1:1:2 (1aC2/1bC1/2B) ratios and plasmid cDNAs encoding GluN1 and the two distinct GluN2 isoforms were transfected at a 1:2:2 ratio. To prevent NMDA receptor-mediated cytotoxicity, the antagonists D,L-2-amino-5-phosphonovalerate (200 µM) and 7-chlorokynurenic acid (200 µM) were added to the culture medium, and experiments were performed approximately 24 hours following transfection.
Whole-cell voltage-clamp recordings (Axopatch 200B; Molecular Devices, Sunnyvale, CA) were performed at room temperature (20°C) at a holding potential of –60 mV (unless otherwise stated). Recording electrodes with open-tip resistance of 2–4 MΩ were made from thin-wall glass micropipettes (TW150F-4; World Precision Instruments, Sarasota, FL) pulled using a horizontal puller (P-1000; Sutter Instruments, Novato, CA). The electrodes were filled with internal solution containing 110 mM D-gluconic acid, 110 mM CsOH, 30 mM CsCl, 5 mM HEPES, 4 mM NaCl, 0.5 mM CaCl2, 2 mM MgCl2, 5 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid, 2 mM NaATP, and 0.3 mM NaGTP (pH 7.35 with CsOH). The extracellular recording solution was composed of 150 mM NaCl, 10 mM HEPES, 3 mM KCl, 0.5 mM CaCl2, and 0.01 mM EDTA (pH 7.4 with NaOH). Holding potentials were not corrected for the liquid junction potential, which was measured to be +10.1 ± 0.6 mV (n = 4). Rapid solution exchange was achieved on lifted cells with a two-barrel theta-glass pipette controlled by a piezoelectric translator (MXPZT-300; Siskiyou Corporation, Grants Pass, OR) and the 10%–90% open-tip solution exchange times were 0.6–0.8 ms. Only cells with current responses of less than 1000 pA and series resistance of less than 10 MΩ were used for data analyses.
Data Analysis.
Concentration-response data were analyzed with GraphPad Prism (GraphPad Software, La Jolla, CA). Agonist concentration-response data for individual oocytes were fitted to the following Hill equation:
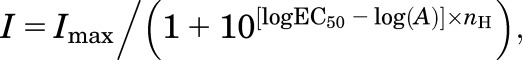 |
where Imax is the maximum current in response to the agonist, nH is the Hill slope, (A) is the agonist concentration, and EC50 is the agonist concentration that produces half-maximum response. Antagonist concentration-response data were fitted to the following equation:
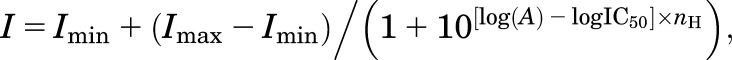 |
where Imin is the minimum current in response to the agonist plus a saturating concentration of antagonist. For graphical presentation, data points from individual oocytes were normalized to the maximum current response to glutamate plus glycine in the same recording and averaged.
Whole-cell voltage-clamp recordings using HEK293 cells were analyzed using Axograph (www.axograph.com). The deactivation time courses of current responses (each averaged from at least five sweeps) were fitted using: Itotal = Ifast exp (−time/τfast) + Islow exp (−time/τslow), where τfast and τslow are the deactivation time constants for the fast and slow components, respectively, and Ifast and Islow are the current amplitudes of the fast and slow components, respectively. Weighted deactivation time constants were calculated using: τweighted = (τfast Ifast + τslow Islow)/(Ifast + Islow). In FRET-FLIM experiments, the fluorescence decay of photon counts in the region of interest was analyzed essentially as the deactivation time course of current amplitudes using either monoexponential or two-exponential functions, and the weighted decay constants were also calculated using the aforementioned function. Data are provided as mean ± S.E.M. unless otherwise stated.
Results
Assembly of NMDA Receptors with Two Distinct GluN1 Isoforms.
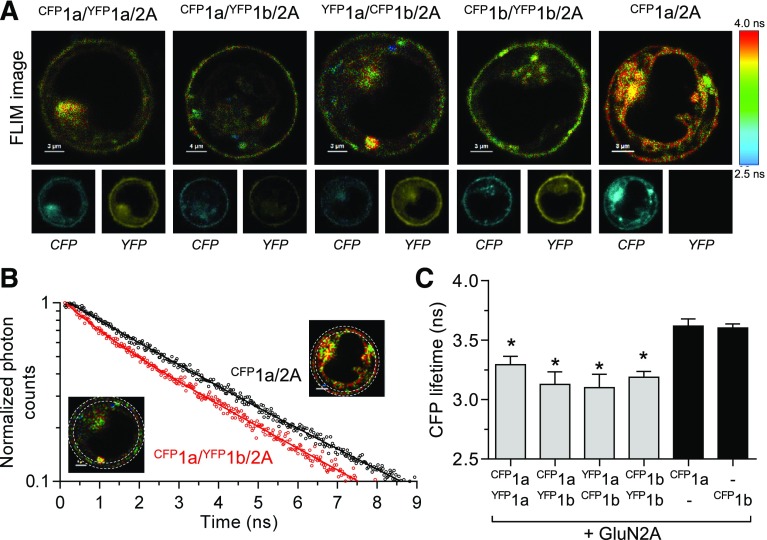
To determine if GluN1-1a and GluN1-1b isoforms coassemble in NMDA receptors, we fused CFP or YFP to their extracellular amino-terminal domains. Upon coexpression with GluN2A, we identified NMDA receptors containing both CFP- and YFP-tagged subunits in the region corresponding to the surface membrane of live HEK293 (Fig. 1A). The localization of CFP- and YFP-tagged GluN1 subunits at the cell surface indicates that they are assembled with GluN2A (Monyer et al., 1992). To determine whether CFP- and YFP-tagged GluN1 subunits were assembled in the same receptor we used FLIM to quantify FRET. FRET only occurs when the donor (CFP) and acceptor (YFP) pair of fluorescent proteins are in close proximity (i.e., <10 nm) and the fluorescence lifetime of the donor is accelarated by energy transfer. The lifetime of CFP fluorescence in the absence of FRET (i.e., donor-only τD) was determined from cells coexpressing CFP-tagged GluN1-1a (CFP1a) and GluN2A using a monoexponential fit, revealing a donor-only τD of 3.62 ns (Fig. 1; Table 1). Coexpression of CFP1a and YFP1b subunits together with GluN2A can, in theory, result in three receptor populations, namely donor-only (τD) CFP1a/2A, donor/acceptor (τDA) CFP1a/YFP1b/2A receptors, and YFP1b/2A with no CFP fluorescence. The CFP lifetime in the membrane of CFP1a/YFP1b/2A-expressing cells was analyzed using a two-exponential fit and the amplitude-weigted average lifetime (τweighted) was 3.13 ns with τD of 3.63 ns and τDA of 1.24 ns (Fig. 1; Table 1). The significant decrease in τweighted for CFP1a/YFP1b/2A-expressing cells compared with CFP1a/GluN2A-expressing cells indicates that FRET occurs as a result of coassembly of CFP1a and YFP1b subunits into CFP1a/YFP1b/2A receptors. Importantly, the amplitude of the donor/acceptor component was 21% of the donor-only component (Table 1), suggesting that a robust fraction of the NMDA receptors contained both CFP- and YFP-tagged GluN1 subunits. Similar results were obtained for cells expressing different combinations of CFP- and YFP-tagged GluN1-1a and GluN1-1b isoforms, thereby demonstrating that GluN1-1a and GluN1-1b isoforms readily coassemble in GluN2A-containing NMDA receptors (Fig. 1C; Table 1). Independent on the coexpressed GluN1-1a and GluN1-1b isoforms, the fraction of donor/acceptor component relative to the donor-only component was 17%–22% and the FRET efficiency calculated from τD and τDA was 66%–68% (Table 1). The consistent results among the different conditions indicates a lack of preference for assembly of GluN1-1a and GluN1-1b isoforms into either 1a/1a, 1a/1b, or 1b/1b pairs, and that FRET is specific to intrareceptor interactions and unaffected by relative expression levels. To provide additional control for intrareceptor versus inter-receptor FRET, we coexpressed CFP1a/2A with YFP fused to the amino-terminal domain of the GluA2 AMPA receptor subunit (YFPGluA2). Coexpression of CFP1a/2A and YFPGluA2 did not change the lifetime of CFP fluorescence compared with cells expressing CFP1a/2A alone, consistent with a lack of inter-receptor FRET (Table 1). In summary, the FRET-FLIM experiments demonstrate that GluN1-1a and GluN1-1b isoforms readily coassemble into NMDA receptors expressed at the cell surface, albeit the functional properties of these receptors are yet to be characterized.
Fig. 1.
Detection of triheteromeric GluN1-1a/GluN1-1b/GluN2A NMDA receptors using FRET-FLIM. (A) Images of CFP fluorescence lifetime (FLIM images) for HEK293 cells expressing combinations of GluN2A with GluN1 isoforms tagged with CFP or YFP at the amino-terminal domain. The corresponding images of fluorescence intensities for CFP and YFP emission are shown below. The pixel color corresponds to the measured CFP lifetime as indicated by the color scale. The cells were replated approximately 1 to 2 hours before measurements using Hanks’ balanced salt solution without divalent cations to detach the cells and give the cells a rounded appearance, which facilitates localization of the cell membrane using confocal microscopy. Cells coexpressing CFP-tagged GluN1-1a and GluN2A (CFP1a/2A) were used to determine the fluorescence lifetime of CFP in the absence of YFP acceptor (i.e., donor only). (B) Representative decays of CFP fluorescence at the cell surface of cells expressing donor and acceptor (CFP1a/YFP1b/2A) and donor only (CFP1a/2A). The decay of CFP1a/2A follows a monoexponential time course, whereas the decay of CFP1a/YFP1b/2A follows a two-exponential time course consistent with the assembly of a population of triheteromeric 1a/1b/2A receptors and FRET occurring between CFP1a and YFP1b subunits. The insets illustrate the region of interest (i.e., the cell surface membrane) used for the fluorescence lifetime measurements. (C) Summary of the weighted CFP lifetimes (i.e., weighted time constants of fluorescence decay) with the black bars indicating the CFP lifetime of donor only in CFP1a/2A and CFP1b/2A. Coassembly of CFP- and YFP-tagged GluN1 subunits is demonstrated by reduced CFP lifetimes compared with donor only. *indicates significantly different from CFP1a/2A (P < 0.05; one-way analysis of variance with Tukey’s post hoc test). Data are mean ± S.D. from 9 to 16 cells and decay time constants are shown in Table 1.
TABLE 1.
Detection of triheteromeric NMDA receptors using FRET-FLIM
Fluorescence lifetime of CFP in HEK293 cells expressing CFP- and YFP-tagged GluN1-1a and GluN1-1b isoforms together with GluN2A. Regions corresponding to the cell surface were selected and CFP lifetimes were analyzed with monoexponential fits for cells expressing CFP-tagged GluN1 (i.e., donor only) or two-exponential fits for cells expressing both CFP- and YFP-tagged GluN1 (i.e., donor/acceptor). The τD and τDA values represent the lifetimes for donor only and donor/acceptor, respectively; % DA is the amplitude of the donor/acceptor component relative to the donor-only component; τweighted is the weighted time constant calculated as described in Materials and Methods. FRET efficiencies (E) are calculated as 1 − (τDA/τD). Dashes indicates data not available. Data are presented as mean ± S.E.M., and n is the number of cells used to generate the data.
| τD | τDA | % DA | τweighted | n | E | |
|---|---|---|---|---|---|---|
| ns | ns | ns | ||||
| CFP1a/2A | 3.62 ± 0.01 | — | — | 3.62 ± 0.01 | 14 | — |
| CFP1b/2A | 3.60 ± 0.01 | — | — | 3.60 ± 0.01 | 9 | — |
| CFP1a/YFP1a/2A | 3.72 ± 0.02 | 1.26 ± 0.03 | 17 ± 1 | 3.29 ± 0.02* | 16 | 0.66 |
| CFP1a/YFP1b/2A | 3.63 ± 0.03 | 1.24 ± 0.06 | 21 ± 1 | 3.13 ± 0.03* | 10 | 0.66 |
| YFP1a/CFP1b/2A | 3.67 ± 0.02 | 1.18 ± 0.07 | 22 ± 1 | 3.10 ± 0.03* | 10 | 0.68 |
| CFP1b/YFP1b/2A | 3.66 ± 0.01 | 1.24 ± 0.03 | 19 ± 1 | 3.19 ± 0.01* | 12 | 0.66 |
| CFP1a/2A + YFPGluA2 | 3.63 ± 0.02 | — | — | 3.63 ± 0.01 | 10 | — |
indicates significantly different from τweighted for CFP1a/2A (P < 0.05; one-way analysis of variance with Tukey’s post hoc test). Only τweighted values were compared.
Selective Cell-Surface Expression of NMDA Receptors with Two Distinct GluN1 Isoforms.
For expression of triheteromeric NMDA receptors containing two distinct GluN1 isoforms without accompanying coexpression of diheteromeric NMDA receptors, we adapted a method previously described for expression of triheteromeric NMDA receptors containing two distinct GluN2 subunits (Hansen et al., 2014). This method relies on engineered C-terminal peptide tags that prevent trafficking of diheteromeric NMDA receptors to the cell surface, while allowing cell-surface expression of triheteromeric NMDA receptors with a defined subunit composition. The engineered C-terminal peptide tags are composed of a peptide linker (L4) and the leucine zipper motifs from GABAB receptor subunits (LZ1 and LZ2), followed by dilysine (KKXX) endoplasmic reticulum (ER) retention/retrieval motifs that prevent trafficking to the cell surface unless masked by heterodimeric coiled-coil formation between LZ1 and LZ2 (Hansen et al., 2014) (Fig. 2, A and B). In this study, the engineered C-terminal peptide tags (C1 and C2) are fused to the C-terminus of GluN1 isoforms, thereby enabling expression of triheteromeric NMDA receptors with a defined composition of GluN1 subunits.
Fig. 2.
Selective expression of triheteromeric GluN1-1a/GluN1-1b/GluN2 NMDA receptors. (A) Linear representation of the polypeptide chain of GluN1-1a and GluN1-1b isoforms with the amino-terminal domain (ATD), the agonist binding domain (ABD) formed by S1 and S2, the transmembrane domain formed by M1, M2, M3, and M4, and the intracellular CTD. Alternative splicing can produce the GluN1-1b isoform that differs from GluN1-1a by the insertion of 21 amino acids encoded by exon 5 (N1 cassette). GluN1-1aC1 and GluN1-1aC2 (1aCX) as well as GluN1-1bC1 and GluN1-1bC2 (1bCX) were generated by fusing engineered C1 and C2 peptide tags to the CTD. (B) Coexpression of GluN2A with 1aC2 and 1bC1 can produce three populations of functional NMDA receptors. Diheteromeric 1aC2/1aC2/2A and 1bC1/1bC1/2A receptors are prevented from trafficking to the cell surface due the presence of unmasked dilysine (KKXX) ER retention signals in the CTD, whereas heterodimeric coiled-coil formation between C1 and C2 tags in triheteromeric 1aC2/1bC1/2A receptors masks the ER retention signals and enable trafficking to the cell surface. (C) Representative two-electrode voltage-clamp recordings of responses from recombinant NMDA receptors expressed in Xenopus oocytes activated by 100 µM glutamate plus 50 µM glycine. Some 1aC2/1aC2/2A and 1bC1/1bC1/2A receptors may escape ER retention, and therefore contribute to the total NMDA receptor-mediated currents. 1aC2 and 1bC1 subunits with mutations in the agonist binding pocket that abolish glycine binding (indicated as FATL) render any receptor containing these subunits nonfunctional. Coexpression of 1aC2, 1bC1-FATL, and GluN2A therefore generates functional receptors that are escaped 1aC2/1aC2/2A receptors, while coexpression of 1aC2-FATL, 1bC1, and GluN2A generates functional receptors that are escaped 1bC1/1bC1/2 receptors. (D) Coexpression of 1aC2 and 1bC1 (as well as GluN2A or GluN2B) produced robust current responses on days 2–4 after cRNA injection, whereas current responses from 1aC2/1aC2/2 and 1bC1/1bC1/2 receptors that may have escaped ER retention remained small compared with the total currents. Data are mean ± S.D., and each bar is from six oocytes. (E) The efficiency of selective expression of NMDA receptors containing two distinct GluN1 isoforms can be assessed by determining the functional contribution of escaped receptors using the FATL mutations. The fractional escape current, calculated as the sum of escape currents divided by the total current, were always smaller than 10% on days 2–4 after cRNA injection for GluN1 isoforms in combination with either GluN2A or GluN2B. Data are mean ± S.D.
To evaluate the efficiency by which the C1 and C2 tags control GluN1 subunit composition, GluN1-1aC2 (1aC2), GluN1-1bC1 (1bC1), and GluN2 subunits were initially coexpressed in Xenopus oocytes. In theory, the presence of C1 and C2 in the same receptor would mask the retention signals and exclusively allow cell-surface expression of 1aC2/1bC1/2 receptors (Fig. 2B). However, some 1aC2/1aC2/2 and 1bC1/1bC1/2 receptors may escape ER retention. To determine the functional contribution of these escaped receptors to the overall NMDA receptor-mediated currents, we generated 1aC2 and 1bC1 subunits with two mutations in the agonist binding pocket that abolish glycine binding (F484A + T518L in GluN1-1a and F505A + T539L in GluN1-1b, hereinafter denoted as FATL) and render any receptor containing these subunits nonfunctional (Kvist et al., 2013). Coexpression of 1aC2, 1bC1-FATL, and GluN2 only generates functional receptors that are escaped 1aC2/1aC2/2 receptors, while coexpression of 1aC2-FATL, 1bC1, and GluN2 only generates functional receptors that are escaped 1bC1/1bC1/2 receptors (Fig. 2C). Thus, the efficiency of selective expression of triheteromeric NMDA receptors can be assessed by determining the functional contribution of escaped receptors using the FATL mutations (Fig. 2, C–E). We determined the maximal response to saturating concentrations of glutamate + glycine from oocytes coexpressing 1aC2 + 1bC1 + GluN2 (total current), 1aC2 + 1bC1-FATL + GluN2 (escape current), and 1aC2-FATL + 1bC1 + GluN2 (escape current). On days 2–4 after cRNA injection, the fractional escape current, calculated as the sum of escape currents divided by the total current, were typically smaller than 10% (Fig. 2, D and E). This result could be reproduced using distinct GluN1 isoforms in combination with either GluN2A or GluN2B, demonstrating the efficiency of the method for expression of triheteromeric 1aC2/1bC1/2A and 1aC2/1bC1/2B NMDA receptors in Xenopus oocytes (Fig. 2E). The FATL controls would detect if C1 and C2 tags interact between receptors, rather than within one receptor as intended. For example, if 1aC2/1aC2/2 and 1bC1/1bC1/2 receptors interact via the C1 and C2 tags to enable surface expression, then this would manifest in current responses for cells coexpressing 1aC2 + 1bC1-FATL + GluN2 or 1aC2-FATL + 1bC1 + GluN2. The results also suggest that the inclusion of one copy of GluN1 with FATL mutations in the NMDA receptor is sufficient to render this receptor nonfunctional, consistent with the requirement of simultaneous agonist binding to all GluN1 and GluN2 subunits for NMDA receptor activation (Benveniste and Mayer, 1991; Clements and Westbrook, 1991).
Activation and Deactivation of Triheteromeric 1a/1b/2A and 1a/1b/2B Receptors.
NMDA receptors that contain two GluN1-1a isoforms (1a/1a/2) have increased agonist potency and display slower deactivation of glutamate-activated responses compared with receptors that contain two GluN1-1b isoforms (1b/1b/2) (Prybylowski et al., 2000; Rumbaugh et al., 2000; Vance et al., 2012; Swanger et al., 2015). GluN1-1b expression has been shown to shorten the time course of excitatory postsynaptic currents, thereby creating the possibility that neurons can tune NMDA receptor signaling by shifting the ratio of expressed GluN1-1a and GluN1-1b isoforms in a spatiotemporal manner (Prybylowski et al., 2000; Rumbaugh et al., 2000; Swanger et al., 2015). However, the agonist potencies and deactivation kinetics for NMDA receptors that contain one GluN1-1a and one GluN1-1b isoform (1a/1b/2) are unresolved.
We determined glutamate and glycine potencies at 1a/1b/2A and 1a/1b/2B receptors as well as the respective diheteromeric 1a/2 and 1b/2 receptors expressed in Xenopus oocytes using two-electrode voltage-clamp recordings (Fig. 3, A and B; Table 2). Glutamate potencies were intermediate at 1aC2/1bC1/2A and 1aC2/1bC1/2B receptors compared with the respective 1a/2 and 1b/2 receptors. The same trend was observed for glycine potencies, albeit the differences between 1a/2 and 1b/2 receptors were less pronounced for glycine compared with glutamate activation (Fig. 3, E and F; Table 2). Importantly, the presence of the C1 and C2 tags did not change glutamate potencies in 1aC1/1aC2/2 and 1bC1/1bC2/2 compared with wild-type 1a/2 and 1b/2, respectively (Table 2).
Fig. 3.
Activation and deactivation of triheteromeric NMDA receptors. (A and B) Glutamate concentration-response data measured using two-electrode voltage-clamp recordings of responses from triheteromeric 1aC2/1bC1/2 and diheteromeric 1a/2 and 1b/2 NMDA receptors expressed in Xenopus oocytes. Responses were activated in the continuous presence of 100 µM glycine. Data are mean ± S.D. from five to eight oocytes. See Table 2 for EC50 values. (C and D) Representative whole-cell patch-clamp recordings of responses from NMDA receptors expressed in HEK293 cells. Responses are activated by a brief application (3–5 ms) of 1 mM glutamate in the continuous presence of 100 µM glycine and normalized to the peak amplitude. See Table 3 for rise times and deactivation time constants. (E and F) Glycine concentration-response data measured using two-electrode voltage-clamp recordings of NMDA receptor responses activated in the continuous presence of 100 µM glutamate. Data are mean ± S.D. from five to six oocytes. See Table 2 for EC50 values. (G and H) Representative whole-cell patch-clamp recordings of responses from NMDA receptors expressed in HEK293 cells. Responses are activated by a brief application (3–5 ms) of 100 µM glycine in the continuous presence of 1 mM glutamate and normalized to the peak amplitude. See Table 3 for rise times and deactivation time constants.
TABLE 2.
Concentration-response data for glutamate and glycine
Data were measured using two-electrode voltage-clamp recordings of responses from NMDA receptors expressed in Xenopus oocytes. Glutamate concentration-response data were determined in the continous presence of 100 µM glycine, and glycine concentration-response data were determined in the continous presence of 100 µM glutamate. Data are presented as mean ± S.E.M., and n is the number of oocytes used to generate the data. Statistical tests were performed using log EC50 values, and GluN2A-containing receptors were not compared with GluN2B-containing receptors.
| Glutamate |
Glycine |
|||||
|---|---|---|---|---|---|---|
| EC50 | Hill Slope | n | EC50 | Hill Slope | n | |
| µM | µM | |||||
| 1a/2A | 3.42 ± 0.06# | 1.53 | 6 | 0.86 ± 0.03# | 1.68 | 6 |
| 1aC2/1aC1/2A | 3.41 ± 0.13# | 1.46 | 8 | |||
| 1aC2/1bC1/2A | 4.16 ± 0.12*,# | 1.47 | 6 | 1.07 ± 0.02*,# | 1.47 | 6 |
| 1bC2/1bC1/2A | 5.40 ± 0.14* | 1.37 | 6 | |||
| 1b/2A | 5.11 ± 0.19* | 1.48 | 6 | 1.33 ± 0.01* | 1.51 | 5 |
| 1a/2B | 1.41 ± 0.12# | 1.63 | 5 | 0.29 ± 0.01 | 1.64 | 6 |
| 1aC2/1aC1/2B | 1.32 ± 0.06# | 1.52 | 6 | |||
| 1aC2/1bC1/2B | 2.09 ± 0.04*,# | 1.54 | 6 | 0.33 ± 0.01# | 1.58 | 5 |
| 1bC2/1bC1/2B | 2.62 ± 0.04* | 1.49 | 6 | |||
| 1b/2B | 2.93 ± 0.17* | 1.52 | 6 | 0.29 ± 0.01 | 1.57 | 6 |
indicates significantly different from 1a/2 and # indicates significantly different from 1b/2 (P < 0.05; one-way ANOVA with Tukey’s post hoc test).
To evaluate the deactivation time course of triheteromeric 1aC2/1bC1/2A and 1aC2/1bC1/2B receptors, we adapted the method for expression in HEK293 cells to enable rapid-perfusion whole-cell patch-clamp recordings. The key requirement for fidelity of the method in HEK293 cells is to maintain a relatively low NMDA receptor expression, presumably to minimize crowding of the ER and Golgi compartments with receptors containing unmasked ER retention/retrieval signals. To achieve this, we developed DNA constructs for simultaneous low GluN1 expression and high expression of EGFP (see Materials and Methods). These constructs are useful for patch-clamp electrophysiological experiments that require low NMDA receptor expression but high EGFP expression for identification of transfected cells. We compared the deactivation time courses of 1aC1/1bC2/2A and 1aC1/1bC2/2B receptors as well as the respective diheteromeric 1a/2 and 1b/2 receptors. The weighted deactivation time constant (τw) following brief (3–5 ms) activation by 1 mM glutamate in the continuous presence of 100 µM glycine was intermediate for 1aC2/1bC1/2A (41 ms) compared with 1a/2A (49 ms) and 1b/2A (32 ms) (Fig. 3C; Table 3). The intermediate τw was also observed for glutamate deactivation of 1aC2/1bC1/2B (279 ms) compared with 1a/2B (511 ms) and 1b/2B (151 ms) (Fig. 3D; Table 3). The deactivation time course following brief (3–5 ms) activation by 100 µM glycine in the continuous presence of 1 mM glutamate was also evaluated. The τw for glycine deactivation was intermediate for 1aC2/1bC1/2A (190 ms) compared with 1a/2A (223 ms) and 1b/2A (159 ms) as well as for 1aC2/1bC1/2B (563 ms) compared with 1a/2B (748 ms) and 1b/2B (457 ms) (Fig. 3, G and H; Table 3).
TABLE 3.
Time courses of macroscopic NMDA receptor responses
Responses from NMDA receptors expressed in HEK293 were measured using whole-cell patch-clamp receordings. Cells were activated by a brief application (3–5 ms) of either 1 mM glutamate in the continuous presence of 100 µM glycine or 100 µM glycine in the continuous presence of 1 mM glutamate. Rise times are for 10%–90% of the response amplitude and deactivation time constants were determined using two-exponential fits to obtain the τfast, τslow, and fitted percentage of the fast component (% fast). Weighted time constants (τweighted) were calculated as described in Materials and Methods. Data are presented as mean ± S.E.M., and n is the number of cells used to generate the data. Only τweighted values were compared, and GluN2A-containing receptors were not compared with GluN2B-containing receptors.
| Rise Time | τfast | τslow | % fast | τweighted | n | |
|---|---|---|---|---|---|---|
| ms | ms | ms | ms | |||
| Glutamate activation | ||||||
| 1a/2A | 6.1 ± 0.3 | 36 ± 2 | 141 ± 24 | 80 ± 6 | 49 ± 2# | 9 |
| 1aC2/1bC1/2A | 6.1 ± 0.5 | 32 ± 3 | 139 ± 15 | 89 ± 3 | 41 ± 4 | 9 |
| 1b/2A | 5.3 ± 0.3 | 26 ± 3 | 85 ± 55 | 81 ± 8 | 32 ± 2* | 9 |
| 1a/2B | 13.6 ± 1.0 | 214 ± 22 | 844 ± 53 | 52 ± 3 | 511 ± 23# | 9 |
| 1aC2/1bC1/2B | 13.3 ± 0.6 | 148 ± 15 | 550 ± 65 | 65 ± 5 | 279 ± 17*,# | 9 |
| 1b/2B | 11.8 ± 0.8 | 85 ± 8 | 316 ± 12 | 71 ± 4 | 151 ± 8* | 9 |
| Glycine activation | ||||||
| 1a/2A | 10.6 ± 0.7 | 139 ± 8 | 443 ± 38 | 71 ± 5 | 223 ± 8# | 7 |
| 1aC2/1bC1/2A | 9.0 ± 1.0 | 111 ± 24 | 537 ± 143 | 66 ± 12 | 190 ± 16 | 6 |
| 1b/2A | 12.1 ± 0.5 | 123 ± 8 | 411 ± 74 | 83 ± 5 | 159 ± 9* | 7 |
| 1a/2B | 14.9 ± 1.5 | 410 ± 49 | 1194 ± 185 | 47 ± 9 | 748 ± 34# | 7 |
| 1aC2/1bC1/2B | 15.0 ± 1.6 | 371 ± 35 | 1254 ± 261 | 69 ± 10 | 563 ± 30*,# | 6 |
| 1b/2B | 12.6 ± 0.7 | 253 ± 54 | 859 ± 249 | 46 ± 13 | 457 ± 23* | 7 |
indicates significantly different from 1a/2; #indicates significantly different from 1b/2 (P < 0.05; one-way ANOVA with Tukey’s post hoc test).
In summary, these data corroborate the previously described accelerated deactivation of diheteromeric 1b/2 receptors compared with 1a/2 receptors (Rumbaugh et al., 2000; Vance et al., 2012; Swanger et al., 2015). Furthermore, the results establish that the inclusion of one copy of GluN1-1b, which includes residues encoded by exon 5, is sufficient to decrease agonist potency and to accelerate the deactivation time course, resulting in functional properties that are intermediate to those of 1a/2 and 1b/2 receptors.
Proton Inhibition of Triheteromeric 1a/1b/2A and 1a/1b/2B Receptors.
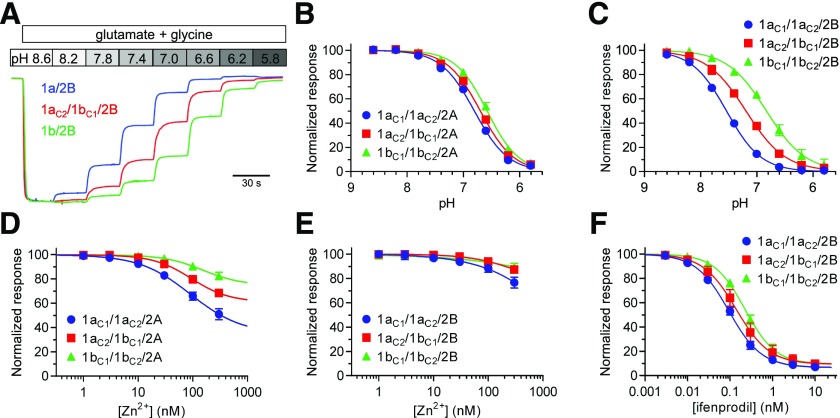
Neuronal NMDA receptors are inhibited by extracellular protons with a proton IC50 near physiological pH (7.2–7.4), and can therefore respond to small changes in extracellular pH under physiological conditions (Giffard et al., 1990; Traynelis and Cull-Candy, 1990, 1991; Vyklický et al., 1990). Residues encoded by exon 5 in the GluN1 subunit attenuate proton inhibition since proton sensitivity is diminished for NMDA receptors with two copies of the GluN1-1b isoform (Traynelis et al., 1995, 1998). We examined proton inhibition of triheteromeric 1a/1b/2A and 1a/1b/2B receptors expressed in Xenopus oocytes using two-electrode voltage-clamp recordings. Proton pIC50 (i.e., pH that produce 50% inhibition) was 6.70 for 1aC2/1bC1/2A, which is significantly different from both 1a/2A (6.74) and 1b/2A (6.47), and proton pIC50 was intermediate for 1aC2/1bC1/2B (7.19) compared with 1a/2B (7.49) and 1b/2B (6.71) (Fig. 4, A–C; Table 4). Thus, the inclusion of one copy of GluN1-1b reduces proton inhibition, albeit this reduction is less pronounced for GluN2A-containing receptors compared with GluN2B-containing NMDA receptors.
Fig. 4.
Pharmacological properties of triheteromeric NMDA receptors. (A) Representative two-electrode voltage-clamp recordings showing differences in proton inhibition of triheteromeric 1aC2/1bC1/2B and diheteromeric 1a/2B and 1b/2B NMDA receptors. (B and C) Proton concentration-inhibition data measured using two-electrode voltage-clamp recordings of responses from GluN2A- and GluN2B-containing NMDA receptors. Responses were activated by 100 µM glutamate in the continuous presence of 100 µM glycine and inhibited by increasing concentrations of protons (i.e., lower pH). Data are mean ± S.D. from four to six oocytes. (D and E) Extracellular Zn2+ concentration-inhibition data for responses from GluN2A- and GluN2B-containing NMDA receptors. Responses were activated by 100 µM glutamate in the continuous presence of 100 µM glycine and the oocytes were voltage clamped at −20 mV to minimize low-affinity voltage-dependent channel block by Zn2+. Data are mean ± S.D. from four to six oocytes. (F) Ifenprodil concentration-inhibition data for responses from GluN2B-containing NMDA receptors activated by 100 µM glutamate in the continuous presence of 100 µM glycine. Data are mean ± S.D. from five to six oocytes. See Table 3 for IC50 values.
TABLE 4.
Concentration-response data for allosteric NMDA receptor modulators
Data were measured using two-electrode voltage-clamp recordings of responses from NMDA receptors expressed in Xenopus oocytes. Percentage of inhibition was obtained from the fitted maximal inhibition at saturating antagonist concentrations. Responses were activated by 100 µM glutamate plus 100 µM glycine. Maximal proton inhibition was 100%. Spermine potentation is the percentage of enhancement of responses by 100 µM spermine. Data are presented as mean ± S.E.M., and n is the number of oocytes. Statistical tests were performed using log IC50 values, and GluN2A-containing receptors were not compared with GluN2B-containing receptors.
| Proton Inhibition |
IC50 | Inhibition | Hill Slope | n | |||
|---|---|---|---|---|---|---|---|
| pIC50 | Hill Slope | n | |||||
| pH | nM | % | |||||
| Zn2+ inhibition | |||||||
| 1a/2A | 6.74 ± 0.01# | 1.5 | 4 | ||||
| 1aC2/1aC1/2A | 6.80 ± 0.00# | 1.4 | 6 | 91 ± 7# | 65 ± 2# | 0.9 | 5 |
| 1aC2/1bC1/2A | 6.70 ± 0.01*,# | 1.5 | 5 | 95 ± 4# | 39 ± 2*,# | 1.3 | 5 |
| 1bC2/1bC1/2A | 6.58 ± 0.01* | 1.6 | 6 | 167 ± 13* | 26 ± 3* | 1.1 | 5 |
| 1b/2A | 6.47 ± 0.01* | 1.7 | 4 | ||||
| Ifenprodil inhibition | |||||||
| 1a/2B | 7.49 ± 0.01# | 1.5 | 4 | ||||
| 1aC2/1aC1/2B | 7.54 ± 0.01# | 1.4 | 4 | 94 ± 3# | 94 ± 1 | 1.1 | 5 |
| 1aC2/1bC1/2B | 7.19 ± 0.01*,# | 1.2 | 5 | 143 ± 13*,# | 91 ± 1 | 1.1 | 6 |
| 1bC2/1bC1/2B | 6.83 ± 0.01* | 1.2 | 4 | 225 ± 10* | 91 ± 1 | 1.2 | 6 |
| 1b/2B | 6.71 ± 0.03* | 1.2 | 6 | ||||
indicates significantly different from 1a/2 and # indicates significantly different from 1b/2 (P < 0.05; one-way ANOVA with Tukey’s post hoc test).
Allosteric Inhibition of Triheteromeric 1a/1b/2A and 1a/1b/2B Receptors.
Extracellular Zn2+ is a GluN2A-selective negative allosteric modulator with a high-affinity binding site located in the amino-terminal domain of the GluN2A subunit (Romero-Hernandez et al., 2016). Similarly, ifenprodil is a GluN2B-selective negative allosteric modulator, albeit this noncompetitive antagonist binds with high affinity to a site located at the interface between the amino-terminal domains of the GluN1 and GluN2B subunits (Karakas et al., 2011). High-affinity inhibition by extracellular Zn2+ and ifenprodil appears to be mediated by a mechanism in which binding of these ligands enhances proton inhibition and thereby reduces receptor function (Pahk and Williams, 1997; Mott et al., 1998; Traynelis et al., 1998; Choi and Lipton, 1999; Erreger and Traynelis, 2008; Bhatt et al., 2013). Since residues encoded by exon 5 in the GluN1 subunit are key determinants of proton inhibition, we evaluated inhibition of triheteromeric NMDA receptors by Zn2+ and ifenprodil at physiological pH 7.4. Extracellular Zn2+ inhibited diheteromeric 1aC1/1aC2/2A with greater potency and efficacy compared with diheteromeric 1bC1/1bC2/2A, whereas triheteromeric 1aC2/1bC1/2A was inhibited with intermediate potency and efficacy (Fig. 4D; Table 4). Extracellular Zn2+ up to 300 nM only minimally affected responses from GluN2B-containing NMDA receptors (Fig. 4E), consistent with the absence of a high-affinity binding site in the GluN2B amino-terminal domain (Rachline et al., 2005; Karakas et al., 2009). Ifenprodil potency was intermediate for 1aC2/1bC1/2B (143 nM) compared with 1aC1/1aC2/2B (94 nM) and 1bC1/1bC2/2B (225 nM), but maximal inhibition was unaffected by residues encoded by exon 5 in GluN1 (Fig. 4F; Table 4). These results suggest that one copy of GluN1-1b reduces inhibition of 1a/1b/2 receptors by both extracellular Zn2+ and ifenprodil. Intermediate inhibition of 1a/1b/2 receptors would be consistent with a mechanism of action in which proton inhibition is enhanced by extracellular Zn2+ and ifenprodil since 1a/1b/2 receptors display intermediate proton sensitivity (Fig. 4, A–C; Table 4).
Potentiation of Triheteromeric 1a/1b/2B Receptors by Extracellular Polyamines.
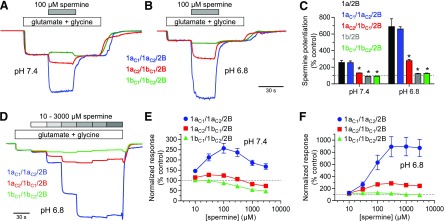
Binding of extracellular polyamines, such as spermine, to GluN2B-containing NMDA receptors have been proposed to enhance receptor function by relieving proton inhibition (Traynelis et al., 1995; Kashiwagi et al., 1996, 1997; Gallagher et al., 1997). Spermine potentiation is therefore strongly influenced by residues encoded by exon 5 in GluN1, and 1a/2B receptors, which are highly sensitive to proton inhibition, are strongly potentiated by spermine compared with 1b/2B receptors (Rumbaugh et al., 2000). We evaluated potentiation of GluN2B-containing NMDA receptors by spermine at physiological pH 7.4 and acidic pH 6.8 using two-electrode voltage-clamp recordings at a holding potential of −20 mV to minimize voltage-dependent channel block by spermine. Spermine (100 µM) potentiated responses from 1a/2B by 2.6- and 6.9-fold at pH 7.4 and 6.8, respectively (Fig. 5, A–C). Responses from 1aC2/1bC1/2B were more modestly enhanced by spermine (1.3-fold potentiation at pH 7.4 and 2.8-fold at pH 6.8), and the strong potentiation by spermine was markedly reduced at 1b/2B (0.9-fold potentiation at pH 7.4 and 1.2-fold at pH 6.8) (Fig. 5, A–C). Concentration-response data showed that 100 µM spermine produces maximal enhancement of 1a/2B at physiological pH 7.4, whereas channel block by spermine counteracts the potentiating effect at higher concentrations (Fig. 5, D–F). These results suggest that 1a/1b/2B receptors are potentiated by extracellular spermine despite the presence of one copy of residues encoded by exon 5 in GluN1-1b.
Fig. 5.
Potentiation of triheteromeric NMDA receptors by spermine. (A and B) Representative two-electrode voltage-clamp recordings showing potentiation of GluN2B-containing NMDA receptors by extracellular spermine at pH 7.4 and 6.8. Responses were activated by 100 µM glutamate in the continuous presence of 100 µM glycine. Oocytes were voltage-clamped at −20 mV to minimize voltage-dependent channel block by spermine. (C) Summary of potentiation of GluN2B-containing NMDA receptors by 100 µM spermine at pH 7.4 and 6.8. Data are normalized to control in the absence of spermine (100%) and are shown as mean ± S.D. from six to eight oocytes. *indicates significantly different from 1a/2B at the same pH (P < 0.05; one-way analysis of variance with Tukey’s post hoc test). (D) Representative two-electrode voltage-clamp recordings showing the concentration dependence of potentiation by spermine for GluN2B-containing NMDA receptors at pH 6.8. Responses were activated by 100 µM glutamate in the continuous presence of 100 µM glycine. (E and F) Spermine concentration-response data for responses from GluN2B-containing NMDA receptors at pH 7.4 and 6.8. Data are mean ± S.D. from five to six oocytes.
Contribution of Channel Pore Residues at the N Site to Block by Extracellular Mg2+.
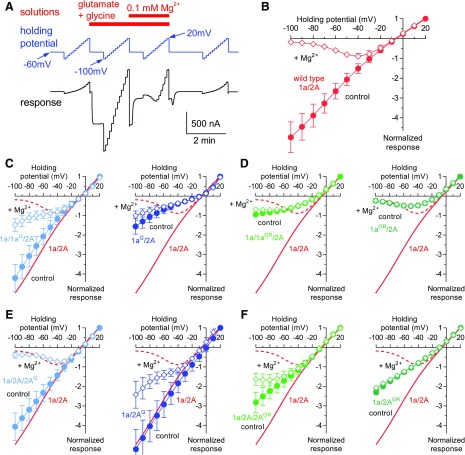
We have previously described an approach to manipulate a single GluN2 subunit in NMDA receptors (Hansen et al., 2014), which has provided insight into interactions between subunits and their influence on receptor function (Sun et al., 2017). The method to manipulate only one of the two GluN1 subunits in the NMDA receptor creates additional possibilities for experimental design in studies of receptor structure and function. To explore this idea, we evaluated the contribution of residues in the ion channel pore of GluN1 and GluN2A subunits to Ca2+ permeability and voltage-dependent block by extracellular Mg2+.
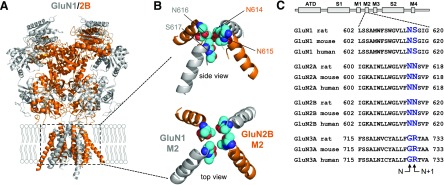
Key determinants of channel block by Mg2+ reside at a narrow constriction of the channel pore formed by residues in the membrane reentrant loop M2 (i.e., the N site) (Burnashev et al., 1992; Wollmuth et al., 1996, 1998; Sobolevsky et al., 2002) (Fig. 6). The residue at the position of the N site is an asparagine (N) in both GluN1 and GluN2 subunits (Fig. 6C). However, the narrow constriction in GluN1/2 receptors is formed by the N site asparagine in GluN1 and the asparagine residue adjacent to the N site (i.e., N + 1 site) in GluN2.
Fig. 6.
Key determinants of channel permeation and block in NMDA receptor subunits. (A) Crystal structure of the GluN1/2B NMDA receptor (PDB ID 4TLM) (Lee et al., 2014). The intracellular CTD is omitted from the structure. (B) Isolated views of the M2 membrane reentrant loops from the crystal structure with residues at the N and N + 1 sites highlighted as spheres. (C) Sequence alignment of residues in the M2 membrane reentrant loops of rat, mouse, and human GluN1-1a, GluN2A, GluN2B, and GluN3A subunits with residues at the N and N + 1 sites highlighted.
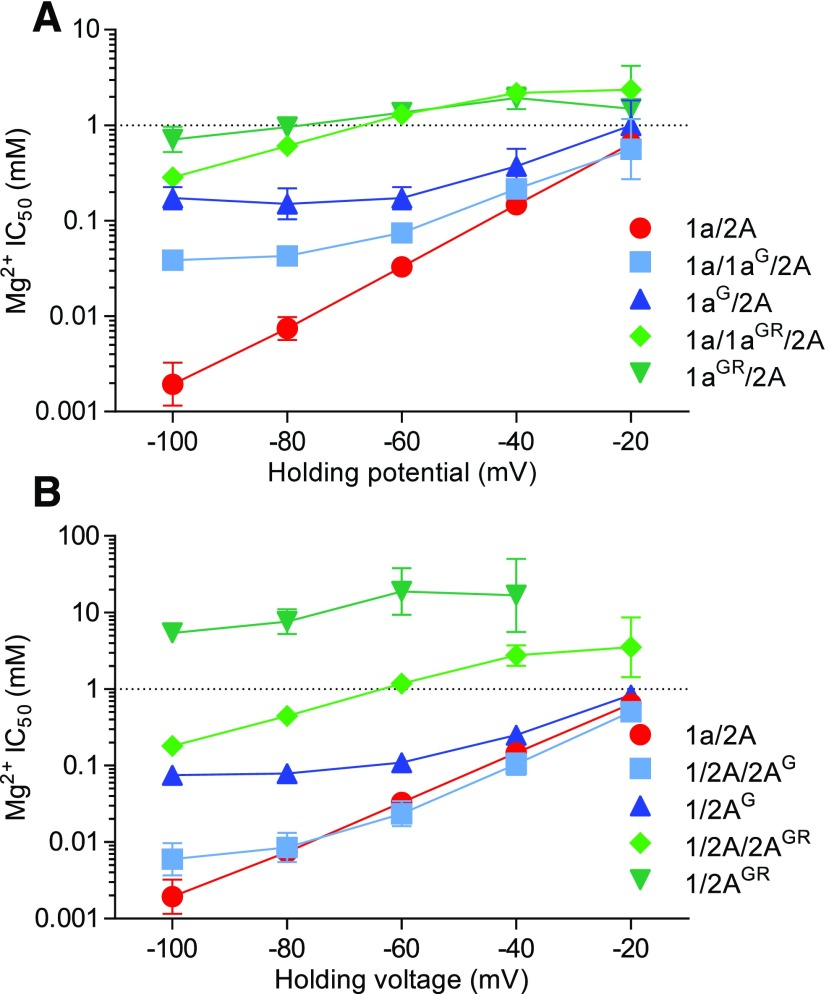
We evaluated Mg2+ block of NMDA receptors with a glycine residue at the N site in either one GluN1 (N616G) or one GluN2A (N614G) subunit (1a/1aG/2A or 1a/2A/2AG receptors). Mg2+ block of these triheteromeric receptors were compared with wild-type 1a/2A and receptors with glycine residues at both N sites in either GluN1 or GluN2A subunits (1aG/2A or 1a/2AG receptors). The current-voltage relationship of NMDA receptor responses was determined in the absence and presence of 0.1 mM Mg2+ using two-electrode voltage-clamp recordings (Fig. 7), and IC50 values for Mg2+ block were determined at holding potentials from −100 to −20 mV (Fig. 8). One copy of glycine at the N site in GluN1 diminished Mg2+ block and significantly increased Mg2+ IC50 at −60 mV by 2.2-fold from 34 ± 2 µM (n = 12) at wild-type 1a/2A to 75 ± 3 µM (n = 12) at 1a/1aG/2A (Figs. 7C and 8A; see Supplemental Table 1 for statistical tests). This effect was increased for 1aG/2A receptors, which contain two copies of glycine at the N site; the current-voltage relationship of 1aG/2A revealed some inward rectification under these experimental conditions, and the Mg2+ IC50 at −60 mV was significantly increased by 5.3-fold to 177 ± 19 µM (n = 6) at 1aG/2A compared with wild-type 1a/2A (Figs. 7C and 8A; Supplemental Table 1). By contrast, one or two copies of glycine at the N site in GluN2A (i.e., 1a/2A/2AG and 1a2AG) were less effective at reducing Mg2+ block; the Mg2+ IC50 at −60 mV was unaffected for 1a/2A/2AG (25 ± 3 µM; n = 12) and significantly increased by 3.3-fold for 1a/2AG (110 ± 4 µM; n = 4) compared with wild-type 1a/2A (Figs. 7E and 8B; Supplemental Table 1). Thus, glycine residues at the N site in GluN1 and GluN2A more strongly affect Mg2+ block of receptors with two copies compared with one copy of the substitution. Furthermore, glycine residues at the N site more effectively reduce Mg2+ block when introduced in GluN1 compared with GluN2A. This asymmetric effect is consistent with previous observations that key determinants of Mg2+ block are formed by the N site asparagine in GluN1 and the asparagine residue adjacent to the N site (i.e., N + 1 site) in GluN2 (Burnashev et al., 1992; Wollmuth et al., 1996, 1998; Sobolevsky et al., 2002).
Fig. 7.
Current-voltage relationships for Mg2+ block of NMDA receptors with mutated pore residues. (A) Protocols for solution applications and holding potentials shown with the corresponding NMDA receptor responses measured using two-electrode voltage-clamp recordings to determine the current-voltage relationships for Mg2+ block of NMDA receptors. (B) Current-voltage relationship of responses from wild-type 1a/2A receptors activated by 100 µM glutamate in the continuous presence of 100 µM glycine in the absence of extracellular Mg2+ (control) and in the presence of 0.1 mM Mg2+. Responses are normalized to the response amplitude at +20 mV and are shown as mean ± S.D. from 10 oocytes. (C and D) Current-voltage relationships for block by 0.1 mM Mg2+ of NMDA receptors with one or two copies of glycine (G) at the N site or GR at the N and N + 1 sites in GluN1. Data from (B) for wild-type 1a/2A are shown as red lines. Responses are normalized to the response amplitude at +20 mV and are shown as mean ± S.D. from 6 to 12 oocytes. (E and F) Current-voltage relationships for block by 0.1 mM Mg2+ of NMDA receptors with one or two copies of G at the N site or GR at the N and N + 1 sites in GluN2A. Data are mean ± S.D. from 6 to 12 oocytes.
Fig. 8.
Voltage dependence of NMDA receptor inhibition by extracellular Mg2+. (A and B) IC50 values for Mg2+ block plotted as a function of holding potential for NMDA receptors with one or two copies of glycine (G) at the N site or GR at the N and N + 1 sites in either GluN1 or GluN2A. Responses were activated by 100 µM glutamate in the continuous presence of 100 µM glycine and inhibition by increasing concentrations of Mg2+ was measured using two-electrode voltage-clamp recordings. Data are mean ± S.D. from 6 to 12 oocytes. See Supplemental Table 1 for values and statistical analyses.
Effects of Pore Residues from GluN3 on Mg2+ Block of GluN1/2 Receptors.
Coexpression of GluN3 with GluN1 and GluN2 subunits produces NMDA receptors with decreased Ca2+ permeability and diminished Mg2+ block, albeit it remains to be convincingly demonstrated whether these receptors are triheteromeric GluN1/2/3 receptors and/or a mixed population of GluN1/2 and GluN1/3 receptors (e.g., see (Das et al., 1998; Perez-Otano et al., 2001; Chatterton et al., 2002). The N site residue is glycine (G) and the N + 1 site residue is arginine (R) in GluN3 subunits (Fig. 6C), and diheteromeric GluN1/3 receptors are characterized by low Ca2+ permeability and the lack of block by extracellular Mg2+ (Chatterton et al., 2002; Sasaki et al., 2002; Smothers and Woodward, 2007; Madry et al., 2010).
To determine the influence of pore residues at the N and N + 1 sites in GluN3 on permeation and block in GluN1/2 receptors, we evaluated Mg2+ block of NMDA receptors with glycine-arginine (GR) residues at the N and N + 1 sites in either one GluN1 (N616G + S617R) or one GluN2A (N614G + N615R) subunit (1a/1aGR/2A or 1a/2A/2AGR receptors). These 1a/1aGR/2A or 1a/2A/2AGR receptors were designed to mimic the narrow constriction of the channel pore in triheteromeric GluN1/2/3 receptors, in which one GluN3 subunit replace either one GluN1 or one GluN2 subunit. Mg2+ block of these triheteromeric receptors was compared with wild-type 1a/2A and receptors with substitutions in either two GluN1 or two GluN2A subunits (1aGR/2A or 1a/2AGR receptors). One copy of GR residues in GluN1 strongly diminished Mg2+ block and significantly increased Mg2+ IC50 at −60 mV by 39-fold to 1300 ± 60 µM (n = 6) at 1a/1aGR/2A compared with wild-type 1a/2A (Figs. 7D and 8A; Supplemental Table 1). This effect was not changed for receptors with two copies of GR residues in GluN1, since the Mg2+ IC50 at −60 mV was similarly increased by 41-fold to 1370 ± 100 µM (n = 6) at 1aGR/2A compared with wild-type 1a/2A (Figs. 7D and 8A; Supplemental Table 1). One or two copies of GR residues in GluN2A (i.e., 1a/2A/2AGR and 1a2AGR) also resulted in strong reduction of Mg2+ block; the Mg2+ IC50 at −60 mV was significantly increased by 36-fold for 1a/2A/2AGR (1200 ± 80 µM; n = 11) and by 700-fold for 1a/2AGR (23.4 ± 7.1 mM; n = 6) compared with wild-type 1a/2A (Figs. 7F and 8B; Supplemental Table 1). One copy of GR residues in GluN1 therefore reduces Mg2+ block to virtually the same extent as two copies, whereas one copy of GR residues in GluN2A produces a lower reduction of Mg2+ block compared with two copies. Thus, these results further highlight the asymmetric contribution of pore residues in GluN1 and GluN2A to channel block. In addition, these results demonstrate that a single copy of pore residues from GluN3 subunits strongly attenuates Mg2+ block of NMDA receptors, independently of whether the residues are introduced to one GluN1 or one GluN2 subunit.
Contribution of Channel Pore Residues to Ca2+ Permeability.
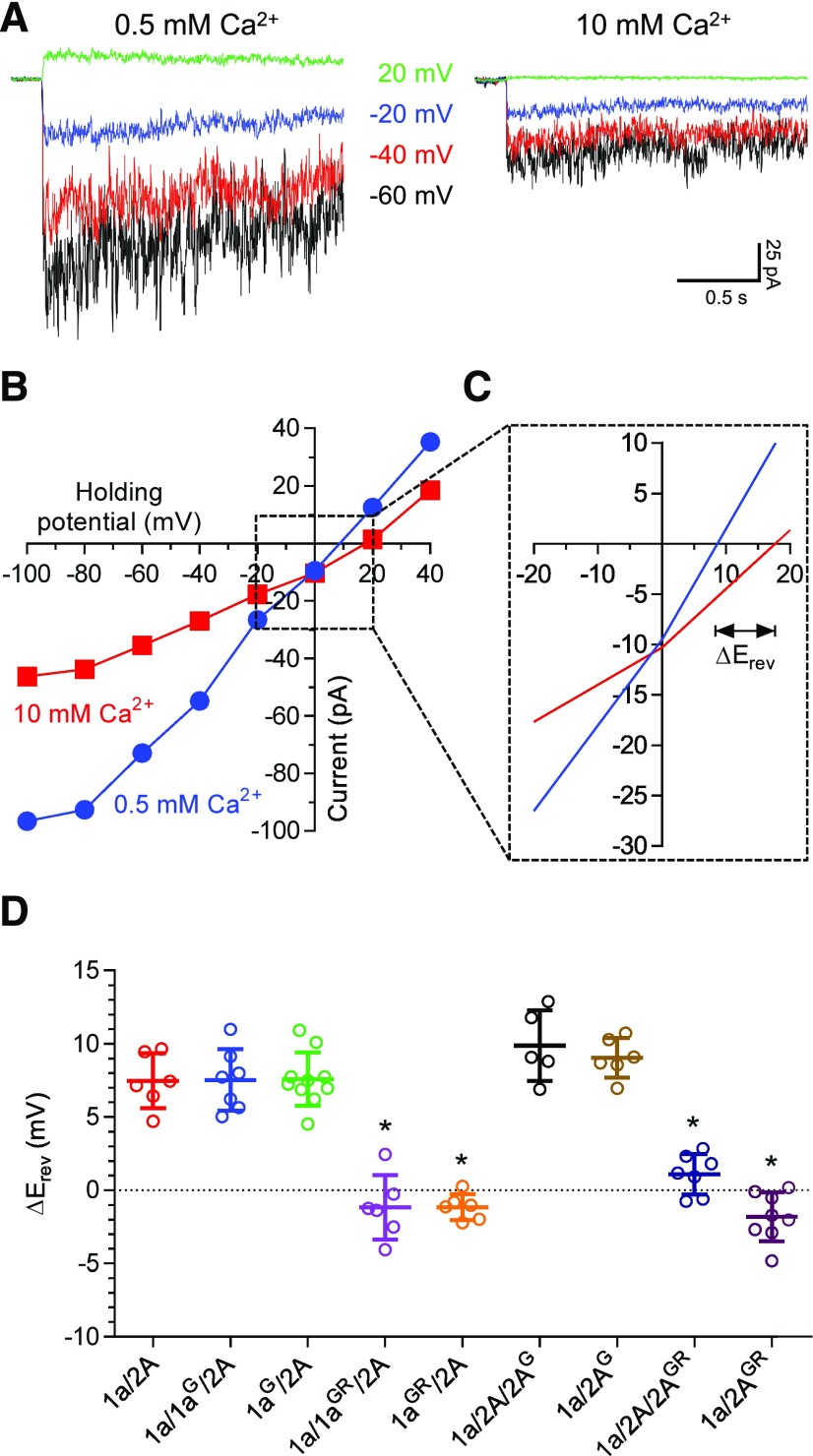
We also determined the impact of G and GR mutations at the N and N + 1 sites on Ca2+ permeability of NMDA receptors expressed in HEK293 cells using whole-cell patch-clamp recordings. We determined the current-voltage relationships of responses activated by 100 µM glutamate plus 100 µM glycine in the presence of 0.5 or 10 mM external Ca2+, since the change in reversal potential (ΔErev) is dependent on Ca2+ permeability (Fig. 9, A and B). The reversal potential increases by 7.5 ± 0.8 mV (n = 6) for wild-type 1a/2A receptors when external Ca2+ is increased from 0.5 and 10 mM, consistent with high Ca2+ permeability (Fig. 9, C and D). However, the response amplitude of wild-type 1a/2A is inhibited in high external Ca2+ (Fig. 9, A and B), consistent previous observations from single-channel recordings of a Ca2+-dependent decrease in channel conductance (Premkumar and Auerbach, 1996; Wyllie et al., 1996; Premkumar et al., 1997; Dravid et al., 2008). Glycine at the N site in GluN1 has no effects on ΔErev for 1a/1aG/2A (7.5 ± 0.8 mV; n = 7) and 1aG/2A (7.6 ± 0.6 mV; n = 10) compared with wild-type 1a/2A, suggesting that Ca2+ permeability is unchanged. Glycine at the N site in GluN2A increases ΔErev for 1a/1a/2AG (9.9 ± 1.1 mV; n = 5) and 1a/2AG (9.0 ± 0.5 mV; n = 6), suggesting that the Ca2+ permeability is increased (Fig. 9D). By contrast, GR residues in GluN1 results in ΔErev values close to 0 mV for 1a/1aGR/2A (−1.2 ± 0.9 mV; n = 6) and 1aGR/2A (−1.2 ± 0.4 mV; n = 6), indicating that a single copy of GR residues in GluN1 is sufficient to abolish Ca2+ permeability (Fig. 9D). Similarly, GR residues in GluN2A results in ΔErev of 1.1 ± 0.5 mV (n = 7) for 1a/1a/2AGR and −1.8 ± 0.6 mV (n = 8) for 1a/2AGR (Fig. 9D). These results using ΔErev values to assess Ca2+ permeability suggest that one copy of GR residues in GluN1 reduces Ca2+ permeability to the same extent as two copies, whereas two copies of GR residues in GluN2A appear to result in a stronger reduction in Ca2+ permeability compared with one copy. Similar to the effects in Mg2+ block, a single copy of pore residues from GluN3 virtually abolishes Ca2+ permeability of NMDA receptors, independently of whether the residues are introduced to one GluN1 or one GluN2 subunit.
Fig. 9.
Ca2+ permeability of NMDA receptors with mutated pore forming residues. (A) Representative whole-cell patch-clamp recordings of responses from HEK293 cells expressing wild-type 1a/2A receptors. Responses are activated by 100 µM glutamate in the continuous presence of 100 µM glycine at different holding potentials and with either 0.5 or 10 mM Ca2+ in the extracellular recording solution. (B) Representative current-voltage relationships of wild-type 1a/2A receptor responses in 0.5 and 10 mM extracellular Ca2+. (C) Closer view of the current-voltage relationships used to determine the shift in reversal potential (ΔErev) resulting from changing the concentration of extracellular Ca2+. (D) Summary of changes in reversal potential (ΔErev) upon increasing extracellular Ca2+ for NMDA receptors with one or two copies of glycine (G) at the N site or GR at the N and N + 1 sites in either GluN1 or GluN2A. *indicates significantly different from 1a/2A (P < 0.05; one-way ANOVA with Tukey’s post hoc test). Data are mean ± S.D.
Discussion
We demonstrate that GluN1-1a and GluN1-1b isoforms can form functional triheteromeric 1a/1b/2A and 1a/1b/2B receptors and describe a method for selective expression of recombinant NMDA receptors containing two different GluN1 isoforms in Xenopus oocytes and HEK293 cells. We reveal intermediate deactivation kinetics and pharmacological properties of 1a/1b/2A and 1a/1b/2B receptors compared with the respective diheteromeric NMDA receptor subtypes. The intermediate properties of triheteromeric NMDA receptors with two distinct GluN1 isoforms contrast the dominant effect of GluN2A on the function of triheteromeric GluN1/2A/2B receptors (Tovar et al., 2013; Hansen et al., 2014; Sun et al., 2017), which account for a large portion of NMDA receptors in the adult cortex and hippocampus (Sheng et al., 1994; Luo et al., 1997; Rauner and Köhr, 2011). During development of the CNS, the prevalence of diheteromeric GluN1/2B receptors with slow deactivation kinetics is decreased due to increased expression of GluN2A and increased assembly of triheteromeric GluN1/2A/2B receptors with faster deactivation kinetics. This developmental shift in subunit composition from GluN1/2B to GluN2A-containing receptors therefore accelerates NMDA receptor-mediated synaptic currents. Triheteromeric NMDA receptors containing other combinations of GluN2 subunits, such as GluN1/2A/2C and GluN1/2B/2D, are also widely expressed in the CNS (Chazot et al., 1994; Cathala et al., 2000; Piña-Crespo and Gibb, 2002; Brickley et al., 2003; Jones and Gibb, 2005; Brothwell et al., 2008; Swanger et al., 2015; Perszyk et al., 2016). The intermediate properties of triheteromeric NMDA receptors containing two different GluN1 isoforms create the possibility that neurons can also tune NMDA receptor signaling by shifting the ratio of expressed GluN1-1a and GluN1-1b isoforms.
In addition to enabling functional evaluation of triheteromeric NMDA receptor subtypes, the method to control subunit composition also provides the ability to introduce mutations in a single subunit. To demonstrate this, we evaluated the contribution of channel pore residues to Mg2+ block and Ca2+ permeability. We introduced a glycine substitution at the N site of one GluN1-1a (1aG) or one GluN2A (2AG) subunit in triheteromeric 1a/1aG/2A and 1a/2A/2AG receptors and compared the properties of these receptors to those of wild-type 1a/2A and receptors with substitutions in both GluN1 (1aG/2A) or both GluN2A (1a/2AG) subunits. This approach to control the mutation copy number (or mutation dosage) clearly exposed the previously described asymmetric contribution of N site residues in GluN1 and GluN2 to Mg2+ block (Burnashev et al., 1992; Wollmuth et al., 1996, 1998; Sobolevsky et al., 2002). This asymmetric contribution would have been less discernable if only diheteromeric 1aG/2A and 1a/2AG receptors were compared with wild-type 1a/2A receptors. In addition, we demonstrate that a single copy of pore residues from GluN3 subunits provides strong relief of Mg2+ block and abolishes Ca2+ permeability, independent of whether the GluN3 residues are introduced into one GluN1 or one GluN2 subunit in the GluN1/2 receptor. There is currently a lack of published evidence to show whether GluN3 substitutes one GluN1 or one GluN2 subunit to form triheteromeric GluN1/2/3 receptors and these findings do not distinguish between the two potential subunit stoichiometries. However, the findings provide, for the first time, important clues to permeation and block of putative triheteromeric GluN1/2/3 receptors, which have not yet been functionally characterized as a homogenous receptor population in heterologous or native expression systems. Thus, examination of functional receptor properties and mutation copy number (0, 1, or 2 mutations) can reveal important new insights to structure-function relationships of NMDA receptors.
The approach to control the mutation copy number provides a powerful tool to determine the functional impact of disease-related de novo mutations identified in NMDA receptor subunits of patients with neurological conditions (reviewed in Burnashev and Szepetowski, 2015; Yuan et al., 2015; Hu et al., 2016). These patients are heterozygous for the disease-related de novo mutation and only a fraction of their NMDA receptors will possess two copies of the mutation, whereas a larger fraction presumably will have only one copy of the mutation. Here, we provide an approach to evaluate the relationship between NMDA receptor function and the copy number of GluN1 mutations. Understanding such relationships could be useful to rationalize the link between de novo GluN1 mutations and symptoms in various neurological conditions.
In summary, we reveal additional complexity in NMDA receptor signaling created from diversity in subunit composition by demonstrating that coexpression of GluN1-1a and GluN1-1b results in the formation of functional triheteromeric receptors with properties intermediate to those of the respective diheteromeric NMDA receptors. This new insight suggests a mechanism by which neurons could tune NMDA receptor signaling by shifting the ratio of expressed GluN1-1a and GluN1-1b isoforms, although evidence for expression of NMDA receptors containing two distinct GluN1 isoforms in vivo is currently lacking. We describe a method to selectively express triheteromeric NMDA receptors containing two distinct GluN1 isoforms in heterologous expression systems, which can enable new studies on receptor structure and function, mechanisms of allosteric modulation, and functional consequences of GluN1 mutations. This work expands the repertoire of innovative approaches to study NMDA receptors, thereby facilitating new discoveries related to NMDA receptor signaling in key brain functions as well as in CNS disorders.
Acknowledgments
The authors thank Gina C. Bullard for excellent technical assistance and the staff in the BioSpectroscopy Core Research Laboratory at University of Montana for the use of equipment, critical comments, and discussions.
Abbreviations
- CFP
cyan fluorescent protein
- CNS
central nervous system
- CTD
C-terminal domain
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- FLIM
fluorescence lifetime imaging
- FRET
Förster resonance energy transfer
- GR
glycine-arginine
- NMDA
N-methyl-d-aspartate
- ORF
open reading frame
- YFP
yellow fluorescent protein
Authorship Contributions
Participated in research design: Yi, Zachariassen, Dorsett, Hansen.
Conducted experiments: Yi, Zachariassen, Dorsett, Hansen.
Performed data analysis: Yi, Zachariassen, Dorsett, Hansen.
Wrote or contributed to the writing of the manuscript: Yi, Zachariassen, Dorsett, Hansen.
Footnotes
This work was supported by the National Institutes of Health [Grants P20GM103546, R01NS097536, and S10OD021806].
The authors declare no conflict of interest.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. (1994) Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol 347:150–160. [DOI] [PubMed] [Google Scholar]
- Barbosa C, Peixeiro I, Romão L. (2013) Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9:e1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste M, Mayer ML. (1991) Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys J 59:560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt JM, Prakash A, Suryavanshi PS, Dravid SM. (2013) Effect of ifenprodil on GluN1/GluN2B N-methyl-D-aspartate receptor gating. Mol Pharmacol 83:9–21. [DOI] [PubMed] [Google Scholar]
- Brickley SG, Misra C, Mok MH, Mishina M, Cull-Candy SG. (2003) NR2B and NR2D subunits coassemble in cerebellar Golgi cells to form a distinct NMDA receptor subtype restricted to extrasynaptic sites. J Neurosci 23:4958–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothwell SL, Barber JL, Monaghan DT, Jane DE, Gibb AJ, Jones S. (2008) NR2B- and NR2D-containing synaptic NMDA receptors in developing rat substantia nigra pars compacta dopaminergic neurones. J Physiol 586:739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnashev N, Schoepfer R, Monyer H, Ruppersberg JP, Günther W, Seeburg PH, Sakmann B. (1992) Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257:1415–1419. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Szepetowski P. (2015) NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol 20:73–82. [DOI] [PubMed] [Google Scholar]
- Cathala L, Misra C, Cull-Candy S. (2000) Developmental profile of the changing properties of NMDA receptors at cerebellar mossy fiber-granule cell synapses. J Neurosci 20:5899–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, et al. (2002) Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 415:793–798. [DOI] [PubMed] [Google Scholar]
- Chazot PL, Coleman SK, Cik M, Stephenson FA. (1994) Molecular characterization of N-methyl-D-aspartate receptors expressed in mammalian cells yields evidence for the coexistence of three subunit types within a discrete receptor molecule. J Biol Chem 269:24403–24409. [PubMed] [Google Scholar]
- Choi YB, Lipton SA. (1999) Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron 23:171–180. [DOI] [PubMed] [Google Scholar]
- Clements JD, Westbrook GL. (1991) Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron 7:605–613. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T, Premkumar LS, Takasu M, Crandall JE, Dikkes P, Conner DA, Rayudu PV, Cheung W, et al. (1998) Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393:377–381. [DOI] [PubMed] [Google Scholar]
- Dravid SM, Prakash A, Traynelis SF. (2008) Activation of recombinant NR1/NR2C NMDA receptors. J Physiol 586:4425–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Bennett MV, Zukin RS. (1993) Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc Natl Acad Sci USA 90:6731–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand GM, Gregor P, Zheng X, Bennett MV, Uhl GR, Zukin RS. (1992) Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc Natl Acad Sci USA 89:9359–9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF. (2008) Zinc inhibition of rat NR1/NR2A N-methyl-D-aspartate receptors. J Physiol 586:763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MJ, Huang H, Grant ER, Lynch DR. (1997) The NR2B-specific interactions of polyamines and protons with the N-methyl-D-aspartate receptor. J Biol Chem 272:24971–24979. [DOI] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. (2009) Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 459:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffard RG, Monyer H, Christine CW, Choi DW. (1990) Acidosis reduces NMDA receptor activation, glutamate neurotoxicity, and oxygen-glucose deprivation neuronal injury in cortical cultures. Brain Res 506:339–342. [DOI] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Yuan H, Traynelis SF. (2014) Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron 81:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. (2013) Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol 84:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. (1993) Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron 10:943–954. [DOI] [PubMed] [Google Scholar]
- Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. (2016) Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci 132:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M, et al. (1993) Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem 268:2836–2843. [PubMed] [Google Scholar]
- Jones S, Gibb AJ. (2005) Functional NR2B- and NR2D-containing NMDA receptor channels in rat substantia nigra dopaminergic neurones. J Physiol 569:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. (2009) Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J 28:3910–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. (2011) Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 475:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson U, Sjödin J, Angeby Möller K, Johansson S, Wikström L, Näsström J. (2002) Glutamate-induced currents reveal three functionally distinct NMDA receptor populations in rat dorsal horn—effects of peripheral nerve lesion and inflammation. Neuroscience 112:861–868. [DOI] [PubMed] [Google Scholar]
- Kashiwagi K, Fukuchi J, Chao J, Igarashi K, Williams K. (1996) An aspartate residue in the extracellular loop of the N-methyl-D-aspartate receptor controls sensitivity to spermine and protons. Mol Pharmacol 49:1131–1141. [PubMed] [Google Scholar]
- Kashiwagi K, Pahk AJ, Masuko T, Igarashi K, Williams K. (1997) Block and modulation of N-methyl-D-aspartate receptors by polyamines and protons: role of amino acid residues in the transmembrane and pore-forming regions of NR1 and NR2 subunits. Mol Pharmacol 52:701–713. [DOI] [PubMed] [Google Scholar]
- Kvist T, Greenwood JR, Hansen KB, Traynelis SF, Bräuner-Osborne H. (2013) Structure-based discovery of antagonists for GluN3-containing N-methyl-D-aspartate receptors. Neuropharmacology 75:324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH. (1994) Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J Neurosci 14:3180–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Lü W, Michel JC, Goehring A, Du J, Song X, Gouaux E. (2014) NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. (1997) The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B). Mol Pharmacol 51:79–86. [DOI] [PubMed] [Google Scholar]
- Madry C, Betz H, Geiger JR, Laube B. (2010) Potentiation of glycine-gated NR1/NR3A NMDA receptors relieves Ca2+-dependent outward rectification. Front Mol Neurosci 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJ, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA. (2011) An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One 6:e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Thomas AA. (2002) Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem J 367:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. (1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12:529–540. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. (1992) Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256:1217–1221. [DOI] [PubMed] [Google Scholar]
- Mott DD, Doherty JJ, Zhang S, Washburn MS, Fendley MJ, Lyuboslavsky P, Traynelis SF, Dingledine R. (1998) Phenylethanolamines inhibit NMDA receptors by enhancing proton inhibition. Nat Neurosci 1:659–667. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. (2003) Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron 40:581–594. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Axel R, Shneider NA. (1992) Alternative splicing generates functionally distinct N-methyl-D-aspartate receptors. Proc Natl Acad Sci USA 89:8552–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paarmann I, Frermann D, Keller BU, Villmann C, Breitinger HG, Hollmann M. (2005) Kinetics and subunit composition of NMDA receptors in respiratory-related neurons. J Neurochem 93:812–824. [DOI] [PubMed] [Google Scholar]
- Pahk AJ, Williams K. (1997) Influence of extracellular pH on inhibition by ifenprodil at N-methyl-D-aspartate receptors in Xenopus oocytes. Neurosci Lett 225:29–32. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. (2013) NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 14:383–400. [DOI] [PubMed] [Google Scholar]
- Paupard MC, Friedman LK, Zukin RS. (1997) Developmental regulation and cell-specific expression of N-methyl-D-aspartate receptor splice variants in rat hippocampus. Neuroscience 79:399–409. [DOI] [PubMed] [Google Scholar]
- Perez-Otano I, Schulteis CT, Contractor A, Lipton SA, Trimmer JS, Sucher NJ, Heinemann SF. (2001) Assembly with the NR1 subunit is required for surface expression of NR3A-containing NMDA receptors. J Neurosci 21:1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RE, DiRaddo JO, Strong KL, Low CM, Ogden KK, Khatri A, Vargish GA, Pelkey KA, Tricoire L, Liotta DC, et al. (2016) GluN2D-containing N-methyl-D-aspartate receptors mediate synaptic transmission in hippocampal interneurons and regulate interneuron activity. Mol Pharmacol 90:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña-Crespo JC, Gibb AJ. (2002) Subtypes of NMDA receptors in new-born rat hippocampal granule cells. J Physiol 541:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar LS, Auerbach A. (1996) Identification of a high affinity divalent cation binding site near the entrance of the NMDA receptor channel. Neuron 16:869–880. [DOI] [PubMed] [Google Scholar]
- Premkumar LS, Qin F, Auerbach A. (1997) Subconductance states of a mutant NMDA receptor channel kinetics, calcium, and voltage dependence. J Gen Physiol 109:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prybylowski K, Rumbaugh G, Wolfe BB, Vicini S. (2000) Increased exon 5 expression alters extrasynaptic NMDA receptors in cerebellar neurons. J Neurochem 75:1140–1146. [DOI] [PubMed] [Google Scholar]
- Rachline J, Perin-Dureau F, Le Goff A, Neyton J, Paoletti P. (2005) The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J Neurosci 25:308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauner C, Köhr G. (2011) Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. J Biol Chem 286:7558–7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H. (2016) Molecular basis for subtype specificity and high-affinity zinc inhibition in the GluN1-GluN2A NMDA receptor amino-terminal domain. Neuron 92:1324–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S. (2000) Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol 83:1300–1306. [DOI] [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong HK, Gong X, Chan SF, Zhang D, et al. (2002) Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol 87:2052–2063. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. (2001) An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci 21:3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. (1994) Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368:144–147. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. (2007) Pharmacological characterization of glycine-activated currents in HEK 293 cells expressing N-methyl-D-aspartate NR1 and NR3 subunits. J Pharmacol Exp Ther 322:739–748. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rooney L, Wollmuth LP. (2002) Staggering of subunits in NMDAR channels. Biophys J 83:3304–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert DG, Testa CM, Penney JB, Jr, Young AB. (1993) Alternatively spliced isoforms of the NMDAR1 glutamate receptor subunit: differential expression in the basal ganglia of the rat. Neurosci Lett 152:161–164. [DOI] [PubMed] [Google Scholar]
- Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S. (1992) Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem Biophys Res Commun 185:826–832. [DOI] [PubMed] [Google Scholar]
- Sun W, Hansen KB, Jahr CE. (2017) Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron 94:58–64 e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SA, Vance KM, Pare JF, Sotty F, Fog K, Smith Y, Traynelis SF. (2015) NMDA receptors containing the GluN2D subunit control neuronal function in the subthalamic nucleus. J Neurosci 35:15971–15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, McGinley MJ, Westbrook GL. (2013) Triheteromeric NMDA receptors at hippocampal synapses. J Neurosci 33:9150–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. (1998) Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci 18:6163–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Cull-Candy SG. (1990) Proton inhibition of N-methyl-D-aspartate receptors in cerebellar neurons. Nature 345:347–350. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Cull-Candy SG. (1991) Pharmacological properties and H+ sensitivity of excitatory amino acid receptor channels in rat cerebellar granule neurones. J Physiol 433:727–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. (1995) Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268:873–876. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. (2010) Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62:405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Hansen KB, Traynelis SF. (2012) GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol 590:3857–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. (1998) Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol 79:555–566. [DOI] [PubMed] [Google Scholar]
- Vyklický L, Jr, Vlachová V, Krůsek J. (1990) The effect of external pH changes on responses to excitatory amino acids in mouse hippocampal neurones. J Physiol 430:497–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Sakmann B. (1998) Adjacent asparagines in the NR2-subunit of the NMDA receptor channel control the voltage-dependent block by extracellular Mg2+. J Physiol 506:13–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmuth LP, Kuner T, Seeburg PH, Sakmann B. (1996) Differential contribution of the NR1- and NR2A-subunits to the selectivity filter of recombinant NMDA receptor channels. J Physiol 491:779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie DJ, Béhé P, Nassar M, Schoepfer R, Colquhoun D. (1996) Single-channel currents from recombinant NMDA NR1a/NR2D receptors expressed in Xenopus oocytes. Proc Biol Sci 263:1079–1086. [DOI] [PubMed] [Google Scholar]
- Yi F, Traynelis SF, Hansen KB. (2017) Selective cell-surface expression of triheteromeric NMDA receptors. Methods Mol Biol 1677:145–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. (2009) Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 29:12045–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Low CM, Moody OA, Jenkins A, Traynelis SF. (2015) Ionotropic GABA and glutamate receptor mutations and human neurologic diseases. Mol Pharmacol 88:203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariassen LG, Katchan L, Jensen AG, Pickering DS, Plested AJ, Kristensen AS. (2016) Structural rearrangement of the intracellular domains during AMPA receptor activation. Proc Natl Acad Sci USA 113:E3950–E3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zheng X, Paupard MC, Wang AP, Santchi L, Friedman LK, Zukin RS, Bennett MVL. (1994) Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. Proc Natl Acad Sci USA 91:10883–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Carrozza DP, Williams K, Pritchett DB, Molinoff PB. (1995) Expression of mRNAs encoding subunits of the NMDA receptor in developing rat brain. J Neurochem 64:531–539. [DOI] [PubMed] [Google Scholar]