Abstract
Redox homeostasis plays multiple roles in essentially all aspects of cellular function, and hence, reliable methods for measuring cellular or tissue redox status are key elements in understanding the redox related signal pathways. However, in the free radical biology field, there are many controversies on the methods to measure reactive oxygen species. In this chapter we describe our experience in measuring superoxide, hydrogen peroxide, and a general redox status using redox-sensitive green fluorescence proteins (roGFPs) in human melanoma cells.
Keywords: Reactive oxygen species, Superoxide, Hydrogen peroxide, Redox-sensitive, GFP, ro1GFP, ro2GFP, Dihydroethidium (DHE), Dichlorofluorescin diacetate (DCFDA), Fluorometer, Laser microscopy
1. Introduction
Quantitatively measuring the cellular reactive oxygen species (ROS) or cellular redox status is critical for understanding the role of redox in the cellular function and human diseases, including cardiovascular disease, aging, and cancer [1, 2]. ROS include superoxide, hydrogen peroxide and other peroxides, and hydroxyl radical or ion. These molecules can serve as signals for normal physiological reactions; however, when in excess, they will also oxidize lipids, proteins and DNA, and cause cellular damage, resulting in DNA mutation, disease conditions, or apoptosis. Cellular redox balance is mainly maintained by two interactive systems: glutathione system and thioredoxin system [3].
Based on the biochemical characteristics, cellular superoxide and hydrogen peroxide levels can be measured in the laboratory via fluorescence probes, among an extensive list the most commonly used includes dihydroethidium (DHE) [4], dichlorofluorescin diacetate (DCFDA) [5], and AmplexRed. DCFDA was believed specific for hydrogen peroxide while DHE was for superoxide. New evidence challenged the specificity of DCFDA [6, 7]. In addition, auto-oxidation and autofluorescence seem to be a challenge that was not previously fully recognized. These problems led to continuing effort of seeking new probes. Recent years have witnessed developing and thriving of new protein-based probes including redox-sensitive GFPs (roGFPs), redox-sensitive YFPs (rxYFP), and HyPer [8]. The roGFP probes were fused to various organelle targeting peptides, and hence, various versions suitable for detecting redox status for whole cell, mitochondria, or endoplasmic reticulum were developed [9, 10, 11]. The probes that we obtained from Dr. Remington’s laboratory include ro1GFP, ro2GFP, pRA306 (mitochondria-targeting ro1GFP, mito-ro1GFP) and pRA305 (mitochondria-targeting ro2-GFP, mito-ro2GFP) [11, 12]. The ro1GFP and ro2GFP differ on amino acid modifications and green fluorescence peaks (emission at 528 nm) excited at 405 nm and 488 nm [11]. The fluorescence ratio (ex405/ex488, or ex488/ex405) indicates relative redox status of the cells. Stable cell lines expressing these probes were established in human melanoma cells and enabled us to monitor the redox changes in real time following stimulation. In this study we used mito-ro1GFP as an example.
2. Materials
Melanoma cell line: SK-Mel28.
5 % FBS (fetal bovine serum).
5 % NBS (newborn bovine serum).
1 % penicillin–streptomycin.
Eagle’s modified minimum essential media (EMEM).
Glass bottom 96-well plates.
DCFDA (Sigma D6883).
DMSO.
DHE (Sigma D7008).
PBS.
N-acetyl cycstein (NAC).
Arsenic trioxide (ATO).
3. Methods
3.1 Growth Media and Measuring Solution Preparation
Prepare Cell Growth Media solution by combining 5 % FBS (fetal bovine serum), 5 % NBS (newborn bovine serum) 1 % penicillin–streptomycin in Eagle’s modified minimum essential media (EMEM).
Prepare DCFDA solution: DCFDA 5 mg is dissolved in 513 µL of DMSO to make 20 mM stock. Prefer freshly prepared stock solution, although the compound can be stable at −20 °C for months.
Prepare DHE stock solution: DHE 10 mg is dissolved in 528 µL of DMSO to make 60 mM stock. Keep in dark, and store at −20 °C.
3.2 Measuring Redox Status Using DCFDA and a Fluorometer
Day 1, seed melanoma cells in 96-well plate, about 10,000 cells per well.
Day 2, treat cells with stimulator or inhibitor in triplicates, leave 6 wells of control cells without treatment.
Day 3 or after any designated incubation period, remove media, wash cells with 1× PBS once.
Make 10 µM DCFDA working solution by diluting 20 mM DCFDA stock solution 1:2000 into 1× PBS (seeNote1), mix well. Add 100 to each well, except for 3 untreated wells that will be used as blanking control. Add 1× PBS to these 3 wells. Incubate in dark for 10 min, immediately proceed to reading (seeNote2).
Use a BioTek Synergy fluorometer to measure fluorescence at 528 nm with excitation wavelength at 485 nm (seeNote3).
Calculating relative fluorescence: average reading per treatment or control groups minus the average of the blanking well readings will generate the raw experiment results. Increase of fluorescence indicates a more oxidized condition.
3.3 Measuring Superoxide Levels Using DHE and a Fluorometer
Follow steps 1–3 described above to prepare attached cells for measuring.
Make 30 µM DHE working solution by diluting 60 mM DHE stock solution 1:2000 into 1× PBS, mix well. Add 100 to each well, except for 3 untreated wells that will be used as blanking control. Add 1× PBS (no DHE) to these 3 wells. Incubate in dark for 30 min to 1 h, proceed to reading.
Use a BioTek Synergy fluorometer to measure fluorescence at 480 nm with excitation wavelength at 360 nm.
Calculate raw reading as described in step 6 for DCFDA measurement. Increase of fluorescence indicates an increase in superoxide level.
3.4 Measuring Redox Status Using roGFP and a Fluorometer
Establish stable clones expressing ro1GFP, ro2GFP, mito-ro1GFP, and mito-ro2GFP in SK-Mel28 cells according to standard protocol.
On Day 1, seed roGFP cells in 96-well plate, 15,000 cells per plate. Leave at least 3 empty wells for blanking purpose (no cells). Use glass-bottom plates instead of plastic bottom plates to minimize light refraction.
On Day 2, treat cells with arsenic trioxide (ATO, 5 µM) or N-acetyl cysteine (NAC, 0.5 mM), triplicate treatments and control wells with no treatment,
On Day 3, remove media and add 100 µL of 1× PBS into each well. Use a BioTek fluorometer to measure emission at 528 nm when excited at 410 nm; change the excitation wavelength to 485 nm and measure emission at 528 nm again.
Calculate the raw data: Subtract the blank from the average 528 nm fluorescence reading at each excitation wavelength. Use 528 nm fluorescence (ex 410) divided by 528 nm fluorescence (ex 485) as the final parameter for measuring overall cellular redox status (410/485 ratio) (seeNote4).
3.5 Measuring Redox Status Using roGFP and Laser Confocal Microscopy
Establish roGFP-expressing cell lines as above. Seed cells as at 5000–10,000 per well on Day 1. Again use glass-bottom plates instead of plastic bottom to minimum light refraction.
Day 2: treat cells as described above (ATO, 5 µM; NAC, 0.5 mM).
Day 3 or after any designated incubation period, take fluorescence photos (emission at 528 nm) using a multiphoton laser microscope (32-channel Meta detector of Zeiss LSM 510 Meta NLO microscopy system). Emission at 528 nm was photographed when excited at 488 nm and 810 nm (equivalent to 405 nm excitation), respectively.
Use ImageJ to quantitate fluorescence of cells and calculate the ratio.
Steps for ImageJ quantification: (a) whole image fluorescence: open the image in ImageJ program → in the main menu, click on Analyze → Set Measurements, select “area”, “mean gray value” and “integrated density” → go to main menu, click Edit → Options → Colors, choose “Foreground” White, “Background” Black, and “Selection” Green, click “OK”. Go back to main menu, Analyze → Measure. Results window will appear, and the area, total fluorescence reading, and mean gray value will show. Select a small area with no cells (either use square or circle selection tools on the main menu), repeat Analyze → Measure, this is the blanking. To ensure accuracy, choose 3 or more areas as blanking area and use average mean value. Use the mean value from the total image to subtract the blanking mean value; this is the actual mean value. Use this mean value to times the total area of the image, this is the actual total fluorescence in the whole image (Tables 1 and 2). (b) measuring fluorescence from individual cells: Click square or circle selection tool in main menu; set up measurements as above; select one cell on the image; Click Analyze → Measure, the Results window will pop out. To ensure accuracy, randomly choose 5 or more cells for measurements; measure blanking background as described above; use the mean value of the 5 cells to subtract the mean value of background to obtain raw mean value. Add up the total area of the 5 cells; use the area sum to times raw mean value to obtain total fluorescence in these 5 cells; use this number divided by 5 to obtain average fluorescence reading from individual single cells (seeNote5).
Table 1.
Fluorescence readings from the initial photos
| Untreated control cells | ATO 5 µM, 72 h | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| ex. 810 nm | ex. 488 nm | ex. 810 nm | ex. 488 nm | |||||||||
| Area | Mean | IntDen | Area | Mean | IntDen | Area | Mean | IntDen | Area | Mean | IntDen | |
| Whole | 21,024 | 10.99 | 2,31,092 | 21,315 | 2.38 | 50,696 | 21,608 | 11.86 | 2,56,181 | 21,316 | 1.74 | 37,067 |
| Blank 1 | 341 | 5.71 | 1946 | 366 | 1.09 | 398 | 354 | 7.54 | 2670 | 75 | 1.44 | 108 |
| Blank 2 | 214 | 5.65 | 1210 | 432 | 0.99 | 429 | 238 | 6.52 | 1552 | 147 | 1.01 | 149 |
| Blank 3 | 412 | 5.54 | 2284 | 320 | 1.00 | 320 | 100 | 6.26 | 626 | 140 | 1.09 | 152 |
| Cell 1 | 642 | 31.02 | 19,913 | 435 | 5.52 | 2402 | 620 | 36.96 | 22,918 | 216 | 11.6 | 2506 |
| Cell 2 | 506 | 26.31 | 13,313 | 638 | 15.22 | 9708 | 326 | 44.87 | 14,626 | 311 | 7.26 | 2259 |
| Cell 3 | 452 | 24.04 | 10,867 | 690 | 6.94 | 4792 | 253 | 43.82 | 11,086 | 586 | 4.87 | 2856 |
| Cell 4 | 376 | 25.15 | 9457 | 561 | 5.69 | 3191 | 359 | 40.03 | 14,369 | 535 | 3.26 | 1743 |
| Cell 5 | 402 | 16.89 | 6788 | 550 | 4.17 | 2296 | 364 | 28.25 | 10,283 | 177 | 8.02 | 1420 |
Table 2.
Details on calculation of fluorescence ratio
| Control | ATO | |||
|---|---|---|---|---|
| 810 nm | 488 nm | 810 nm | 488 nm | |
| Mean-whole image | 10.99 | 2.38 | 11.86 | 1.74 |
| Actual mean-whole image | 5.36 | 1.35 | 5.09 | 0.56 |
| Area-whole | 21,024 | 21,315 | 21,608 | 21,316 |
| Total fluorescence-whole | 1,12,618.56 | 28,846.30 | 1,09,912.69 | 11,936.96 |
| Mean-blank | 5.63 | 1.03 | 6.77 | 1.18 |
| Mean-5 cells | 24.682 | 7.51 | 38.79 | 7.00 |
| Mean-5 cells actual | 19.05 | 6.48 | 32.01 | 5.82 |
| Total area-5 cells | 2378 | 2874 | 1922 | 1825 |
| Total fluorescence-5 cells | 45,297.73 | 18,627.35 | 61,528.35 | 10,625.20 |
| Fluorescence per cell | 9059.55 | 3725.47 | 12,305.67 | 2125.03 |
| 810/488-whole | 3.90 | 9.21 | ||
| 810/488 per cell | 2.43 | 5.79 | ||
| Fold of increase-whole | 2.36 | ATO/Ctrl | ||
| Fold of increase-per cell | 2.38 | ATO/Ctrl | ||
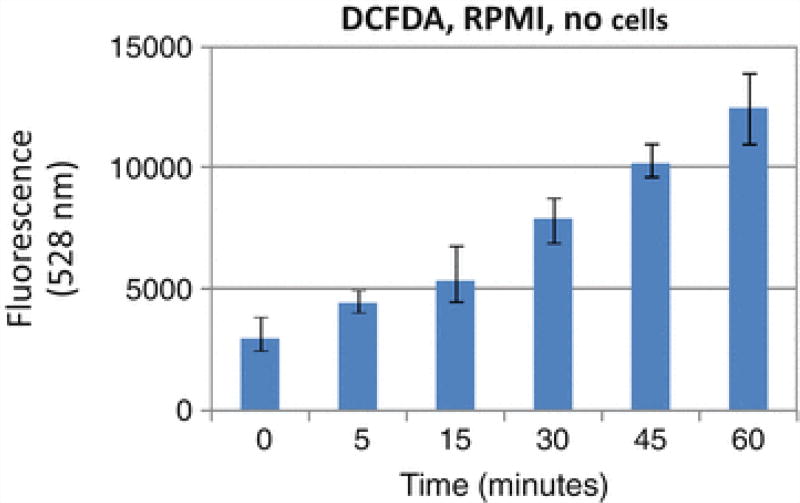
Fig. 1.
Auto-oxidation of DCFDA in RPMI media
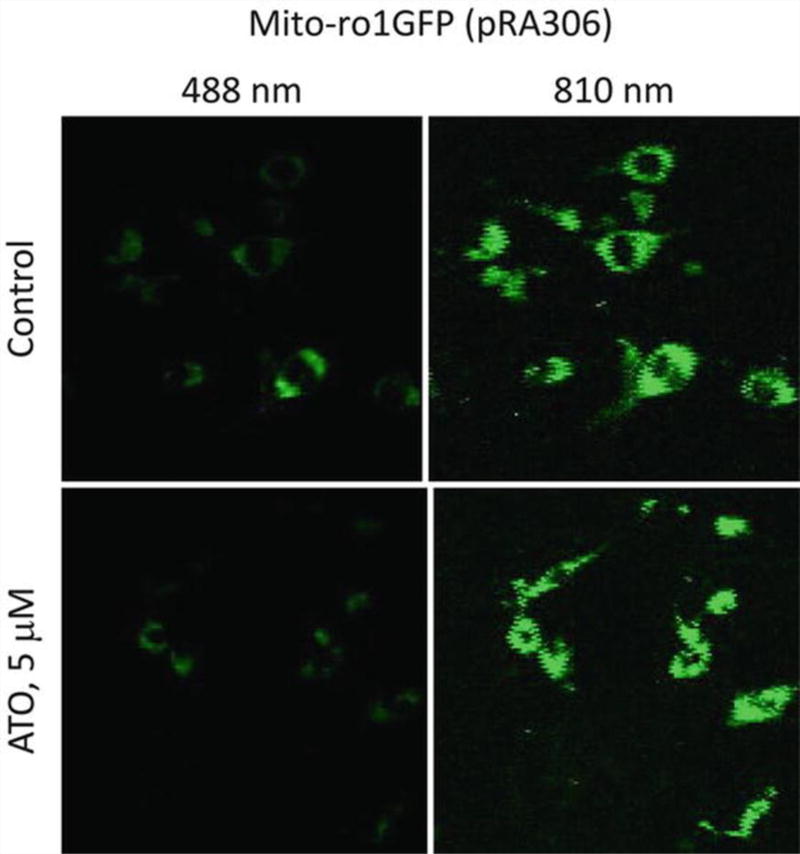
Fig. 2.
SK-Mel-28 melanoma cells stably expressing a mito-ro1GFP plasmid pRA306 were examined by a laser confocal microscope at 528 nm for green fluorescence. Top panels: no-treatment control cells, bottom panels: 5 µM arsenic trioxide (ATO) treated for 24 h. Left panels: excited at 488 nm; right panels: excited at 810 nm
Acknowledgments
This study is supported by Oxnard Foundation and Waltmar Foundation to F.L.M., and Department of Medicine Allan Hubbell Education Funds (University of California Irvine) to F.L.S.
Footnotes
PBS must be used for measuring DCFDA fluorescence. We tested DCFDA in RPMI media (without phenol red, without serum and antibiotics). DCFDA will be oxidized in the media alone without cells (Fig. 1). Autofluorescence in the absence of H2O2was also reported in the literature.
Because of the reason listed in Note1, we do not recommend to load this dye before stimulation. In our experience, once loaded into cells, DCFDA will fluoresce autonomously which will dramatically mask the effect of the stimulants. Essentially 30 min after being loaded, the reading will reach such a high level that it is difficult to distinguish between control and stimulated conditions. We strongly recommend to measure fluorescence 15 min after being loaded.
The filters we used for DFC are 528 nm with 20 nm of band width (emission) and 485 nm with band width of 20 nm (excitation). For DHE they are 530 nm with band width of 25 nm (excitation) and 620 nm with 40 nm band width (emission).
In order to use fluorometer method, the cell clones must be homogenous and express high fluorescence, or else the detection may not be sensitive enough to distinguish the fluorescence from the background. In addition, the fluorometer method is not suitable to measure redox status when apoptosis occurs because of the autofluorescent nature of the apoptotic cells. Overall from our experience, the microscopy method is more accurate and reliable in measuring the roGFP fluorescence.
The results from the whole image are usually consistent with measurements from the individual cells (Table 2). Here we show an example of mito-ro1GFP cells (untreated control cells and 5 µM ATO-treated cells). Figure 2 shows the original image from the microscope. As expected, ATO-induced oxidation led to an increased fluorescence at 810 nm and decreased fluorescence at 488 nm, consequently, the ratio of 810/488 nm fluorescence emission increased from 3.90 to 9.21 for the whole image, and 2.43 to 5.79 on a per cell basis (raw data in Table 1, detailed calculation in Table 2).
Contributor Information
Feng Liu-Smith, Department of Epidemiology, Chao Family Comprehensive Cancer Center, University of California Irvine School of Medicine, B200 Sprague Hall, 839 Health Science Road, Irvine, CA, 92697, USA.
Tatiana B. Krasieva, Beckman Laser Institute and Medical Clinic, University of California Irvine, Irvine, CA, 92697, USA
Jing Liu, The State Key Laboratory of Medical Genetics and School of Life Sciences, Central South University, Changsha, Hunan Province, China.
Jiankang Liu, Institute of Mitochondrial Biology and Medicine, The Key Laboratory of Biomedical Information Engineering of Ministry of Education, Xi'an Jiaotong University School of Life Science and Technology, Xi'an, Shaangxi Province, China.
Frank L. Meyskens, Jr, Department of Medicine, Chao Family Comprehensive Cancer Center, University of California Irvine School of Medicine, B200 Sprague Hall, 839 Health Science Road, Irvine, CA, 92697, USA.
References
- 1.Ziech D, Franco R, Pappa A, Panayiotidis MI. Reactive oxygen species (ROS)—induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 2011;711(1–2):167–173. doi: 10.1016/j.mrfmmm.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Zinkevich NS, Gutterman DD. ROS-induced ROS release in vascular biology: redox-redox signaling. Am J Physiol Heart Circ Physiol. 2011;301(3):H647–H653. doi: 10.1152/ajpheart.01271.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalinina EV, Chernov NN, Saprin AN. Involvement of thio-, peroxi-, and glutaredoxins in cellular redox-dependent processes. Biochemistry (Mosc) 2008;73(13):1493–1510. doi: 10.1134/s0006297908130099. [DOI] [PubMed] [Google Scholar]
- 4.Peshavariya HM, Dusting GJ, Selemidis S. Analysis of dihydroethidium fluorescence for the detection of intracellular and extracellular superoxide produced by NADPH oxidase. Free Radic Res. 2007;41(6):699–712. doi: 10.1080/10715760701297354. [DOI] [PubMed] [Google Scholar]
- 5.Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J Immunol. 1983;130(4):1910–1917. [PubMed] [Google Scholar]
- 6.Myhre O, Andersen JM, Aarnes H, Fonnum F. Evaluation of the probes 2′,7′-dichlorofluorescin diacetate, luminol, and lucigenin as indicators of reactive species formation. Biochem Pharmacol. 2003;65(10):1575–1582. doi: 10.1016/s0006-2952(03)00083-2. [DOI] [PubMed] [Google Scholar]
- 7.Rota C, Chignell CF, Mason RP. Evidence for free radical formation during the oxidation of 2′–7′-dichlorofluorescin to the fluorescent dye 2′–7′-dichlorofluorescein by horseradish peroxidase: possible implications for oxidative stress measurements. Free Radic Biol Med. 1999;27(7–8):873–881. doi: 10.1016/s0891-5849(99)00137-9. [DOI] [PubMed] [Google Scholar]
- 8.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13(5):621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 9.Birk J, Ramming T, Odermatt A, Appenzeller-Herzog C. Green fluorescent protein-based monitoring of endoplasmic reticulum redox poise. Front Genet. 2013;4:108. doi: 10.3389/fgene.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon MB, Remington SJ. Redox-sensitive green fluorescent protein: probes for dynamic intracellular redox responses. A review. Methods Mol Biol. 2008;476:51–65. doi: 10.1007/978-1-59745-129-1_4. [DOI] [PubMed] [Google Scholar]
- 11.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279(13):13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 12.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279(21):22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]