Abstract
In aerobic organisms, mitochondrial F1Fo-ATP-synthase is the major site of ATP production. Beside this fundamental role, the protein complex is involved in shaping and maintenance of cristae. Previous electron microscopic studies identified the dissociation of F1Fo-ATP-synthase dimers and oligomers during organismic aging correlating with a massive remodeling of the mitochondrial inner membrane. Here we report results aimed to experimentally proof this impact and to obtain further insights into the control of these processes. We focused on the role of the two dimer assembly factors PaATPE and PaATPG of the aging model Podospora anserina. Ablation of either protein strongly affects mitochondrial function and leads to an accumulation of senescence markers demonstrating that the inhibition of dimer formation negatively influences vital functions and accelerates organismic aging. Our data validate a model that links mitochondrial membrane remodeling to aging and identify specific molecular components triggering this process.
Keywords: aging, F1Fo-ATP-synthase, membranes, mitochondria, remodeling
INTRODUCTION
Mitochondria are eukaryotic organelles involved in a number of essential processes including the synthesis of iron/sulfur clusters, biosynthesis of lipids and amino acids, energy transduction and generation of ATP. In aerobic organisms, most of this universal cellular energy carrier is generated by oxidative phosphorylation (OXPHOS) at the respiratory chain. The protein complexes and supercomplexes of the respiratory chain are located in invaginations of the inner membrane (IM), the so-called cristae, which occur in different shapes (e.g. tubular, lamellar) 1,2,3,4. At their basis, the cristae junctions (CJ), cristae are connected to the outer membrane (OM), which surrounds the organelle.
F1Fo-ATP-synthase is an IM-bound protein complex that is essential for the electrochemical-driven generation of ATP. The individual proteins of the complex are encoded by the nuclear and by mitochondrial DNA (mtDNA). Mutations in the corresponding genes lead to F1Fo-ATP-synthase deficiency, mitochondrial dysfunction 5 and severe neurodegenerative pathologies like Alzheimer’s and Parkinson’s disease 6,7,8 and have a strong impact on biological aging 9,10,11. Beside its enzymatic role, the F1Fo-ATP-synthase complex plays a key role in shaping cristae structure 12,13,14. In particular, rows of F1Fo-ATP-synthase dimers at the tips of the cristae appear to be critical for maintaining their typical convex curvature 15,16. Recent studies identified the impact of another protein complex, the mitochondrial contact site and cristae organizing system (MICOS) 17, in controlling IM ultrastructure 3,18,19,20. This complex is localized at the CJ. Despite the distant localization of the two complexes, interactions between them, or at least of components of them, were demonstrated. It was found that Mic10, a core component of MICOS, interacts with the dimeric F1Fo-ATP-synthase and stabilizes the complex 21,22. The mechanisms regulating these interactions are largely unknown but are of key relevance for the remodeling of mitochondrial ultrastructure that is observed as adaptive response to physiological changes 23,24.
Pronounced changes of the mitochondrial ultrastructure were previously demonstrated to occur during aging of the aging model organism Podospora anserina. Mitochondria from juvenile cultures predominantly contain well-structured lamellar cristae with rows of F1Fo-ATP-synthase dimers at their tips. During aging, the invaginations of the IM recede and the membrane curvature becomes concave forming a system of vesicles in the matrix space. This process of membrane reorganization goes along with a dissociation of F1Fo-ATP-synthase dimers. Occasionally, vesicles in the matrix of old mitochondria are found in close contact with the OM at contact sites. It was suggested that at late age the OM bursts at these sites and releases mitochondrial content to the cytosol inducing programmed cell death (PCD) 25. The relevance of this series of cellular events observed during P. anserina aging is supported by genetic manipulations of the molecular machinery involved in the control of PCD, the final phase in the life cycle of P. anserina 26,27. Strikingly, overexpression of a gene coding for the mitochondrial peptidyl prolyl-cis,trans-isomerase, cyclophilin D (CYPD), which is involved in the control of the mitochondrial permeability transition pore (mPTP) opening 28, leads to accelerated aging. In this overexpression strain mitochondria of young chronological age (6-days) display a vesicular ultrastructure that is characteristic for old wild-type mitochondria of 18-days of age 29. The molecular components involved in this process and its regulation were unknown.
In the yeast Saccharomyces cerevisiae, it was shown that dimer and oligomer formation is mediated by the interaction of two dimer assembly factors, subunit e (Atp21, Su e) and subunit g (Atp20, Su g) of F1Fo-ATP-synthase 30,31. Deletion of a gene coding for either one of these proteins leads to a loss of dimers and oligomers without influencing the catalytic ATP-synthase activity of the enzyme but inducing a strong alteration of mitochondrial ultrastructure 30,32. Surprisingly, the downregulation of the gene coding for subunit e or g in HeLa cells affects not only the stability of dimers and oligomers but also the catalytic activity of the enzyme and mitochondrial morphology 33.
Here we report novel data from a study aimed to generate mechanistic insights into the impact of mitochondrial ultrastructure on biological aging. We focused on PaAtpe and PaAtpg encoding the two putative F1Fo-ATP-synthase dimer assembly factors of P. anserina and investigated their age-related expression. Moreover, we analyzed the impact of the ablation of the two dimer assembly factors on biological aging in detail. Our data extend a model describing the age-related mitochondrial membrane remodeling including the role of F1Fo-ATP-synthase dimers and dimer assembly factors PaATPE and PaATPG.
RESULTS AND DISCUSSION
The abundance of F1Fo-ATP-synthase dimers decreases during aging
Previous work revealed an essential role of F1Fo-ATP-synthase dimers in the formation of mitochondrial cristae 12,13,14,16 and a decrease of mitochondria with lamellar cristae accompanied by an increase of those with a vesicular ultrastructure during aging of P. anserina 25,29. To elucidate the biochemical basis of these microscopic observations, we analyzed mitochondria from batches isolated from 6-days and 16-days old cultures of the P. anserina wild type that previously were used for cryo-electron microscopy 25 and subjected them to BN-PAGE analysis (Fig. 1A). We observed a 20% decrease of F1Fo-ATP-synthase dimers in 16-days old cultures compared to 6-days old cultures. This effect is likely to be underestimated because of the elimination of impaired mitochondria during aging by autophagy, a quality control pathway that increases during aging 34,35. Apart from a decrease in abundance of F1Fo-ATP-synthase dimers, a marked change of supercomplex amount S1 (I1III2IV1) and S0 (I1III2IV0) is observed during aging (Fig. 1A). In cells of 6-days old cultures S1 predominates while this supercomplex is decreased in 16-days cultures. In contrast, an increase of S0 is observed during aging (Fig. 1A).
Figure 1. FIGURE 1: Age-dependent dimer decrease and analysis of expression of PaAtpe and PaAtpg in P. anserina.
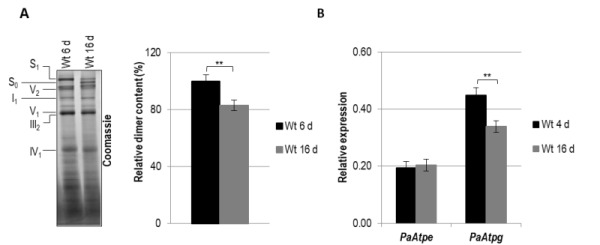
(A) Representative BN-PAGE analysis and quantification of mitochondrial protein extracts (V2, F1Fo-ATP-synthase dimers) from three independent 6-days and 16-days old wild-type strains. F1Fo-ATP-synthase dimers were normalized to Coomassie stained gel. Relative dimer content of 6-days old wild-type strains was set to 100%. Mitochondrial protein complexes were stained with Coomassie Blue and complexes and supercomplexes S1, S0, V2, I1, V1, III1 and IV1 are indicated.
(B) Transcript analysis of PaAtpe and PaAtpg in three young (4 d) and old (16 d) wild type cultures, respectively. RNA was isolated and gene expression was determined by qRT-PCR analyses. The relative expression was normalized to the expression of PaPorin coding for a mitochondrial outer membrane protein. Error bars represent the standard deviation and the P-values were determined by Student´s t test.
Next, we investigated the age-associated expression of PaAtpe and PaAtpg, coding for two potential factors involved in the control of F1Fo-ATP-synthase dimer formation. A comparative transcript analysis of these genes of young (4-days) and senescent (16-days) P. anserina cultures revealed a reduction of the PaAtpg transcript level but no significant alteration of PaAtpe transcripts (Fig. 1B), suggesting that the age-related regulation or stability of the two putative dimer-specific genes differs. Interestingly, in the young wild type, transcript levels of PaAtpe are considerably lower than those of PaAtpg. These observations let us speculate about different regulatory mechanisms and probably different functions of the two dimer assembly factors. It appears that PaATPE is a limiting but central factor in dimer formation. This idea is supported by different studies with yeast demonstrating that dimer assembly factors bind to the Fo part of ATP-synthase in a sequential manner. First Su e binds to the Fo part followed by the association of Su g 31,36. It was shown that the presence of Su e is required for stabilization of Su g whereas Su g is not necessary for binding of Su e 37. Thus, Su e appears to play a key role in ATP-synthase dimer formation while Su g acts as a protein supporting Su e in this function 30,31.
Overall, the observed changes in F1Fo-ATP-synthase dimer quantity and alterations of supercomplexes S1 to S0 indicate mitochondrial membrane reorganization processes during aging of P. anserina. These biochemical data are in concordance with the observations of earlier cryo-electron microscopy studies 25,29.
Ablation of PaATPE and PaATPG affects dimer formation and mitochondrial ultrastructure
Next, we focused on the role of PaATPE and PaATPG in the formation of F1Fo-ATP-synthase dimers. In yeast and HeLa cells the lack of one of the homologues is sufficient to inhibit F1Fo-ATP-synthase dimer formation 30,31,33. In both model organisms, the deletion of either gene strongly affects mitochondrial ultrastructure. In yeast, onion-like structures of the IM are found 12. In HeLa cells, the IM forms arch-like or longitudinal cristae 33.
We generated P. anserina strains in which either PaAtpe (P. anserina accession number: Pa_1_3740) or PaAtpg (P. anserina accession number: Pa_1_2480) were replaced by a bifunctional selection marker that mediates phleomycin and blasticidin resistance 38. Strains were verified by Southern blot analysis using a specific Phleo and PaAtpe or PaAtpg probe (Fig. 2A). Subsequent analysis of mitochondrial extracts from each of the two deletion strains by BN-PAGE revealed the absence of F1Fo-ATP-synthase dimers (Fig. 2B) and demonstrated that the P. anserina proteins encoded by these two genes are indeed involved in the dimerization of F1Fo-ATP-synthase monomers.
Figure 2. FIGURE 2: Deletion of PaAtpe or PaAtpg affects F1Fo-ATP-synthase dimer formation and mitochondrial inner membrane ultrastructure.
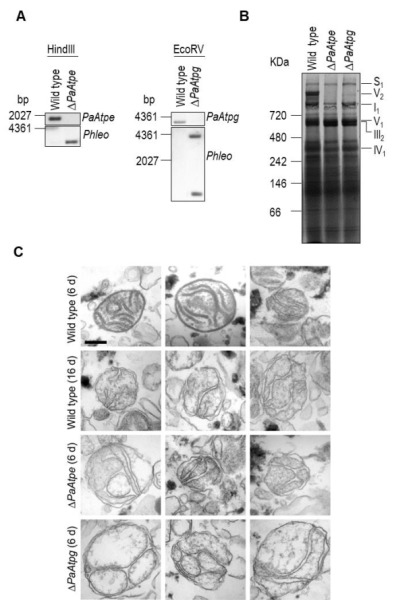
(A) Southern blot analysis of HindIII- and EcoRV-digested genomic DNA (gDNA) of wild-type, ΔPaAtpe and ΔPaAtpg strains. The digested gDNA was hybridized with a specific PaAtpe, PaAtpg and Phleo probe, respectively, and demonstrated a successful deletion of PaAtpe and PaAtpg.
(B) Representative BN-PAGE analysis of mitochondrial protein extracts from 6-days old wild-type, ΔPaAtpe and ΔPaAtpg strains. Mitochondrial complexes including S1, V2, I1, V1, III1 and IV1 were visualized by Coomassie staining.
(C) EM of isolated mitochondria of wild-type and deletion strains of different age (6 days and 16 days old). The morphology of at least 50 organelles per strain and growth condition was analyzed. Representative images are shown at the same magnification. Scale bar: 0,2 µm.
Subsequently, we analyzed whether the lack of F1Fo-ATP-synthase dimers affects mitochondrial ultrastructure. Electron microscopy of mitochondria isolated from young (6-days) and old (16-days) wild-type strains revealed pronounced remodeling of the IM during aging (Fig. 2C), corresponding to what was described earlier in a cryo-electron microscopy study 29. While well-separated lamellar cristae with a typical convex curvature at the tips could be readily found in preparations of young wild type mitochondria, mitochondria from old cultures were generally devoid of lamellar cristae and the inner membrane frequently formed septa and vesicular structures (reticulate ultrastructure). Mitochondria isolated from PaAtpe and PaAtpg deletion strains of young chronological age (i.e. 6-days) strikingly resemble old wild type mitochondria (16-days). Importantly, typical lamellar cristae could not be found in mutant or old wild type mitochondria (Fig. 2C). These results clearly demonstrate a direct impact of F1Fo-ATP-synthase dimers on the formation of lamellar cristae and aging-dependent mitochondrial membrane reorganization.
Defects in F1Fo-ATP-synthase dimerization affects mitochondrial function and leads to accelerated aging
In a series of subsequent experiments, we concentrated on the impact of F1Fo-ATP-synthase dimerization on aging of P. anserina. We compared well-known senescent markers in the two deletion strains and the wild type of defined chronological age. Apart from the recently identified ultrastructural reorganization of the IM, changes in mitochondrial morphotype 39, reorganization of the mitochondrial DNA (mtDNA) 40,41,42,43,44, an increase in hydroxide peroxide (H2O2) release from cultures 39,45 and impairments in respiration 46 were analyzed as hallmarks of aging.
Light microscopy analysis identified an accelerated change of mitochondrial morphotypes in the two investigated deletion strains. In contrast to the wild type, in which mitochondria from 3-days old cultures are mainly of a filamentous morphotype, mitochondria of the same chronological age of the two deletion mutants display a punctate morphotype that is characteristic for mitochondria of old (i.e. 16-days old) wild-type strains (Fig. 3A).
Figure 3. FIGURE 3: Impairments of mitochondrial functions affect growth and lifespan in deletion mutants.
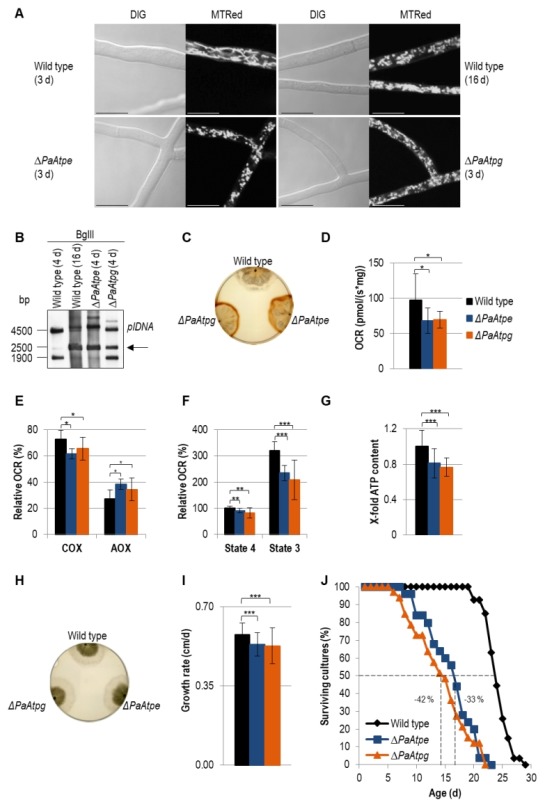
(A) Comparison of mitochondrial morphology of 3-days and 16-days old wild-type strains with those of ΔPaAtpe and ΔPaAtpg (3 d) by fluorescence microscopy. Mitochondria were stained with MitoTracker® Red and analyzed by confocal fluorescence microscopy. Scale bars: 10 µm.
(B) Representative Southern blot analysis of BglII-digested gDNA of wild-type (4-days and 16-days old), ΔPaAtpe and ΔPaAtpg strains (4-days old). The 2500 bp age-specific plDNA (arrow) fragment was detected with a specific plDNA probe.
(C) Release of ROS species H2O2 from wild-type (6-days n=3), ΔPaAtpe (6-days n=3) and ΔPaAtpg strains (6-days n = 3) visualized by DAB precipitation.
(D) Oxygen consumption (basal respiration) of 6-days old deletion mutant mycelium compared to respiration of 6-days old wild type mycelium (n = 3-4 biological replicates with a total number of 12-16 technical replicates).
(E) Relative COX- and AOX-dependent oxygen consumption rate (OCR) of 6-days old wild-type and 6-days old deletion strains mycelium after treatment with KCN (COX-inhibitor) or SHAM (AOX-inhibitor).
(F) Relative complex I-dependent OCR of 6-days old wild-type mitochondria compared to mitochondria from 6-days old deletion mutants (n = 4-8 biological replicates with a total number of 15-28 technical replicates). State 4 OCR of mitochondria from wild type was set to 100%.
(G) ATP production of 6-days old wild-type and 6-days old deletion mutant mitochondria after removal of samples during state 3 respiration measurements. Samples of three biological replicates and two technical replicates, respectively, were measured three times by a luminescence-based assay.
(H) Phenotypic analysis of indicated strains growing on M2 medium for 4 days.
(I) Growth rate of wild type (0.58 ± 0.05 cm/d, n = 27), ΔPaAtpe (0.53 ± 0.05 cm/d, n = 25) and ΔPaAtpg (0.53 ± 0.08 cm/d, n =3 3) growing on M2 medium. Growth rate was determined from day 3 to 6 of growth.
(J) Lifespan analysis of wild type (mean lifespan = 23.3 d, n = 27), ΔPaATPe (mean lifespan = 15.1 d, n = 25,) and ΔPaATPg (mean lifespan = 13.5 d, n = 33) growing on M2 medium. The error bars represent the standard deviation and the P-values were determined by Student´s t test.
Analysis of mtDNA revealed an accelerated amplification of a mitochondrial plasmid-like DNA (plDNA). In 4-days old PaAtpe and PaAtpg deletion strains this senescent marker is already strongly amplified. In the wild type, plDNA is almost absent from 4-days old cultures and found in strains of much older age (e.g. 16-days) (Fig. 3B).
Next, we analyzed H2O2 release 39. Incubation of wild-type and deletion strains of the same chronological age (6-days) with a diaminobenzidin (DAB) staining solution revealed an increase of a brown precipitate demonstrating a strong increase in H2O2 release by ΔPaAtpe and ΔPaAtpg cultures (Fig. 3C).
In order to link these different age-dependent changes to mitochondrial functions, we measured respiration of P. anserina deletion strains. Clearly, total oxygen consumption of deletion strains is reduced in comparison to the wild type (Fig. 3D). Furthermore, we determined respiration of wild-type, ΔPaAtpe and ΔPaAtpg strains in the presence and absence of respiration inhibitors potassium cyanide (KCN), inhibiting complex IV (COX), and salicylhydroxamic acid (SHAM), inhibiting the alternative oxidase AOX (Fig. 3E). Notably, in the two deletion strains alternative respiration is slightly increased as compensatory response to OXPHOS impairments and increased mitochondrial stress 46,47,48. These data are in concordance with studies in yeast in which mutations of several subunits of F1Fo-ATP-synthase like Atp4 49 or Su e and Su g 50,51 lead to a destabilization of of the complex and a reduced COX activity.
Determination of the relative oxygen consumption rate (OCR) via complex I of isolated mitochondria from wild-type, ΔPaAtpe and ΔPaAtpg strains revealed that state 4 (ADP-independent) as well as state 3 (ADP-dependent) respiration is significantly decreased in deletion mutants. State 3 respiration is most strongly affected (Fig. 3F). An impaired respiration often results in an inefficient ATP production. To test F1Fo-ATP-synthase activity, we removed part of the samples used for state 3 respiration measurements and determined ATP production in this state using a luciferin-luciferase assay. We found that ATP content in deletion strains is significantly reduced compared to the wild type (Fig. 3G) indicating that monomeric F1Fo-ATP-synthase is not as efficient as the dimeric form. Thus, the dimerization of F1Fo-ATP-synthase is not only important for F1Fo-ATP-synthase dimerization and mitochondrial ultrastructure but also for energy transduction. Similar effects were also observed in yeast and HeLa cells. In HeLa cells, downregulation of dimer-specific genes leads to an increase of doubling-times and a reduction of OXPHOS capacity 33. Yeast cells in which either one of the two dimer assembly factors is ablated display an abnormal mitochondrial morphology, a decreased growth rate on non-fermentable carbon sources and reduced respiration 12,30.
The effect of reduced ATP generation in PaAtpe and PaAtpg deletion strains is not apparent in the first days (e.g. 4 days) of growth. At this time, cultures display the same morphology and radial growth rate as the wild type (Fig. 3H). Later on (days 3 - 6), growth rate of deletion strains becomes slightly, but significantly reduced when compared to the wild type (Fig. 3I). The reduction of the growth rate in the presenescent life phase is another aging marker that together with the described changes in mitochondrial morphology and ultrastructure as well as mitochondria dysfunction, reorganization of mtDNA and the increase in ROS production suggests an acceleration of the aging process in ΔPaAtpe and ΔPaAtpg strains. This conclusion was finally verified by determination of the lifespan of the two deletion strains. In comparison to the wild type, the lifespan of the mutants is strongly reduced by 33% and 42%, respectively (Fig. 3J). Until now, such an effect on biological aging was not reported in any of the different established aging models, including yeast in which F1Fo-ATP-synthase dimerization factors were extensively studied.
Conclusions and perspectives
Here we provide novel data on the identification of individual components involved in the control of mitochondrial inner membrane remodeling and their impact on biological aging. These experimental data extend a model that was proposed from earlier cryo-electron microscopy studies 25 and from data of genetic interventions into the induction of PCD via the overexpression of a gene coding for cyclophilin D, a regulator of the mitochondrial permeability transition (mPT) 29. In this particular strain, it was demonstrated that autophagy is induced to an extent that leads to autophagic cell death 52. Significantly, CYPD is known to interact with the lateral stalk of the F1Fo-ATP-synthase decreasing its activity 53. Moreover, F1Fo-ATP-synthase dimers were demonstrated to be part of the mPTP 54 and that opening of the mPTP correlates with the dissociation of dimers 55. Overall, these and the data presented in our current study highlight the central role of F1Fo-ATP-synthase in mitochondrial biology. However, the regulatory circuits and the mechanisms involved in the different aspects of F1Fo-ATP-synthase function remain to be unraveled in detail. Concerning the role in mitochondrial ultrastructure remodeling, the identification and the characterization of components involved in this process is crucial. Apart from the role of F1Fo-ATP-synthase dimers at the cristae tips, the MICOS complex has been recently identified 18,19,20. Interestingly, it was found that both complexes are necessary for cristae formation. Notwithstanding that the core MICOS protein Mic60 and dimer assembly factors Su e and Su g play antagonistic roles 14, the other core MICOS protein, Mic10, was found to interact with Su e of the F1Fo-ATP-synthase 21,22. Furthermore, the demonstration of a F1Fo-ATP-synthase oligomerization promoting effect of Mic27 21 also suggests an interplay of the MICOS complex with the F1Fo-ATP-synthase dimer. The elucidation of regulation of these crosstalks will certainly be a key aspect of future investigations.
MATERIALS AND METHODS
P. anserina strains and cultivation
In this study wild type ‘s’ 56 and the newly generated deletion strains ΔPaAtpe and ΔPaAtpg were used. Strains were grown on standard cornmeal agar (BMM) at 27°C under constant light 57. For spore germination, standard cornmeal agar (BMM) supplemented with 60 mM ammonium acetate was used and incubated at 27°C in dark for 2 days. For all experiments strains were cultivated on M2 plates for 2 days at 27°C under constant light and transferred in CM liquid medium for 2 days at 27°C under constant light. Flasks were shaken at 160 rounds per minute (rpm). All strains used in this study were derived from monokaryotic ascospores 57.
Cloning procedures and generation of P. anserina mutants
Deletion of PaAtpe and PaAtpg was performed with the method described by Hamann et al., 2005 38. Briefly, a small 5’-flank of genes PaAtpe and PaAtpg, respectively, was amplified with oligonucleotides ATPe_KO1 (5’-GTGGTACCCGCTGTGAGAGCTTCTTC-3’), ATPe_KO2 (5’-GCAAGCTTTTTGAGGAATCTGGGGGC-3’), ATPg_KO1 (5’-CGGGTACCGTGAAGAGCGTATGTTGG-3’) and ATPg_KO2 (5’-GCAAGCTTTTCGAACTTCGGGCACTG-3’). A small 3’-flank of the same genes was amplified with oligonucleotides ATPe_KO3 (5’- GCACTAGTACTGTCCGTCGACGAACT-3’), ATPe_KO4 (5’- AAAGCGGCCGCTCAACGATGTGACATTG-3’), ATPg_KO3 (5’- CTACTAGTTCTTAGCGCAGGGAGGTG-3’) and ATPg_KO4 (5’- CAGCGGCCGCAGGTTGCAACAGTAGTAG-3’). Recognition sites of restriction endonucleases are underlined. The 5’ fragments were digested with KpnI and HindIII and the 3’ fragments with BcuI and NotI and ligated into previously digested plasmid pKO4. The resulting plasmids were termed pAtpeKO1 and pAtpgKO1 and contain a resistance cassette with phleomycin and blasticidin marker genes for fungal and bacterial selection, respectively. The flanked resistance cassettes were digested with NotI and KpnI and used for transformation of Escherichia coli strain KS272 bearing the plasmid pKOBEG 58 and the cosmid 15C5 containing the PaAtpe locus or cosmid 13B10 containing the PaAtpg locus, respectively. Homologous recombination between the flanks of the resistance cassette and cosmid 15C5 and 13B10 leads to generation of ∆Atpe15C5 and ∆Atpg13B10, which contain the two resistance markers surrounded by large flanks. The cosmid was isolated and used for transformation of wild type P. anserina. Selection of positive PaAtpe and PaAtpg deletion strains was performed by growth on medium containing phleomycin. The deletion of PaAtpe and PaAtpg was verified by Southern blot analysis as described below.
Southern blot analysis
Total DNA was isolated according to Lecellier and Silar, 1994 59. DNA restriction, gel electrophoresis and Southern blotting were performed according to standard protocols. For Southern blot hybridization and detection, digoxigenin-labeled hybridization probes (‘DIG DNA Labeling and Detection Kit’) were used according to the manufacturer’s instructions. The PaAtpe-specific hybridization probe was amplified by PCR using the oligonucleotides Pa_1_3740-1 (5’-AATGGCCTCTTCCGGAGTC-3’) and Pa_1_3740-2 (5’-CTCGAGGTCGAAGCTTGAG-3’) corresponding to 511 nucleotides of the gene PaAtpe. The PaAtpg-specific hybridization probe was amplified by PCR using the oligonucleotides Pa_1_2480-3 (5’- TCTTTCGCCCTCCTCTCAC-3’) and Pa_1_2480-4 (5’- AGACGATCTTGGCCACCTC-3’) corresponding to 418 nucleotides of the gene PaAtpg. As a hybridization probe specific for the phleomycin resistance gene (Ble), the 1293 bp BamHI-fragment of the plasmid pKO4 60 was used. As a hybridization probe specific for the plDNA the 2500 bp BamHI-fragment of the plasmid pSP17 61 was used.
Growth rate and lifespan determination
To determine lifespan and growth rate of P. anserina strains used in this study, monokaryotic ascospores were isolated from crosses of respective strains and germinated for 2 days at 27°C in the dark on BMM supplemented with 60 mM ammonium acetate. After germination mycelia were placed on M2 medium 57 and incubated at 27°C under constant light. The lifespan is defined as the period of linear growth in days (d). The growth rate was measured as growth in centimeters per day (cm/d).
Transcript analysis
Plates containing M2 medium covered with cellophane were inoculated with small pieces of mycelium and incubated for 2 days at 27°C under constant light. Total RNA was isolated from scraped fungal mycelium using the Nucleo Spin® RNA Plant-Kit (Macherey-Nagel). Reverse transcription of 1 mg total RNA was performed with the RevertAid™ M-MuLV reverse transcriptase (ThermoScientific) according to the manufacturer’s instruction. Real-time PCR was realized using iQ SYBR Green Supermix (BioRad) followed by the manufacturer’s protocol. For each gene, the efficiency (E) of the primer pairs was calculated based on a real-time PCR with a dilution series of cDNA according to E = 10[-1 ⁄ Slope] 62. The relative expression level (normalized to the level of the porin transcript) was calculated according to the following formula: Relative expression= (E(porin)^CP(porin))/(E(target gene)^CP(target gene)) with: E = PCR efficiency of the respective primer pair and CP = crossing point for each transcript.
Mitochondria isolation
Mitochondria of P. anserina cultures were isolated by differential centrifugation for measurement of mitochondrial oxygen consumption and BN-PAGE analysis 57.
Blue-native polyacrylamide gels (BN-PAGE)
BN-PAGE was performed as previously described 63. For each sample 75-100 µg of mitochondrial protein extracts were solubilized using a digitonin/protein ratio of 3:1 (w/w). Linear gradient gels (4-13%) overlaid with 3.5% stacking gels were used for separation of solubilized mitochondria. Respiratory chain components were subsequently visualized by Coomassie blue staining and assigned as described 64.
Mitochondrial oxygen consumption
Measurement of mitochondrial oxygen consumption was performed at 27°C using two different high-resolution respirometers (Oxygraph-2k series C and G, Oroboros Instruments, Innsbruck, Austria). 200 μg freshly prepared mitochondria were injected into 2 ml air saturated oxygen buffer (0.3 M sucrose, 10 mM KH2PO4, 5 mM MgCl2, 1 mM EDTA, 10 mM KCl, 0.1% BSA; pH = 7.2). To determine the complex I-dependent state 4 respiration (state 4) 20 mM pyruvate and 5 mM malate were added. Thereafter, 1.5 mM ADP was added to determine complex I-dependent state 3 respiration (state 3).
Determination of complex IV- and AOX-dependent oxygen consumption was performed using strains cultivated on M2 medium for 2 days and in CM liquid medium for 2 days as described above. Small pieces of mycelium were subsequently transferred into the respirometer and oxygen consumption was measured in liquid CM medium according to the manufacturer’s instructions. 1 mM KCN was added to inhibit respiration via complex IV. 4 mM salicylhydroxamic acid (SHAM) was added to inhibit respiration via AOX.
Data were analyzed using the manufacturer´s software DatLab 6.
ATP measurement
For determination of ATP production samples were removed after measurement of state 3 respiration. Thereafter samples were boiled in a water bath for 15 minutes. After boiling they were centrifuged for 10 min and 14,000 rpm and transferred into new reaction tubes. The supernatant was diluted (1:30) and ATP amount was measured by using the ATP Bioluminescence Assay Kit (CLS II, Roche) adapted for use in a microtiter plate format. The assay was performed according to the manufacturer’s instructions.
Fluorescence microscopy
P. anserina mycelia were grown on microscope slides with a central depression that is filled with M2 agar medium and incubated for 1-2 days at 27°C and constant light. Hyphae were stained with 1 µM of the mitochondrial specific dye Mito Tracker CMX ROS (Invitrogen) and visualized using a confocal laser scanning microscope (CLSM TCS SP5, Leica).
Electron microscopy
Electron microscopy of chemically fixed isolated mitochondria was performed as described 65.
Hydrogen peroxide release assay
To determine the H2O2 release mycelia were cultivated on M2 medium at 27°C in the dark for 4 days. The plates were flooded with a solution containing 100 mM Tris/HCl pH 6.9 and 2.5 mM diaminobenzidine (DAB) and incubated for 30 min in the dark and 27°C. Subsequently, the solution was poured off and the plates were incubated again for 3 h at 27°C in the dark.
Statistical analysis
All statistical significances were calculated using the Student t-test. P-values <0.05 were considered statistically significant. P <= 0.05: *, P <= 0.01: **, P <= 0.001: ***.
Funding Statement
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG: Os75/17-1) to HDO and via the Cluster of Excellence ‘Macromolecular Complexes’. We thank Alexandra Werner for preparation of P. anserina mitochondria of defined age.
AUTHOR CONTRIBUTIONS: HDO conceived and supervised this study. MS performed electron microscopy and, together with BW, analyzed the data. NGR performed the rest of the experiments and NGR and HDO analyzed data. HDO and NGR wrote the manu-script. BW and MS edited the manuscript. All authors read and approved the final manuscript.
References
- 1.Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793(1):5–19. doi: 10.1016/j.bbamcr.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Cogliati S, Enriquez JA, Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem Sci. 2016;41(3):261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Harner ME, Unger AK, Geerts WJ, Mari M, Izawa T, Stenger M, Geimer S, Reggiori F, Westermann B, Neupert W. An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife. 2016;5:e18853. doi: 10.7554/eLife.18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rampelt H, Zerbes RM, van der Laan M, Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim Biophys Acta. 2017;1864(4):737–746. doi: 10.1016/j.bbamcr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 5.Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. 2012;35(2):211–225. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27(2):306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coskun P, Wyrembak J, Schriner SE, Chen HW, Marciniack C, Laferla F, Wallace DC. A mitochondrial etiology of Alzheimer and Parkinson disease. Biochim Biophys Acta. 2012;1820(5):553–564. doi: 10.1016/j.bbagen.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23(2):64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 10.Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36(5):254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Osiewacz HD, Bernhardt D. Mitochondrial quality control: impact on aging and life span - a mini-review. Gerontology. 2013;59(5):413–420. doi: 10.1159/000348662. [DOI] [PubMed] [Google Scholar]
- 12.Bornhövd C, Vogel F, Neupert W, Reichert AS. Mitochondrial membrane potential is dependent on the oligomeric state of F1Fo -ATP synthase supracomplexes. J Biol Chem. 2006;281(20):13990–13998. doi: 10.1074/jbc.M512334200. [DOI] [PubMed] [Google Scholar]
- 13.Strauss M, Hofhaus G, Schroder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27(7):1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, Cyrklaff M, Neupert W, Reichert AS. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185(6):1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci U S A. 2011;108(34):14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci U S A. 2012;109(34):13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, Nunnari J. Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J Cell Biol. 2014;204(7):1083–1086. doi: 10.1083/jcb.201401006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30(21):4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195(2):323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou JC, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21(4):694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 21.Eydt K, Davies KM, Behrendt C, Wittig I, Reichert AS. Cristae architecture is determined by an interplay of the MICOS complex and the F1Fo ATP synthase via Mic27 and Mic10. Microb Cell. 2017;4(8):259–272. doi: 10.15698/mic2017.08.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rampelt H, Bohnert M, Zerbes RM, Horvath SE, Warscheid B, Pfanner N, van der Laan M. Mic10, a core subunit of the mitochondrial contact site and cristae organizing system, interacts with the dimeric F1Fo-ATP synthase. J Mol Biol. 2017;429(8):1162–1170. doi: 10.1016/j.jmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763(5-6):542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Wai T, Langer T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol Metab. 2016;27(2):105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Daum B, Walter A, Horst A, Osiewacz HD, Kühlbrandt W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc Natl Acad Sci U S A. 2013;110(38):15301–15306. doi: 10.1073/pnas.1305462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamann A, Brust D, Osiewacz HD. Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol Microbiol. 2007;65(4):948–958. doi: 10.1111/j.1365-2958.2007.05839.x. [DOI] [PubMed] [Google Scholar]
- 27.Hamann A, Brust D, Osiewacz HD. Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol. 2008;16(6):276–283. doi: 10.1016/j.tim.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Baines CP. The molecular composition of the mitochondrial permeability transition pore. J Mol Cell Cardiol. 2009;46(6):850–857. doi: 10.1016/j.yjmcc.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brust D, Daum B, Breunig C, Hamann A, Kühlbrandt W, Osiewacz HD. Cyclophilin D links programmed cell death and organismal aging in Podospora anserina. Aging Cell. 2010;9(5):761–775. doi: 10.1111/j.1474-9726.2010.00609.x. [DOI] [PubMed] [Google Scholar]
- 30.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H. Yeast mitochondrial F1Fo -ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17(24):7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner K, Rehling P, Sanjuan Szklarz LK, Taylor RD, Pfanner N, van der Laan M. Mitochondrial F1Fo-ATP synthase: the small subunits e and g associate with monomeric complexes to trigger dimerization. J Mol Biol. 2009;392(4):855–861. doi: 10.1016/j.jmb.2009.07.059. [DOI] [PubMed] [Google Scholar]
- 32.Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brethes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21(3):221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Habersetzer J, Larrieu I, Priault M, Salin B, Rossignol R, Brethes D, Paumard P. Human F1F0 ATP synthase, mitochondrial ultrastructure and OXPHOS impairment: a (super-)complex matter? PLoS One. 2013;8(10):e75429. doi: 10.1371/journal.pone.0075429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philipp O, Hamann A, Servos J, Werner A, Koch I, Osiewacz HD. A genome-wide longitudinal transcriptome analysis of the aging model Podospora anserina. PLoS One. 2013;8(12):e83109. doi: 10.1371/journal.pone.0083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knuppertz L, Hamann A, Pampaloni F, Stelzer E, Osiewacz HD. Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy. 2014;10(5):822–834. doi: 10.4161/auto.28148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner K, Perschil I, Fichter CD, van der Laan M. Stepwise assembly of dimeric F(1)F(o)-ATP synthase in mitochondria involves the small F(o)-subunits k and i. Mol Biol Cell. 2010;21(9):1494–1504. doi: 10.1091/mbc.E09-12-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Everard-Gigot V, Dunn CD, Dolan BM, Brunner S, Jensen RE, Stuart RA. Functional analysis of subunit e of the F1Fo-ATP synthase of the yeast Saccharomyces cerevisiae: importance of the N-terminal membrane anchor region. Eukaryot Cell. 2005;4(2):346–355. doi: 10.1128/EC.4.2.346-355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamann A, Krause K, Werner A, Osiewacz HD. A two-step protocol for efficient deletion of genes in the filamentous ascomycete Podospora anserina. Curr Genet. 2005;48(4):270–275. doi: 10.1007/s00294-005-0018-1. [DOI] [PubMed] [Google Scholar]
- 39.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nystrom T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9(1):99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 40.Stahl U, Lemke PA, Tudzynski P, Kück U, Esser K. Evidence for plasmid like DNA in a filamentous fungus, the ascomycete Podospora anserina. Mol Gen Genet. 1978;162(3):341–343. doi: 10.1007/BF00268860. [DOI] [PubMed] [Google Scholar]
- 41.Cummings DJ, Belcour L, Grandchamp C. Mitochondrial DNA from Podospora anserina. I. Isolation and characterization. Mol Gen Genet. 1979;171(3):229–238. doi: 10.1007/BF00267577. [DOI] [PubMed] [Google Scholar]
- 42.Belcour L, Begel O, Mosse MO, Vierny C. Mitochondrial DNA amplification in senescent cultures of Podospora anserina: variability between the retained, amplified sequences. Curr Genet. 1981;3(1):13–21. doi: 10.1007/BF00419575. [DOI] [PubMed] [Google Scholar]
- 43.Kück U, Stahl U, Esser K. Plasmid-like DNA is part of mitochondrial DNA in Podospora anserina. Curr Genet. 1981;3(2):151–156. doi: 10.1007/BF00365719. [DOI] [PubMed] [Google Scholar]
- 44.Osiewacz HD, Esser K. The mitochondrial plasmid of Podospora anserina: a mobile intron of a mitochondrial gene. Curr Genet. 1984;8(4):299–305. doi: 10.1007/BF00419728. [DOI] [PubMed] [Google Scholar]
- 45.Wiemer M, Osiewacz HD. Effect of paraquat-induced oxidative stress on gene expression and aging of the filamentous ascomycete Podospora anserina. Microb Cell. 2014;1(7):225–240. doi: 10.15698/mic2014.07.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borghouts C, Werner A, Elthon T, Osiewacz HD. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol Cell Biol. 2001;21(2):390–399. doi: 10.1128/MCB.21.2.390-399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorin S, Dufour E, Boulay J, Begel O, Marsy S, Sainsard-Chanet A. Overexpression of the alternative oxidase restores senescence and fertility in a long-lived respiration-deficient mutant of Podospora anserina. Mol Microbiol. 2001;42(5):1259–1267. doi: 10.1046/j.1365-2958.2001.02690.x. [DOI] [PubMed] [Google Scholar]
- 48.Scheckhuber CQ, Houthoofd K, Weil AC, Werner A, De Vreese A, Vanfleteren JR, Osiewacz HD. Alternative oxidase dependent respiration leads to an increased mitochondrial content in two long-lived mutants of the aging model Podospora anserina. PLoS One. 2011;6(1):e16620. doi: 10.1371/journal.pone.0016620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul MF, Velours J, Arselin de Chateaubodeau G, Aigle M, Guerin B. The role of subunit 4, a nuclear-encoded protein of the F0 sector of yeast mitochondrial ATP synthase, in the assembly of the whole complex. Eur J Biochem. 1989;185(1):163–171. doi: 10.1111/j.1432-1033.1989.tb15098.x. [DOI] [PubMed] [Google Scholar]
- 50.Boyle GM, Roucou X, Nagley P, Devenish RJ, Prescott M. Identification of subunit g of yeast mitochondrial F1F0-ATP synthase, a protein required for maximal activity of cytochrome c oxidase. Eur J Biochem. 1999;262(2):315–323. doi: 10.1046/j.1432-1327.1999.00345.x. [DOI] [PubMed] [Google Scholar]
- 51.Saddar S, Dienhart MK, Stuart RA. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem. 2008;283(11):6677–6686. doi: 10.1074/jbc.M708440200. [DOI] [PubMed] [Google Scholar]
- 52.Kramer P, Jung AT, Hamann A, Osiewacz HD. Cyclophilin D is involved in the regulation of autophagy and affects the lifespan of P. anserina in response to mitochondrial oxidative stress. . Front Genet. 2016;7:165. doi: 10.3389/fgene.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giorgio V, Bisetto E, Soriano ME, Dabbeni-Sala F, Basso E, Petronilli V, Forte MA, Bernardi P, Lippe G. Cyclophilin D modulates mitochondrial FoF1-ATP synthase by interacting with the lateral stalk of the complex. J Biol Chem. 2009;284(49):33982–33988. doi: 10.1074/jbc.M109.020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabo I, Lippe G, Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci U S A. 2013;110(15):5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonora M, Morganti C, Morciano G, Pedriali G, Lebiedzinska-Arciszewska M, Aquila G, Giorgi C, Rizzo P, Campo G, Ferrari R, Kroemer G, Wieckowski MR, Galluzzi L, Pinton P. Mitochondrial permeability transition involves dissociation of F1Fo ATP synthase dimers and C-ring conformation. EMBO Rep. 2017;18(7):1077–1089. doi: 10.15252/embr.201643602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizet G. Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina. C R Hebd Seances Acad Sci. 1953;237(15):838–840. [PubMed] [Google Scholar]
- 57.Osiewacz HD, Hamann A, Zintel S. Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol Biol. 2013;965(439-462) doi: 10.1007/978-1-62703-239-1_29. [DOI] [PubMed] [Google Scholar]
- 58.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28(22):E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lecellier G, Silar P. Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr Genet. 1994;25(2):122–123. doi: 10.1007/BF00309536. [DOI] [PubMed] [Google Scholar]
- 60.Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol. 2009;11(7):852–858. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- 61.Borghouts C, Kimpel E, Osiewacz HD. Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene grisea. Proc Natl Acad Sci U S A. 1997;94(20):10768–10773. doi: 10.1073/pnas.94.20.10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 64.Krause F, Scheckhuber CQ, Werner A, Rexroth S, Reifschneider NH, Dencher NA, Osiewacz HD. Supramolecular organization of cytochrome c oxidase- and alternative oxidase-dependent respiratory chains in the filamentous fungus Podospora anserina. J Biol Chem. 2004;279(25):26453–26461. doi: 10.1074/jbc.M402756200. [DOI] [PubMed] [Google Scholar]
- 65.Unger AK, Geimer S, Harner M, Neupert W, Westermann B. Analysis of yeast mitochondria by electron microscopy. Methods Mol Biol. 2017;1567:293–314. doi: 10.1007/978-1-4939-6824-4_18. [DOI] [PubMed] [Google Scholar]


