Abstract
PRDM9 is a zinc finger protein that binds DNA at specific locations in the genome where it trimethylates histone H3 at lysines 4 and 36 at surrounding nucleosomes. In meiosis in many species, including humans and mice where PRDM9 has been most intensely studied, these actions determine the location of recombination hotspots, where genetic recombination occurs. In addition, PRDM9 facilitates the association of hotspots with the chromosome axis, the site of the programmed DNA double strand breaks (DSBs) that give rise to genetic exchange between chromosomes. In the absence of PRDM9 DSBs are not properly repaired. Collectively, these actions determine patterns of genetic linkage and the possibilities for chromosome reorganization over successive generations.
Keywords: PRDM9, meiosis, recombination
PRDM9 FUNCTIONS IN MEIOTIC RECOMBINATION
The DNA-binding, zinc finger, histone methyltransferase PRDM9 determines the location of recombination hotspots in both male and female germ cells in a number of mammalian species. Hayashi and Matsui first described PRDM9 in mice in 2006, naming it “Meisetz”, a protein essential for meiosis but absent from somatic cells [1]. In 2009, Mihola et al. identified PRDM9 as the first, and still only known, mammalian hybrid sterility gene involved in speciation [2], and in 2010, three groups simultaneously identified it as the gene responsible for determining the location of recombination hotspots [3–5]. Since then, there have been multiple confirmations of PRDM9’s role in controlling human recombination hotspots [6–8], and multiple studies have reported its role in controlling hotspot locations in non-human primates [9–13], rodents [14–16], ruminants [17–21], and equids [22].
Recent years have seen considerable advances in our understanding of the molecular details of PRDM9’s interaction with DNA as well as its ability to alter the local chromatin structure by methylating histone H3K4 and H3K36. It has also become apparent that, in addition to trimethylating adjacent nucleosomes, PRDM9 associates hotspots, which originate in DNA loops, with the chromosomal axis, where the DNA double-strand breaks (DSBs) required for genetic exchange are formed, and assures the proper repair of DSBs.
Several reviews describe particular aspects of PRDM9 function [23–25]. Here we provide a broad overview and point out some of the open questions regarding the properties and functions of a protein that plays a key role in the mechanisms of mammalian genetic recombination and that has a major influence on the nature of genetic linkage and how patterns of chromosome reorganization can change over successive generations.
STRUCTURE OF PRDM9
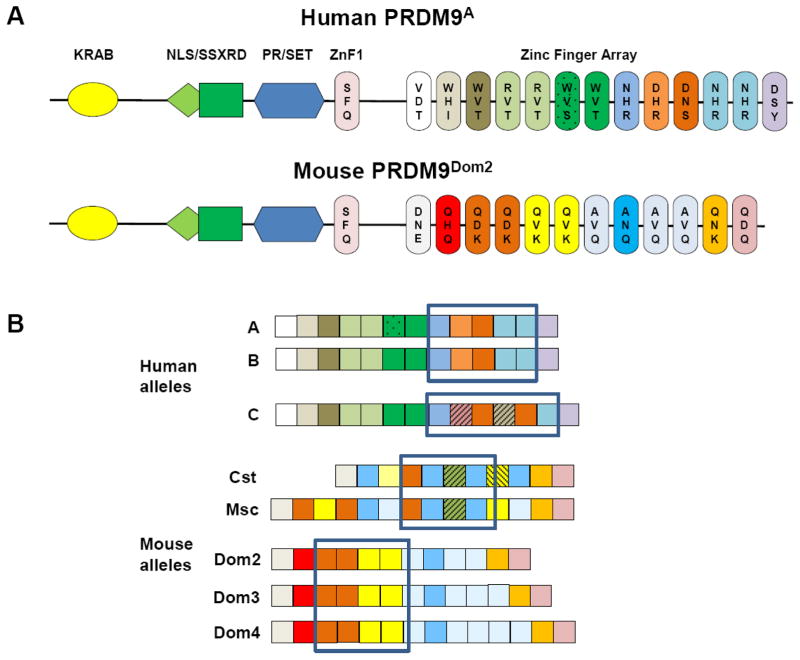
PRDM9 (PR/SET domain containing protein 9) (Figure 1A) contains an N-terminal KRAB domain involved in protein-protein interactions [26, 27], an SSXRD nuclear localization signal, a SET domain providing methyltransferase activity [28–31], and a zinc finger domain containing a single, proximal zinc finger separated from the rest of the terminal C2H2 zinc finger array, which can contain anywhere from 8 to over 20 fingers [5, 7, 8, 17, 18, 32, 33]. PRDM9’s protein structure is highly conserved across species except for the terminal zinc finger array, which is highly polymorphic, both in the number of fingers present and the identity of the three amino acids in each finger that contact DNA and determine PRDM9’s DNA-binding specificity (Figure 1B). As discussed below, the origins of this extreme polymorphism lie in the details of the DNA repair process during genetic recombination, which continually force evolutionary selection of new PRDM9 alleles encoding proteins with altered binding specificities.
Figure 1. Molecular structure of PRDM9.
A. The unique composition of the PRDM9 protein consisting of a KRAB-like, SSXRD, SET, and zinc finger array domains. The amino acid residues at -1, 3, and 6 positions relative to the α helix in each finger are responsible for the contact with DNA and are shown within the fingers. Human A and mouse Dom2 and protein variants are shown. B. Composition of the tandem zinc finger array of the major human and mouse Prdm9/PRDM9 alleles. Individual fingers are presented as colored squares. The colors reflect differences in the amino acid residues at -1,3, and 6 positions. The boxes encompass the fingers that contribute to the principal motif of each allele [36–38, 47]. Human A allele binds a principal motif CCxCCxTxxCC with fingers 8–11 [41]. Mouse Dom2 allele binds a principal motif Gx(T/A)GxT(G/A)CT(G/A)(C/T)(C/T) with fingers 3–7 [47].
PRDM9 FUNCTION IN MEIOSIS
Nearly all that we know about the detailed mechanisms of PRDM9 function come from studies of germline cells from juvenile male mice undergoing the first wave of meiosis; these cells are a particularly rich source of cells in the early leptotene through zygotene stages of meiosis when PRDM9 is active. Cytological assays show PRDM9 first appearing at the preleptotene to leptotene stages, when it localizes to the nucleus, and then disappearing by the end of zygotene [26, 34]. In a typical C57BL/6 germ cell, PRDM9 binds at approximately 4,700 sites, choosing from the ~16,000 sites present on each of the four sets of chromatids that arise following meiotic DNA replication [35]. Each PRDM9 variant binds a set of DNA sites characterized by a common recognition motif, which is determined by the composition of its zinc finger domain [6, 7, 32, 36]. Although PRDM9 uses all of its fingers to bind DNA, only a subset of 4–6 fingers contribute substantially to the motif [37, 38]; the remaining fingers play a secondary role in providing sequence specificity [39] and also act in a non-sequence specific manner to further stabilize binding [36]. In vivo, PRDM9 functions as a multimer [37, 40, 41],; a fact that has implications for hotspot choice in individuals heterozygous for PRDM9 alleles that differ in their binding affinity for DNA [40, 42]. In addition to binding DNA [4, 5], PRDM9 trimethylates histone H3 at lysine-4 [29, 35, 43] and lysine-36 [28, 30, 31, 44] at nearby nucleosomes with its SET domain, reorganizing the local chromatin structure and limiting the space in which the recombination intermediates will be resolved [35]. PRDM9 is also reported to automethylate its own lysine sequences [44]. The biological significance of this activity is as yet unknown; it would be valuable to know whether methylated and unmethylated PRDM9 differ in their ability to bind DNA or methylate histone H3, and particularly whether automethylation affects PRDM9’s stability in vivo, as PRDM9 disappears abruptly at the end of the zygotene stage of meiosis.
Hotspots vary over at least three orders of magnitude in their binding affinity and the likelihood that they will become trimethylated [35, 45], which influences the likelihood that a genetic crossover will eventually occur at that site [35]. Additionally, the ~16,000 sites used in vivo represent fewer than half of the genomic binding sites that can be detected in in vitro binding studies; the critical factor being the prior presence of chromatin modifications that close chromatin and prevent PRDM9 binding in vivo [45]. The choice between used and unused sites is independent of the relative affinity of PRDM9 for DNA at the various sites, indicating that this is not a competition between the binding energies of PRDM9 and factors that close chromatin; instead, prior chromatin modifications appear to gate whether PRDM9 binding sites can be used in vivo [45].
Recently, one study [46], which used a custom antibody to PRDM9, reported the presence in mice of a second, distinct class of PRDM9-containing chromosomal sites identified by ChIP-seq. These Class 2 sites do not have the characteristics of recombination hotspots as they do not contain PRDM9 DNA-binding motifs or acquire DNA double-strand breaks (DSBs). Their significance is presently uncertain as the antibody used for their detection also reacts with another, unknown protein present in Prdm9 KO cells that are deficient in PRDM9, and Class 2 sites were not detected in other PRDM9 ChIP-seq experiments using a different anti-PRDM9 antibody [45, 47].
The nucleosomes that PRDM9 methylates become displaced laterally, creating a nucleosome depleted valley [35]. The topoisomerase SPO11 then creates DSBs within or near this valley [48]. These DSBs initiate the inter-homolog DNA exchange that gives rise to genetic recombinants [43, 48, 49]. In doing so, a homeostatic mechanism [50] mediated by the protein kinase ATM [51] assures that only ~250–400 of the ~4,700 methylated hotspots acquire a DSB. The factors determining which H3K4me3 sites acquire a DSB are as yet poorly understood.
Since hotspot locations are determined by the preferred DNA binding sites of whichever PRDM9 allele(s) are present, the initial contact of PRDM9 with DNA almost certainly occurs in the DNA loops rather than along the growing chromosomal axis. However, DNA cleavage by SPO11 [52] occurs along the chromosomal axis in concert with TOPOVIB [53], MEI4 [54] and IHO1 [55]. The necessary step of associating activated hotspots with the chromosomal axis where DSB formation occurs likely requires additional machinery, and four proteins that bind to the KRAB domain of PRDM9—EWSR1, CDYL, EHMT2 [26], and CXXC1 [26, 27]—have now been implicated. EHMT2 and CDYL are thought to bind PRDM9 early in meiosis, possibly defining the dimensions of hotspots by limiting the lateral extent of nucleosome trimethylation [26]. Later, EWSR1 complexes with PRDM9 and interacts with the meiosis-specific cohesin REC8 to associate hotspots with the chromosome axis [26]. Yeast two-hybrid data shows that CXXC1 is also able to interact with IHO1, an essential component of the meiotic DSB machinery localized on the chromosome axis [27]. It is still an open question whether additional proteins are involved or if the affinities of EWSR1 and CXXC1 for both PRDM9 and axis proteins are enough to stabilize location of hotspots on the chromosome axis.
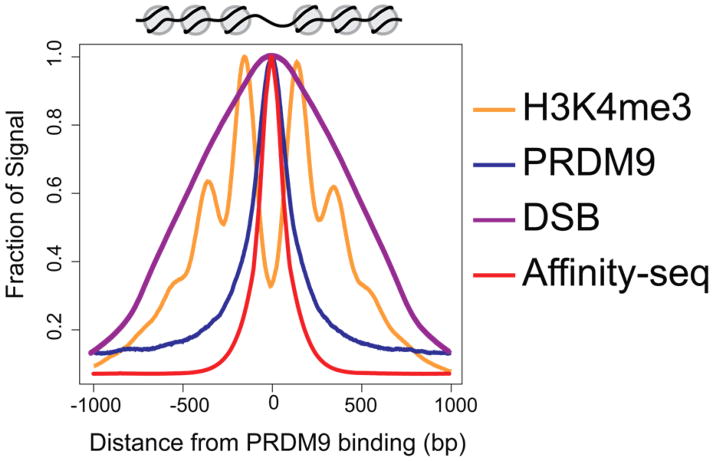
In the widely accepted molecular model of the recombination process [56], the 5′ strand on each side of the DSB break is resected, generating single-stranded 3′ tails that invade the homologous DNA sequence on another chromatid, thus generating the Holliday Junctions that mediate the exchange of DNA between homologs. The extent of Holliday Junction migration away from the initial site of DSB formation is limited to the region of PRDM9-methylated nucleosomes, which, in turn, limits the final location of the genetic crossover that eventuates [35] (Figure 2).
Figure 2. PRDM9 binding, nucleosome modification, repositioning, and DSB initiation.
PRDM9 binding sites can be determined in vitro (Affinity-seq, red [45]) and in vivo (PRDM9 ChIP-seq, blue [47]). Its subsequent histone trimethylation at nearby nucleosomes (H3K4me3, blue; H3K36me3, orange [35]) limits the space in which double strand break initiation will occur (DMC1, purple [49]). The nucleosome repositioning after PRDM9 trimethylation of H3K4 and H3K36 is presented above the main panel.
In the absence of PRDM9, meiotic cells make DSBs at the remaining H3K4me3 sites, particularly those occurring in the open chromatin regions of other regulatory elements, such as gene promoters [49]. These ectopic DSBs are not repaired successfully, and cells containing them undergo pachytene arrest and apoptosis. The consequence is sterility resulting from the complete failure of both sperm and egg production. Since this failure to repair DSBs occurs in both juvenile and adult mice, it is unlikely to be related to the differences in DNA repair processes seen at the two ages [57]. This sequence of events implies that the presence of H3K4me3 alone is not sufficient to distinguish hotspots from other H3K4me3 sites and that additional factors must be involved. Obvious possibilities are the additional presence of H3K36me3, as H3K4me3 and H3K36me3 only co-occur in germ cells at hotspots [28], the additional presence of PRDM9 and its interaction with other proteins, or some combination of these factors.
EVOLUTION OF PRDM9
The repair process of the DNA sequences resected from the initiating chromatid uses the sequence of its partner as the template. Considering this directionally oriented repair, it was predicted that mutations inactivating hotspots would be under strong positive selection, with the eventual disappearance of hotspots finally resulting in loss of recombination and fertility [58]. Since contemporary species obviously reproduce, this prediction gave rise to the “hotspot paradox”, which generated much debate. The eventual discovery of PRDM9 offered a resolution of this dilemma with the realization that mutations in PRDM9’s zinc finger array can rescue sterility by creating an entirely new set of hotspots in a single step. That the evolutionary erosion of hotspots occurs and is repaired by originating new PRDM9 alleles has now been confirmed experimentally in mice [3, 47, 59, 60] and in humans [61], and is assumed to be the evolutionary pressure driving the exceptional diversity of PRDM9 alleles observed in many species.
Human populations vary considerably in their PRDM9 allelic composition [5, 8, 32, 33], generally agreeing with their “out of Africa” origins [62]. African populations have about 50% Allele A, 13% allele C, with a wide variety of minor alleles constituting the remainder [32], while non-African populations are virtually monomorphic with ~90% alleles A and B (mostly A), which only differ in a serine/threonine substitution that does not affect binding specificity [5, 7, 8] (Figure 1B). Neanderthal and Denisovan samples contain PRDM9 alleles closely related to rare contemporary alleles that are limited to Africans [11, 61]. Non-human primates are highly diverse, with extensive variation in the amino acids determining DNA binding specificity [9–11].
Extensive surveys of wild mouse populations have reported well over a hundred alleles that differ in their zinc finger domains [15, 16]. Among inbred strains of laboratory mice, those derived from the Mus musculus domesticus subspecies largely carry either the Dom2 or Dom3 allele, which differ in the presence of two vs. three copies of one of the fingers [5, 63], a difference that is sufficient to create a substantial difference in binding specificity (Figure 1B). Laboratory inbred strains derived from M.m. musculus carry the Msc allele and those derived from M.m. castaneus the Cst allele (Figure 1B).
Extending earlier work on PRDM9 evolution [64–66], newer research [67, 68] used the exceptional diversity in PRDM9’s zinc finger array that arises as a consequence of its role in genetic recombination to infer when PRDM9 has functioned to specify hotspot locations during its evolutionary history. All species whose PRDM9 is thought to have a function in recombination possess the canonical domain structure with intact KRAB, SSXRD, and SET domains as well as the zinc finger array. Absence of any of the domains appears to result in loss of recombination-activating function, suggesting that each domain plays an essential role. The canonical structure with high zinc finger polymorphism is present in contemporary descendants of jawless fishes (Agnathans), the most primitive vertebrates. It is also present in some bony fishes, turtles, snakes, lizards, coelacanths, and nearly all mammals that have been examined with the exception of canids. It has not been observed in amphibians, birds, and some reptiles. If recombinationally active PRDM9 was indeed present in our most primitive vertebrate ancestors, its sporadic presence in the vertebrate evolutionary tree indicates that PRDM9’s function in recombination has been lost and/or gained multiple times in the course of vertebrate evolution. Primate examples of gains and losses include human PRDM7, a gene duplication with three amino acid substitutions in the SET domain, one of which practically destroys the ability to methylate both H3K4 and H3K36, along with loss of most of the zinc finger array [69], and tarsiers (Tarsius syrichta), which exhibit two frame shift mutations, one of which cancels the other to regenerate an active sequence [10]. In ruminants, multiple gene duplications have created several PRDM9 paralogues, at least two of which appear to be recombinationally active [19, 21, 70]. In considering what the evolutionary pressures involved in these various changes might have been, the most obvious advantage to acquiring PRDM9 is sequestering DSB formation away from promoters and other functional genomic elements [49]. Less clear is what advantages might have accrued to reversing that step.
In what appears to be a remarkable case of horizontal gene transfer, the malarial parasite Plasmodium berghei, possesses a single zinc finger protein (PbZfp) that lacks a SET domain, but is nevertheless required for methylation of histone H3 at K4, K27 and K36, presumably by complexing with another histone methyltransferase [71]. Mice infected with parasites carrying a knockout of PbZfp survive longer than controls and show severely reduced mosquito infectivity. PbZfp-deficient parasites also have a markedly reduced ability to produce plasmodial oocysts, implicating a role in genetic recombination and the possibility that mammalian PRDM9 may have served as the original source of this gene [71].
PRDM9-INDEPENDENT RECOMBINATION
Although PRDM9 has been shown to play a major role in determining the location and fate of recombination hotspots in multiple mammalian species (humans, non-human primates, mice, cattle, sheep, and horses), the notable exception is canids (dogs, wolves, coyotes, and foxes), where PRDM9 has become a pseudogene [72–74]. Despite lacking PRDM9, dogs do have hotspots, whose locations are highly stable over evolutionary time, in contrast to the rapid evolutionary erosion of hotspots in mammals with an active PRDM9 gene [72, 74]. Canine hotspots are not associated with the locations of the H3K4me3 peaks seen in canine spermatocytes; instead they are enriched in CpG-rich regions just upstream of transcription start sites, with a preference for unmethylated CpG islands [74, 75]. These differences reinforce their lack of dependence on PRDM9. Clearly, canids rely on an alternative recombination pathway that involves recombination hotspots but does not include PRDM9-dependent mechanisms. Recent evidence suggests that an alternative pathway can also exist in a mammalian species with an active PRDM9 gene, where it can be relied upon in the absence of PRDM9 [59]. A woman has been identified who is homozygous for a null mutation in PRDM9 and is nevertheless fertile, having borne three normal children [76].
It appears there is a fundamental difference between humans and mice in whether PRDM9 determines the location of the obligate crossover in males at the pseudoautosomal region (PAR) of the X and Y chromosomes. Evidence for both PRDM9 independent recombination at the PAR in mice [49], and PRDM9 dependent recombination at the PAR in humans [6, 77] have been presented. This difference could reflect the differences in DNA sequence – both unique and repeated – of mouse and human PARs [78].
Examining PRDM9’s evolutionary history in a broader context suggests that the manner in which recombination is organized along chromosomes has undergone several major revisions over an evolutionary time scale. There is a major division between organisms that do not have hotspots and in which DSB formation follows synapsis (e.g. Drosophila and C. elegans) and organisms that do have hotspots and in which synapsis depends on prior DSB formation (e.g., budding yeast and vertebrates). Among organisms that employ hotspots but do not rely on PRDM9 (e.g., yeast [79–81], birds [82], and Arabidopsis [83, 84]), their hotspots tend to occur at H3K4me3 sites in open chromatin, such as promoters. Among organisms that employ both hotspots and PRDM9 to determine their locations (e.g. mammals), hotspots are more randomly located and largely sequestered away from genomic functional elements.
PRDM9 AND HYBRID STERILITY
Before PRDM9’s role in recombination was recognized, one study [2] identified it as a hybrid sterility gene that can prevent gene flow between subspecies of Mus musculus. Meiosis in hybrid sterile male mice strongly resembles the meiotic failure seen in Prdm9−/− mutants, involving impairment of double-strand break repair, chromosome asynapsis, and disrupted sex-body formation, with meiotic arrest at the pachytene stage. Nevertheless the two phenomena differ in at least two important aspects; PRDM9 null mutations cause meiotic failure in both sexes, whereas the sterility of M.m. musculus x M.m. domesticus hybrids only affects males and requires an epistatic interaction between PRDM9 and the X chromosome.
Hybrid sterility has been intensely studied in crosses between the M.m. domesticus C57BL/6 (B6) strain of mice carrying the Prdm9Dom2 allele and the M.m. musculus PWD strain carrying the Prdm9Msc allele. Male B6 x PWD F1 hybrids carrying a B6 X chromosome are fertile, but reciprocal PWD x B6 F1 males carrying a PWD X chromosome are sterile [85]. As noted, the effect is sex-specific; female mice of both crosses are fertile although their meiosis still has synaptic problems [86]. The epistatic locus has been localized to the center of the X chromosome, thereby eliminating alternative explanations involving the Y chromosome, imprinted genes, or mitochondrial DNA [87]. This region, designated Hstx2, is a highly repetitive 4.7 Mb region of the X [88] that also has a strong effect on the overall rate of genetic recombination in fertile hybrids [89].
The role of PRDM9 in hybrid sterility is complex. Fertility is at least partially restored if sensitive F1 hybrids carry a null mutation of the Dom2 allele, Prdm9Msc/null, indicating that the presence of the Dom2 allele, but not other Prdm9 alleles, is required for meiotic arrest [42]. In contrast, fertility is also restored if the F1 hybrids carry an additional copy of Prdm9 as a transgene. This requirement is not allele specific; rescue occurs with the Dom3 allele [2], the human C allele [60], and importantly, even with the Dom2 allele itself.
Several models have been offered to explain the molecular basis of hybrid sterility based on the presence of PRDM9 binding sites with sequence differences between the parents, causing these asymmetric hotspots to repair more slowly, increasing asynapsis, and ultimately resulting in subfertility and sterility [59, 60, 90]. As intriguing as these models are, we still lack molecular explanations for the obligatory role of the Hstx2 region of the X chromosome, why the Dom2 allele is so exceptional in causing hybrid sterility, and why sterility is relieved by the presence of additional copies of the Prdm9 gene, including the Dom2 allele.
MEDICAL IMPLICATIONS OF PRDM9 FUNCTION IN MEIOSIS
PRDM9 deficiency has been implicated in two independent reports of azoospermia in Japan [91] [92]. This contrasts with the Pakistani woman with a homozygous null mutation in PRDM9 who was nevertheless fertile [76]. Deciding whether these differences relate to sex or reflect human variation in the presence of an alternative recombination pathway will require further data.
The first instance of a human PRDM9 A-allele influence on genetic diseases was reported for the frequency of human minisatellite rearrangements and chromosomal rearrangements present in Charcot-Marie-Tooth disease arising from nonallelic homologous recombination (NAHR) [8]. Subsequently, these findings were extended to 14 out of 27 recurring disease-associated chromosome rearrangement breakpoints, which coincided with locations of human PRDM9 A-allele hotspots, including Charcot-Marie-Tooth disease, and Hunter and Potocki-Lupski/Smith-Magenis syndromes [6]. Many of these disease-associated breakpoints occurred within low-copy number repeat sequences, suggesting they also arose by NAHR and that presence of particular PRDM9 alleles predisposes to particular chromosomal rearrangement disorders. There is additional evidence for the participation of PRDM9 in chromosome rearrangements at two NAHR hotspots associated with Neurofibromatosis 1 mutations [93].
European mothers of Down syndrome children resulting from nondisjunction in meiosis I combined with lack of crossovers on Chr 21 show an increased frequency of non-A PRDM9 alleles that are predicted to have a reduced frequency of PRDM9 binding sites on Chr 21 [94]. An excess of the PRDM9 C allele was found in the parents of children with B-cell acute lymphoblastic leukemia [95]. This was confirmed with marginal significance in an independent study [96]. The correlation between a particular PRDM9 allele and cancer occurs in somatic cells and the role of PRDM9 in it is not clearly understood.
CONCLUDING REMARKS
The initial discovery of PRDM9’s role in the recombination process—determining the location of recombination hotspots in many mammals, including humans and mice—opened intense study of this protein’s properties and functions. We now understand that during meiosis PRDM9 binds DNA at particular recognition sequences, reorganizes the local chromatin environment by methylating adjacent nucleosomes, marks hotspots for eventual DSB formation, facilitates the translocation of activated hotspots to the chromosome axis where DSB formation occurs, and is then required to assure proper repair of the DNA breaks. As a result, PRDM9 plays a key role in determining patterns of genetic segregation and linkage, as well as influencing the possibilities for chromosome reorganization over successive generations. As fruitful as these studies have been, we are now at a transition point, presented with a new generation of experimental questions and challenges (see Outstanding Questions). Solving them will likely to go beyond providing new and valuable insights into the recombination process. PRDM9 has also become a valuable model for studying the entire family of zinc finger proteins. There are over 800 genes encoding zinc finger proteins in mammalian genomes; they comprise by far the largest class of DNA binding proteins. The experimental strategies developed in the study of PRDM9 and the results obtained using them are likely to illuminate our understandings of an exceptional class of proteins engaged in myriad regulatory functions.
OUTSTANDING QUESTIONS.
PRDM9 does not act in isolation. Rather, it associates with multiple other proteins. At the least, these complexes facilitate translocation of hotspots to the chromosomal axis and participate in DSB repair. What are the identities and roles of these additional proteins in carrying out these and possibly other functions?
-
What labels H3K4me3 sites as hotspots rather than as other genomic regulatory elements such as promoters?
-
Are hotspots marked by:
the presence of PRDM9?
the simultaneous presence of H3K4me3 and H3K36me3 on nearby nucleosomes?
both?
What is the recognition mechanism?
-
-
Are some mammals truly polymorphic for two recombination pathways, one dependent on PRDM9 and the other not, and what is this second pathway?
Is this the explanation for why recombination in the X-Y pseudoautosomal region appears to be PRDM9-dependent in humans but not in mice?
What is the molecular mechanism of PRDM9-dependent meiotic pairing and homologous chromosome synapsis?
What is the molecular basis of PRDM9-dependent hybrid sterility, and what can the answer to this question teach us about fundamental mechanisms of recombination?
-
What are the selective pressures for gain and loss of PRDM9’s function in recombination over evolutionary time?
Are there organisms in which it participates in some aspect of recombination even when it does not specify the location of hotspots?
TRENDS BOX.
PRDM9 influences the possibility of genetic exchange by determining the locations of meiotic recombination hotspots in most mammals.
To do so, it uses its zinc finger array to bind specific DNA sequences.
Pre-existing chromatin structure influences which of PRDM9’s DNA binding sites are available for use in vivo.
-
PRDM9 trimethylates histone 3 at both lysines 4 and 36, a unique activity among mammalian chromatin modifying enzymes.
The extent of histone 3 methylation determines the space in which recombination will occur
PRDM9 reorganizes local nucleosomal structure, creating a nucleosome-free center where the DNA double-strand break required for DNA exchange between chromsomes are formed.
PRDM9 forms complexes with other proteins that associate hotspots with the chromosomal axis and affect the subsequent double-strand break initiation and repair.
PRDM9 alleles influence human chromosomal rearrangements, causing hereditary diseases.
Acknowledgments
We are grateful to the members of our laboratories for their experimental efforts and their continual contibutions to our thinking. This work was supported by NIH grants GM 099640 to KP and CA34196 to The Jackson Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayashi K, Matsui Y. Meisetz, a novel histone tri-methyltransferase, regulates meiosis-specific epigenesis. Cell cycle. 2006;5:615–620. doi: 10.4161/cc.5.6.2572. [DOI] [PubMed] [Google Scholar]
- 2.Mihola O, et al. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- 3.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parvanov ED, et al. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinch AG, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevison LS, et al. The Time Scale of Recombination Rate Evolution in Great Apes. Mol Biol Evol. 2016;33:928–945. doi: 10.1093/molbev/msv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heerschop S, et al. The pioneering role of PRDM9 indel mutations in tarsier evolution. Sci Rep. 2016;6:34618. doi: 10.1038/srep34618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz JJ, et al. Primate evolution of the recombination regulator PRDM9. Nature communications. 2014;5:4370. doi: 10.1038/ncomms5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auton A, et al. A Fine-Scale Chimpanzee Genetic Map from Population Sequencing. Science. 2012 doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groeneveld LF, et al. High diversity at PRDM9 in chimpanzees and bonobos. PloS one. 2012;7:e39064. doi: 10.1371/journal.pone.0039064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capilla L, et al. Proceedings. Biological sciences. The Royal Society; 2014. Genetic recombination variation in wild Robertsonian mice: on the role of chromosomal fusions and Prdm9 allelic background; p. 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buard J, et al. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PloS one. 2014;9:e85021. doi: 10.1371/journal.pone.0085021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kono H, et al. Prdm9 Polymorphism Unveils Mouse Evolutionary Tracks. DNA research : an international journal for rapid publication of reports on genes and genomes. 2014 doi: 10.1093/dnares/dst059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandor C, et al. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS genetics. 2012;8:e1002854. doi: 10.1371/journal.pgen.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L, et al. Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis. PLoS genetics. 2015;11:e1005387. doi: 10.1371/journal.pgen.1005387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahlawat S, et al. Evolutionary dynamics of meiotic recombination hotspots regulator PRDM9 in bovids. Mol Genet Genomics. 2017;292:117–131. doi: 10.1007/s00438-016-1260-6. [DOI] [PubMed] [Google Scholar]
- 20.Ahlawat S, et al. Evidence of positive selection and concerted evolution in the rapidly evolving PRDM9 zinc finger domain in goats and sheep. Anim Genet. 2016;47:740–751. doi: 10.1111/age.12487. [DOI] [PubMed] [Google Scholar]
- 21.Ahlawat S, et al. Zinc Finger Domain of the PRDM9 Gene on Chromosome 1 Exhibits High Diversity in Ruminants but Its Paralog PRDM7 Contains Multiple Disruptive Mutations. PloS one. 2016;11:e0156159. doi: 10.1371/journal.pone.0156159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner CC, Ryder OA. Characterization of Prdm9 in equids and sterility in mules. PloS one. 2013;8:e61746. doi: 10.1371/journal.pone.0061746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paigen K, Petkov P. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 2010;11:221–233. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baudat F, et al. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 25.Keeney S, et al. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parvanov ED, et al. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Molecular biology of the cell. 2017;28:488–499. doi: 10.1091/mbc.E16-09-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai Y, et al. The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma. 2017 doi: 10.1007/s00412-017-0631-z. [DOI] [PMC free article] [PubMed]
- 28.Powers NR, et al. The Meiotic Recombination Activator PRDM9 Trimethylates Both H3K36 and H3K4 at Recombination Hotspots In Vivo. PLoS genetics. 2016;12:e1006146. doi: 10.1371/journal.pgen.1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi K, et al. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, et al. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell reports. 2013;5:13–20. doi: 10.1016/j.celrep.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 31.Eram MS, et al. Trimethylation of Histone H3 Lysine 36 by Human Methyltransferase PRDM9. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.523183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg IL, et al. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fledel-Alon A, et al. Variation in human recombination rates and its genetic determinants. PloS one. 2011;6:e20321. doi: 10.1371/journal.pone.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun F, et al. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma. 2015;124:397–415. doi: 10.1007/s00412-015-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker CL, et al. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome research. 2014;24:724–732. doi: 10.1101/gr.170167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billings T, et al. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome biology. 2013;14:R35. doi: 10.1186/gb-2013-14-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel A, et al. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016;30:257–265. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel A, et al. Structural basis of human PRDM9 allele C specific recognition of its cognate DNA sequence. J Biol Chem. 2017 doi: 10.1074/jbc.M117.805754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker CL, et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS genetics. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker CL, et al. Multimer Formation Explains Allelic Suppression of PRDM9 Recombination Hotspots. PLoS genetics. 2015;11:e1005512. doi: 10.1371/journal.pgen.1005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altemose N, et al. A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis. Elife. 2017:6. doi: 10.7554/eLife.28383. [DOI] [PMC free article] [PubMed]
- 42.Flachs P, et al. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS genetics. 2012;8:e1003044. doi: 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh-Stenta X, et al. Discovery and characterisation of the automethylation properties of PRDM9. Biochem J. 2017;474:971–982. doi: 10.1042/BCJ20161067. [DOI] [PubMed] [Google Scholar]
- 45.Walker M, et al. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics & chromatin. 2015;8:31. doi: 10.1186/s13072-015-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grey C, et al. In vivo binding of PRDM9 reveals interactions with noncanonical genomic sites. Genome research. 2017;27:580–590. doi: 10.1101/gr.217240.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker CL, et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS genetics. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange J, et al. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell. 2016;167:695–708. e616. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brick K, et al. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–240. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keeney S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robert T, et al. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 54.Kumar R, et al. MEI4 - a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J Cell Sci. 2015;128:1800–1811. doi: 10.1242/jcs.165464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanzione M, et al. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat Cell Biol. 2016;18:1208–1220. doi: 10.1038/ncb3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 57.Zelazowski MJ, et al. Age-Dependent Alterations in Meiotic Recombination Cause Chromosome Segregation Errors in Spermatocytes. Cell. 2017 doi: 10.1016/j.cell.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boulton A, et al. The hotspot conversion paradox and the evolution of meiotic recombination. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8058–8063. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smagulova F, et al. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 2016;30:266–280. doi: 10.1101/gad.270009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies B, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lesecque Y, et al. The red queen model of recombination hotspots evolution in the light of archaic and modern human genomes. PLoS genetics. 2014;10:e1004790. doi: 10.1371/journal.pgen.1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingman M, Gyllensten U. Analysis of the complete human mtDNA genome: methodology and inferences for human evolution. J Hered. 2001;92:454–461. doi: 10.1093/jhered/92.6.454. [DOI] [PubMed] [Google Scholar]
- 63.Brunschwig H, et al. Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics. 2012;191:757–764. doi: 10.1534/genetics.112.141036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas JH, et al. Extraordinary molecular evolution in the PRDM9 fertility gene. PloS one. 2009;4:e8505. doi: 10.1371/journal.pone.0008505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeffreys AJ, et al. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:600–605. doi: 10.1073/pnas.1220813110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker Z, et al. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife. 2017:6. doi: 10.7554/eLife.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Latrille T, et al. The Red Queen model of recombination hot-spot evolution: a theoretical investigation. Philos Trans R Soc Lond B Biol Sci. 2017:372. doi: 10.1098/rstb.2016.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blazer LL, et al. PR Domain-containing Protein 7 (PRDM7) Is a Histone 3 Lysine 4 Trimethyltransferase. J Biol Chem. 2016;291:13509–13519. doi: 10.1074/jbc.M116.721472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Padhi A, et al. Ruminant-specific multiple duplication events of PRDM9 before speciation. BMC Evol Biol. 2017;17:79. doi: 10.1186/s12862-017-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gopalakrishnan AM, et al. Multifunctional Involvement of a C2H2 Zinc Finger Protein (PbZfp) in Malaria Transmission, Histone Modification, and Susceptibility to DNA Damage Response. MBio. 2017:8. doi: 10.1128/mBio.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Axelsson E, et al. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome research. 2011 doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munoz-Fuentes V, et al. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PloS one. 2011;6:e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auton A, et al. Genetic Recombination Is Targeted towards Gene Promoter Regions in Dogs. PLoS genetics. 2013;9:e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell CL, et al. A Pedigree-Based Map of Recombination in the Domestic Dog Genome. G3 (Bethesda) 2016 doi: 10.1534/g3.116.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Narasimhan VM, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hinch AG, et al. Recombination in the human Pseudoautosomal region PAR1. PLoS Genet. 2014;10:e1004503. doi: 10.1371/journal.pgen.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helena Mangs A, Morris BJ. The Human Pseudoautosomal Region (PAR): Origin, Function and Future. Current genomics. 2007;8:129–136. doi: 10.2174/138920207780368141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neale MJ, Keeney S. End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae. Methods Mol Biol. 2009;557:183–195. doi: 10.1007/978-1-59745-527-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petes TD, Merker JD. Context dependence of meiotic recombination hotspots in yeast: the relationship between recombination activity of a reporter construct and base composition. Genetics. 2002;162:2049–2052. doi: 10.1093/genetics/162.4.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singhal S, et al. Stable recombination hotspots in birds. Science. 2015;350:928–932. doi: 10.1126/science.aad0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muyt AD, et al. Meiotic recombination and crossovers in plants. Genome Dyn. 2009;5:14–25. doi: 10.1159/000166616. [DOI] [PubMed] [Google Scholar]
- 84.Drouaud J, et al. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome research. 2006;16:106–114. doi: 10.1101/gr.4319006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Forejt J, Ivanyi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet Res. 1974;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- 86.Bhattacharyya T, et al. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E468–477. doi: 10.1073/pnas.1219126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dzur-Gejdosova M, et al. Dissecting the genetic architecture of F1 hybrid sterility in house mice. Evolution. 2012;66:3321–3335. doi: 10.1111/j.1558-5646.2012.01684.x. [DOI] [PubMed] [Google Scholar]
- 88.Bhattacharyya T, et al. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS genetics. 2014;10:e1004088. doi: 10.1371/journal.pgen.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balcova M, et al. Hybrid Sterility Locus on Chromosome X Controls Meiotic Recombination Rate in Mouse. PLoS genetics. 2016;12:e1005906. doi: 10.1371/journal.pgen.1005906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamada S, et al. Genomic and chromatin features shaping meiotic double-strand break formation and repair in mice. Cell Cycle. 2017 doi: 10.1080/15384101.2017.1361065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyamoto T, et al. Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2008;25:553–557. doi: 10.1007/s10815-008-9270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Irie S, et al. Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 2009;30:426–431. doi: 10.2164/jandrol.108.006262. [DOI] [PubMed] [Google Scholar]
- 93.Hillmer M, et al. Consideration of the haplotype diversity at nonallelic homologous recombination (NAHR) hotspots improves the precision of rearrangement breakpoint identification. Hum Mutat. 2017 doi: 10.1002/humu.23319. [DOI] [PubMed] [Google Scholar]
- 94.Oliver TR, et al. Variation in the Zinc Finger of PRDM9 is Associated with the Absence of Recombination along Nondisjoined Chromosomes 21 of Maternal Origin. J Down Syndr Chromosom Abnorm. 2016:2. doi: 10.4172/2472-1115.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hussin J, et al. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome research. 2013;23:419–430. doi: 10.1101/gr.144188.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woodward EL, et al. Allelic variants of PRDM9 associated with high hyperdiploid childhood acute lymphoblastic leukaemia. Br J Haematol. 2014;166:947–949. doi: 10.1111/bjh.12914. [DOI] [PubMed] [Google Scholar]