SUMMARY
The diverse malignant, stromal, and immune cells in tumors affect growth, metastasis and response to therapy. We profiled transcriptomes of ~6,000 single cells from 18 head and neck squamous cell carcinoma (HNSCC) patients, including five matched pairs of primary tumors and lymph node metastases. Stromal and immune cells had consistent expression programs across patients. Conversely, malignant cells varied within and between tumors in their expression of signatures related to cell cycle, stress, hypoxia, epithelial differentiation, and partial epithelial-to-mesenchymal transition (p-EMT). Cells expressing the p-EMT program spatially localized to the leading edge of primary tumors. By integrating single-cell transcriptomes with bulk expression profiles for hundreds of tumors, we refined HNSCC subtypes by their malignant and stromal composition, and established p-EMT as an independent predictor of nodal metastasis, grade, and adverse pathologic features. Our results provide insight into the HNSCC ecosystem and define stromal interactions and a p-EMT program associated with metastasis.
Keywords: Single-cell RNA sequencing, metastasis, head and neck squamous cell carcinoma, epithelial-to-mesenchymal transition, tumor microenvironment
INTRODUCTION
Genomic and transcriptomic studies have revealed driver mutations, aberrant regulatory programs, and disease subtypes for major human tumors (Stratton et al., 2009; Weinberg, 2014). However, these studies relied on profiling technologies that measure tumors in bulk, limiting their ability to capture intra-tumoral heterogeneity. Substantial evidence indicates that intra-tumoral heterogeneity among malignant and non-malignant cells, and their interactions within the tumor microenvironment (TME) are critical to diverse aspects of tumor biology (Meacham and Morrison, 2013; Weinberg, 2014).
Recent advances in single-cell genomics provide an avenue to explore genetic and functional heterogeneity at a cellular resolution (Navin, 2015; Tanay and Regev, 2017). Single-cell RNA-seq (scRNA-seq) studies of human tumors, circulating tumor cells (CTCs) and patient-derived xenografts have revealed new insights into tumor composition, cancer stem cells, and drug resistance. However, scRNA-seq studies have not deeply characterized epithelial tumors, despite their predominance. In these tumors, metastasis to draining lymph nodes (locoregional metastasis) and other organs (distant metastasis) represents a major cause of morbidity and mortality. Metastases are often treated based on molecular and pathologic features of the primary tumor, raising the question of whether they share the same genetics, epigenetics, and vulnerabilities. However, the potentially different composition of primary tumors and metastases hinders the straightforward comparison of bulk tumor profiles. Single-cell expression profiling studies would, in principle, offer a compelling alternative.
Epithelial-to-mesenchymal transition (EMT) has been suggested as a driver of epithelial tumor spread (Gupta and Massague, 2006; Lambert et al., 2017). The process of EMT is fundamental to embryonic development and may be co-opted by malignant epithelial cells to facilitate invasion and dissemination (Thiery et al., 2009; Ye and Weinberg, 2015). EMT markers have been detected on CTCs associated with metastatic disease (Ting et al., 2014; Yu et al., 2013). However, since most EMT studies have focused on laboratory models, the extent and significance of EMT in primary human tumors and metastases remains controversial (Lambert et al., 2017; Nieto et al., 2016). Moreover, while mesenchymal subtypes have been identified for certain tumors (Cancer Genome Atlas, 2015; Cancer Genome Atlas Research, 2011; Verhaak et al., 2010), it remains unclear whether they reflect mesenchymal cancer cells or, alternatively, contributions of non-malignant mesenchymal cell types in the TME.
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous epithelial tumor with strong associations to alcohol and tobacco exposure (Puram and Rocco, 2015). Metastatic disease remains a central challenge, with patients often presenting at advanced stages with LN metastases. Here, we investigate primary HNSCC tumors and matched LNs to better understand intra-tumoral heterogeneity, invasion, and metastasis. Transcriptional profiles for ~6,000 cells from 18 patients revealed expression programs that distinguish diverse malignant, stromal, and immune cells. Malignant cells varied in their expression of cell cycle, stress, hypoxia and epithelial differentiation programs. A subset of cells also expressed a partial EMT (p-EMT) program with extracellular matrix proteins, but lacking classical EMT transcription factors (TFs). p-EMT cells localized to the leading edge of primary tumors in proximity to cancer-associated fibroblasts (CAFs). We used this knowledge of the HNSCC ecosystem to re-evaluate bulk RNA-seq data from The Cancer Genome Atlas (TCGA). This revealed new insight into HNSCC expression subtypes, and established the p-EMT program as an independent predictor of adverse clinical features, including invasion and metastasis.
RESULTS
A single-cell expression atlas of HNSCC primary tumors and metastases
To explore the cellular diversity in HNSCC tumors, we focused on oral cavity tumors, the most common subsite of HNSCC. We generated full-length scRNA-seq profiles for primary tumors from 18 treatment-naïve patients and for matching LN metastasis from five of these patients (Figure 1; Tables S1 and S2). We also acquired whole exome sequencing (WES) and targeted genotyping (SNaPshot) data for these tumors, which demonstrated a range of putative driver mutations and chromosomal aberrations (Figure S1B; Tables S3 and S4), consistent with established HNSCC genetics (Agrawal et al., 2011; Cancer Genome Atlas, 2015; Stransky et al., 2011).
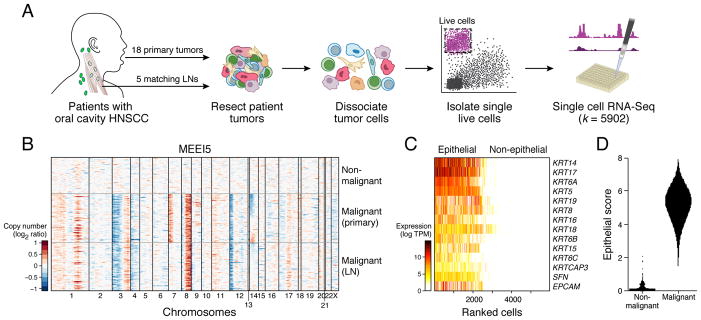
Figure 1. Characterizing intra-tumoral expression heterogeneity in HNSCC by single-cell RNA-seq.
(A) Workflow shows collection and processing of fresh biopsy samples of primary oral cavity HNSCC tumors and matched metastatic LNs for scRNA-seq.
(B) Heat map shows large-scale CNVs for individual cells (rows) from a representative tumor (MEEI5), inferred based on the average expression of 100 genes surrounding each chromosomal position (columns). Red: amplifications; Blue: deletions.
(C) Heatmap shows expression of epithelial marker genes across 5,902 single cells (columns), sorted by the average expression of these genes.
(D) Violin plot shows distributions of epithelial scores (average expression of epithelial marker genes) for cells categorized as malignant or non-malignant based on CNVs.
See Figure S1; Tables S1–S4.
We retained single-cell transcriptomes for 5,902 cells from 18 patients after initial quality controls (Figure S1A). We confidently distinguished 2,215 malignant and 3,363 non-malignant cells by three complementary approaches. First, we inferred large-scale chromosomal copy-number variations (CNVs) in each single cell based on averaged expression profiles across chromosomal intervals (Muller et al., 2016; Patel et al., 2014; Tirosh et al., 2016b). These inferred CNVs, which were consistent with WES (Figures 1B, S1B, and S1C), separated malignant cells from non-malignant cells with normal karyotypes. Second, we distinguished malignant cells by their epithelial origin, which differs from stromal and immune cells in the TME (Figure 1C). We found remarkable concordance between cells with epithelial marker expression and cells with aberrant karyotypes (Figure 1D). Finally, we partitioned cells to preliminary clusters by their global expression patterns. The vast majority of cells were part of clusters with concordant malignant or non-malignant classifications, based on CNV and epithelial marker analyses (Figure S1D).
Landscape of expression heterogeneity in head and neck cancer
Single-cell profiles of non-malignant cells highlighted the composition of the TME. We partitioned the 3,363 non-malignant cells to eight main clusters by their expression states (Figures 2A, S1E, S1F and S2H). We annotated clusters by the expression of known marker genes as T-cells, B/plasma cells, macrophages, dendritic cells, mast cells, endothelial cells, fibroblasts, and myocytes (Figure S1F). Notably, each of the clusters contained cells from different patients, indicating that cell types and expression states in the TME are consistent across HNSCC tumors and do not represent patient-specific subpopulations or batch effects, though they do vary in their proportions.
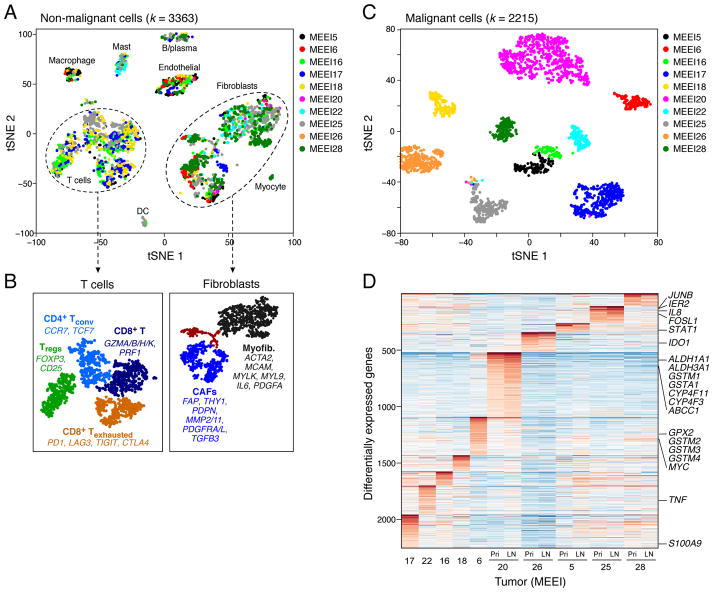
Figure 2. Expression heterogeneity of malignant and non-malignant cells in the HNSCC ecosystem.
(A) t-distributed stochastic neighbor embedding (t-SNE) plot of non-malignant cells from 10 patients reveals consistent clusters of stromal and immune cells across tumors. Clusters are assigned to indicated cell types by differentially expressed genes (see also Figure S1F).
(B) (Left) Zoomed in t-SNE plot of T-cells with distinct naïve-like, regulatory, cytotoxic, and exhausted populations as identified by DBscan clustering. (Right) Zoomed in t-SNE plot of fibroblasts with myofibroblasts, non-activated resting fibroblasts, and activated CAFs, which can be seen to further divide into two sub-clusters. Differentially expressed genes are listed for key subsets.
(C) t-SNE plot of malignant cells from 10 patients (indicated by colors) reveals tumor-specific clusters. Clustering patterns for malignant and non-malignant cells are not driven by transcriptome complexity (see Figure S2H).
(D) Heatmap shows genes (rows) that are differentially expressed across 10 individual primary tumors (columns). For five tumors, expression is also shown for matched LNs. Red: high expression; Blue: low expression. Selected genes are highlighted. Two classical subtype tumors (MEEI6 and MEEI20; see also Figure 6A) preferentially expressed genes associated with detoxification and drug metabolism (e.g. GPX2, GSTMs, CYPs, ABCC1).
See Figures S1 and S2; Table S5.
We found further diversity within both T-cells and fibroblasts through finer clustering, powered by their relatively large numbers in our dataset (Figure 2B). The main T-cell cluster (~1,000 T-cells) can be partitioned into four sub-clusters (Figures 2B and S2A), which we annotated as regulatory T-cells (Tregs), conventional CD4+ T-helper cells (CD4+ Tconv), and two cytotoxic CD8+ T-cell populations (CD8+ T and CD8+ Texhausted). The cytotoxic subsets differed in expression of co-inhibitory receptors (e.g. PD1, CTLA4) and other genes associated with T-cell dysfunction and exhaustion, and thereby defined a putative T-cell exhaustion program in HNSCC (Figures 2B and S2A). Proportions of exhausted CD8+ T-cells varied significantly among patients in our cohort (Figure S2B). These T-cell expression states may inform efforts to understand and predict responses to checkpoint immunotherapies (Mellman et al., 2011).
Despite significant interest, the regulatory states of fibroblasts in human tumors remain obscure. The ~1,500 fibroblasts partitioned into two main subsets (Figure 2B, black and blue), and a third minor subset (Figures 2B, brown, S2C and S2D). One subset expressed classical markers of myofibroblasts, including alpha smooth muscle actin (ACTA2) and myosin light chain proteins (MYLK, MYL9). Myofibroblasts are an established component of the TME and have been linked to wound healing and contracture (Rockey et al., 2013). A second subset expressed receptors, ligands, and extracellular matrix (ECM) genes, including fibroblast activation protein (FAP), podoplanin (PDPN), and connective tissue growth factor (CTGF), that have been associated with CAFs (Madar et al., 2013). The third subset was depleted of markers for myofibroblasts and CAFs and may represent resting fibroblasts. These diverse fibroblast expression states were reproducibly detected across tumors, and may thus represent common features of the HNSCC TME.
Although the cellular identity and origin of CAFs has been ascribed to various lineages (Madar et al., 2013), the subpopulations we detect are highly consistent with a fibroblast identity. Further analysis partitioned CAFs into two subsets (CAF1 and CAF2) with differential expression of immediate early response genes (e.g. JUN, FOS), mesenchymal markers (e.g. VIM, THY1), ligands and receptors (e.g. FGF7, TGFBR2/3), and ECM proteins (e.g. MMP11, CAV1) (Figures S2D and S2E; Table S5). This intra-tumoral fibroblast heterogeneity is consistent with the view that CAFs are involved in complex structural and paracrine interactions in the TME.
In contrast to non-malignant cells, the 2,215 malignant cells clustered according to their tumor of origin (Figures 2C and S2H). Over 2,000 genes were preferentially expressed in individual tumors (Figure 2D). Differentially-expressed genes were enriched within CNVs that vary between tumors (Figure S2F and S2G). Other differences relate to tumor subtypes (see ‘HNSCC subtypes…’, below). For example, genes associated with detoxification and drug metabolism (e.g. GPX2, GSTMs, CYPs, ABCC1) were preferentially expressed by the two classical subtype tumors in our cohort (MEEI6 and MEEI20; Figure 2D). Finally, other differentially expressed genes relate to stress (e.g. JUNB, FOSL1) or immune activation (e.g. IDO1, STAT1, TNF), potentially in response to varied TMEs. Thus, inter-tumoral malignant cell expression heterogeneity reflects differences in genetics, subtypes, and TME between tumors in our cohort.
Intra-tumoral expression heterogeneity of the malignant compartment
We next explored how expression states varied among different malignant cells within the same tumor, focusing on 10 tumors from which the largest numbers of malignant cell transcriptomes were acquired. We used non-negative matrix factorization to uncover coherent sets of genes that were preferentially co-expressed by subsets of malignant cells. For example, we defined six gene signatures that vary among malignant cells of MEEI25 (Figures 3A and S2I; Table S6). Applying the approach to each of the 10 tumors defined a total of 60 gene signatures that coherently vary across individual cells in at least one tumor (Table S6). Next, we used hierarchical clustering to distill these 60 signatures into meta-signatures that reflect common expression programs that vary within multiple tumors (Figures 3B, S3A and S3B; Table S6 and S7). The high concordance between signatures from different tumors suggests that they reflect common patterns of intra-tumoral expression heterogeneity.
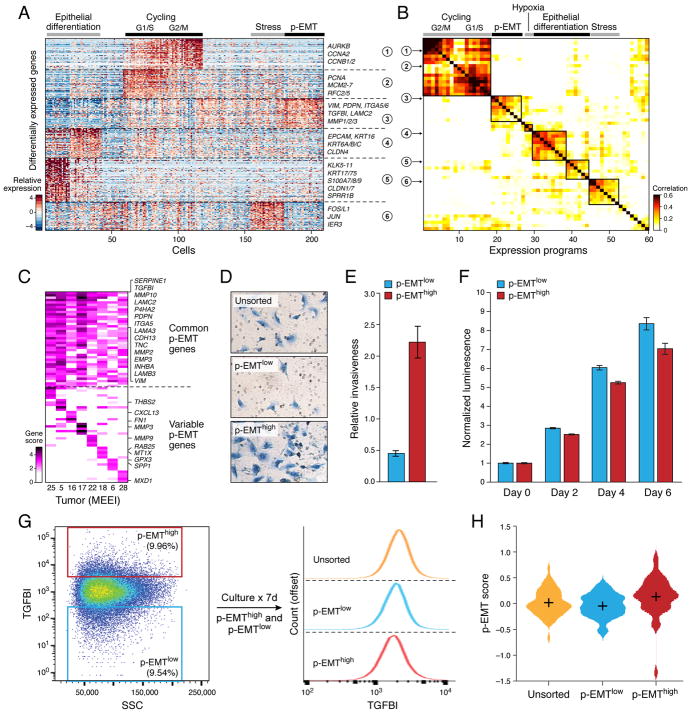
Figure 3. Unbiased clustering reveals a common program of partial EMT (p-EMT) in HNSCC tumors.
(A) Heatmap shows differentially-expressed genes (rows) identified by non-negative matrix factorization (NNMF) clustered by their expression across single cells (columns) from a representative tumor (MEEI25). The gene clusters reveal intra-tumoral programs that are differentially expressed in MEEI25. The corresponding gene signatures are numbered and selected genes indicated (right).
(B) Heatmap depicts pairwise correlations of 60 intra-tumoral programs derived from 10 tumors, as in (A). Clustering identifies seven coherent expression programs across tumors. Rows in the heatmap that correspond to programs derived from MEEI25 are indicated by arrows and numbered as in (A).
(C) Heatmap shows NNMF gene scores (rows) for common (top) and tumor-specific (bottom) genes within the p-EMT program by tumor (columns).
(D) Representative images of SCC9 HNSCC cells sorted by p-EMT marker TGFBI into p-EMThigh and p-EMTlow populations and analyzed by matrigel invasion assay.
(E) Bar plot depicts relative invasiveness of p-EMThigh and p-EMTlow SCC9 cells sorted and analyzed as in (D) (representative experiment; error bars reflect SEM; ANOVA, p<0.005, n=3).
(F) Bar plot depicts relative proliferation of p-EMThigh and p-EMTlow SCC9 cells sorted as in (D) (representative experiment; error bars reflect SEM; ANOVA, p<0.0001, n=4).
(G) (Left) Fluorescence-activated cell sorting plot identifies p-EMThigh and p-EMTlow SCC9 cells isolated based on TGFBI expression. (Right) Histogram (offset) reveals the distribution (x-axis) of TGFBI expression across cells from the respective isolates (p-EMThigh, p-EMTlow, and unsorted; separated by dashed lines). After 7 days in culture, p-EMThigh, p-EMTlow, and unsorted cells have similar distributions of p-EMT marker expression. Additional experiments with the p-EMT marker CXADR demonstrate similar findings (data not shown).
(H) Violin plot depicts p-EMT scores for unsorted, p-EMTlow, and p-EMThigh SCC9 cell sorted and cultured as in (G). Respective isolates largely recapitulate the initial distribution of p-EMT scores.
Seven expression programs were preferentially expressed by subsets of malignant cells in at least two tumors. Two programs (clusters 1,2 in Figures 3A and 3B) reflected the G1/S and G2/M phases of the cell cycle and distinguished cycling cells in each tumor (14–40% of cells in different tumors) (Figure S3A; Table S7). A third program (cluster 6 in Figures 3A and 3B) consisted of JUN, FOS, and immediate early genes implicated in cellular activation and stress responses (Figure S3A; Table S7). A fourth program was enriched for hypoxia-related genes and increased in HNSCC cells cultured in hypoxic conditions (Figures 3B, S3A and S5Q; Table S7).
Two additional programs (clusters 4,5 in Figures 3A and 3B) consisted primarily of epithelial genes, such as EPCAM, cytokeratins (e.g. KRT6, 16, 17, 75), and kallikreins (KLK5-11) (Figure S3A; Table S7). While all malignant cells expressed epithelial markers, many of which were largely uniform across malignant cells (Figures 1C, 1D and S3E), expression of these particular epithelial genes varied coherently across malignant cells (Figure S3D), and may reflect the degree of epithelial differentiation. A final expression program (cluster 3 in Figures 3A and 3B) contained genes associated with ECM and had features of EMT (Figure S3A; Table S7). This program was evident in subsets of cells from seven of the ten tumors examined (Figure S3B).
A partial EMT program in HNSCC
Although EMT programs have been widely considered as potential drivers of drug resistance, invasion, and metastasis, their patterns and significance in human epithelial tumors in vivo remains unclear (Nieto et al., 2016; Thiery et al., 2009; Ye and Weinberg, 2015). We therefore closely examined the ECM program for features of EMT. In addition to ECM genes such as matrix metalloproteinases, laminins and integrins, this program included the EMT markers vimentin (VIM) and integrin α-5 (ITGA5) (Figures 3A, 3C, S3A and S3C; Table S7). Moreover, one of the top scoring genes in this program was TGFβ-induced (TGFBI), implicating the classic EMT regulator TGFβ (Figure S3C).
While the program had key features of classical EMT, it lacked other hallmarks. First, although the signature was accompanied by reduced expression of certain epithelial genes, overall expression of epithelial markers was clearly maintained (Figures S3D and S3E). Second, we did not detect expression of the classical EMT TFs, ZEB1/2, TWIST1/2 and SNAIL1. Only SNAIL2 was detected (in 70% of HNSCC cells), and while its expression correlated with the program across tumors, it did not correlate with the program across individual cells within a tumor (Figure S3F). Recent work suggests that SNAIL2 peaks earlier than other EMT TFs (van Dijk et al., Pre-print, 2017). SNAIL2 is also implicated in a partial EMT response in wound healing (Savagner et al., 2005). We note that EMT is increasingly recognized to be a continuous and variable process (Lambert et al., 2017; Lundgren et al., 2009; Nieto et al., 2016). We therefore suggest that the in vivo program identified here reflects a partial EMT-like state or ‘p-EMT’. Several additional analyses demonstrate that that this p-EMT program is distinct from full EMT programs derived from cell lines and tumor models, as well as from “Mesenchymal” signatures derived from bulk tumor profiles (Figures S4A–D) (Cancer Genome Atlas, 2015; Tan et al., 2014).
In vitro p-EMT cells are dynamic and invasive
We investigated the functional significance of the p-EMT program across five HNSCC cell lines. Expression profiles of 501 cells were largely distinct from human tumors (Figure S3G). However, a subset of cells in SCC9, an oral cavity-derived line, partially recapitulated the in vivo p-EMT program (Figure S3H). When these p-EMThigh cells were isolated by flow cytometry, they demonstrated increased invasiveness (Figures 3D and 3E). They also had a decreased proliferation rate (Figure 3F), consistent with scRNA-seq analysis of patient samples (Figure S4E) and prior EMT studies (Nieto et al., 2016; Ye and Weinberg, 2015).
Prior studies suggested that early stages of EMT may be transitional or metastable (Lambert et al., 2017; Lundgren et al., 2009; Nieto et al., 2016). We therefore considered whether p-EMT might reflect a transient state in dynamic equilibrium with more epithelial subpopulations. To test this, we sorted p-EMThigh and p-EMTlow cells from SCC9, cultured them, and re-assessed marker expression. The two populations remained distinct 4 hours and 24 hours after sorting (t-test, p<0.0001; Figure S4H) but became indistinguishable after 4 days of culture, with both cultures recapitulating the distribution of marker expression in unsorted SCC9 cells (Figures 3G, 3H and S4H). The dynamic nature of this in vitro program raises the possibility that the in vivo p-EMT program may also represent a transient state.
p-EMT cells localize to the leading edge in proximity to CAFs
Taken together, our in vivo profiles and in vitro functional data suggest the p-EMT program is dynamic, invasive, and potentially responsive to TME cues. This led us to investigate the in situ spatial localization of cells expressing this program within HNSCC tumors. We used immunohistochemistry to stain a collection of tumors for the top genes in the p-EMT program (PDPN, LAMC2, LAMB3, MMP10, TGFBI and ITGA5), along with the HNSCC marker p63 (Figures 4A, 4B and S5A–D). These experiments revealed a population of malignant cells that co-stain for p-EMT markers and localize to the leading edge of tumors in close apposition to surrounding stroma. Tumors that lacked the p-EMT program per scRNA-seq did not stain for these markers (Figures S5E–G). In contrast, epithelial differentiation markers (SPRR1B, CLDN4) stained a distinct set of cells at the core of tumors (Figures 4C and S5H–K), consistent with the negative correlation between these programs in scRNA-seq data (Figure 4D).
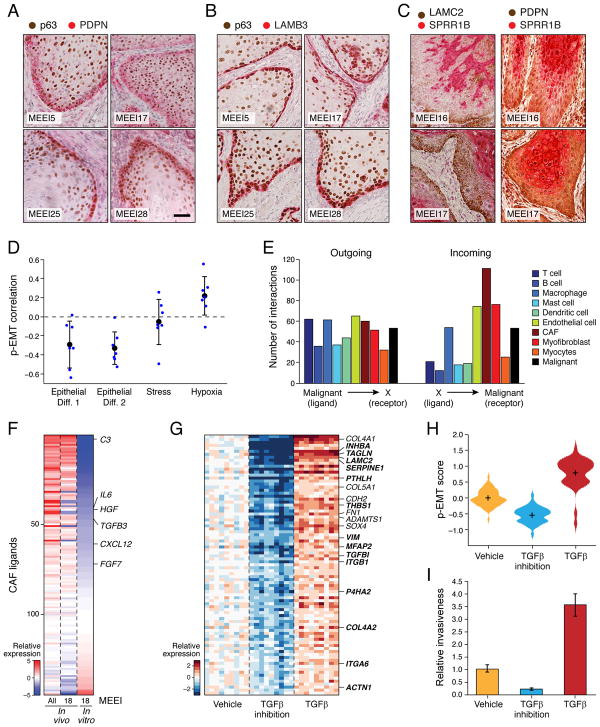
Figure 4. p-EMT cells at the leading edge engage in cross-talk with CAFs.
(A–C) IHC images of representative HNSCC tumors (MEEI5, MEEI16, MEEI17, MEEI25, MEEI28) stained for p-EMT markers (PDPN, LAMB3, LAMC2) and the malignant cell-specific marker p63 (A and B) or the epithelial program marker SPRR1B (C). Scale bar = 100 μM.
(D) Scatter plot shows the Pearson correlation between the p-EMT program and other expression programs underlying HNSCC intra-tumoral heterogeneity (Figure 3). Blue circles depict the correlations within individual tumors; black circles and error-bars represent the average and standard error, respectively, across the different tumors.
(E) Bar plot depicts numbers of putative receptor-ligand interactions between malignant HNSCC cells and indicated cell types. Interaction numbers were calculated based on expression of receptors and corresponding ligands in scRNA-seq data. Outgoing interactions refer to the sum of ligands from malignant cells that interact with receptors on the indicated cell type. Incoming interactions refer to the opposite. CAFs express a significantly greater number of ligands whose receptors are expressed by malignant cells (hypergeometric test, p<0.05).
(F) Heatmap depicts expression of ligands expressed by in vivo and in vitro CAFs. Relative expression is shown for all in vivo CAFs, MEEI18 in vivo CAFs, and in vitro CAFs derived from MEEI18.
(G) Heatmap depicts relative expression of genes that were differentially regulated when SCC9 cells were treated with TGFβ3 or TGFβ pathway inhibitors. Panel includes all genes with significantly higher expression upon TGFβ3 treatment and lower expression upon TGFβ inhibition, relative to vehicle (t-test, p<0.05). Heat intensity reflects relative expression of indicated genes in bulk RNA-seq profiles for nine samples in each group, corresponding to distinct dosage or time points (see STAR Methods). Selected genes are labeled and overlap with the in vivo p-EMT program (bold).
(H) Violin plot depicts distributions of the p-EMT gene expression score across SCC9 cells treated as in (G) and profiled by scRNA-seq. p-EMT scores were increased with TGFβ3 treatment and decreased upon TGFβ inhibition, relative to vehicle (t-test, p<10−16)
(I) Bar plot shows relative invasiveness of SCC9 cells treated as in (G) (representative experiment; error bars reflect SEM; ANOVA, p<0.0001, n=3). In vitro treatment of HNSCC cells with the CAF-related ligand TGFβ causes coherent induction of the p-EMT program and increases invasiveness, while TGFβ inhibition has the opposite effect.
See Figure S5.
The localization of the p-EMT program to the leading edge prompted us to consider interactions with the TME, such as ligand-receptor signaling. We inferred putative tumor-stromal interactions based on high expression of a ligand by one cell type and a corresponding receptor by another cell type (Ramilowski et al., 2015). This predicted “outgoing” signals from malignant cells to the various TME cell types in similar proportions (Figure 4E). Conversely, when we considered “incoming” signals to malignant cells, we found that CAFs expressed notably higher numbers of ligands that correspond to receptors expressed by the malignant cells of the corresponding tumor (hypergeometric test, p<0.05; Figures 4E and S5L). These included interactions that may promote EMT, such as TGFB3-TGFBR2, FGF7-FGFR2 and CXCL12-CXCR7 (Figure 4F) (Moustakas and Heldin, 2016; Ranieri et al., 2016; Yao et al., 2016). Accordingly, when we stained tumors for CAF markers (FAP, PDPN), we found that CAFs were present near p-EMT cells at the leading edge (Figures 4C and S5M).
To evaluate the functional significance of the ligand-receptor interactions, we treated SCC9 cells with TGFβ. Four hours of exposure induced a p-EMT-like program, which was repressed upon inhibition of TGFβ (t-test, p<10−16; Figures 4G and 4H). TGFβ treatment also increased invasiveness and reduced proliferation, while inhibition had opposite effects (ANOVA, p<0.0001; Figures 4I and S5N). In addition, overexpression of TGFBI, a known target of TGFβ and the top p-EMT gene, led to similar effects on invasiveness and proliferation (t-test, p<0.005 and ANOVA, p<0.0001, respectively; Figures S4F and S4G). Conversely, genetic inactivation of TGFBI abrogated the TGFβ response (ANOVA, p<0.0001; Figure S5O and S5P). Although we sought to test CAFs from primary tumors in co-culture, we found that cultured fibroblasts lost expression of typical activation markers and ligands (Figure 4F) and failed to induce a p-EMT response in co-cultured cancer cells (Figure S5R). Taken together, these data suggest that paracrine interactions between CAFs and malignant cells promote a p-EMT program at the leading edge of HNSCC tumors with potential roles in tumor invasion.
Intra-tumoral HNSCC heterogeneity recapitulated in locoregional metastases
To gain further insight into potential determinants of HNSCC spread, we compared LN metastases to primary tumors. Although WES and inferred CNVs revealed some genetic differences between primary and matched LN samples, they did not identify any distinctions that were consistent, possibly due to the small number of individuals studied (Figures S1B, S1C and S6A).
The expression profiles of malignant cells in LNs also largely matched the corresponding primary tumors (Figure 5A). Few differentially-expressed genes were evident for each matched pair, and they were not consistent across the cohort (Figure S6B). The existence of p-EMT high and low subpopulations was also consistent between primary tumors and LNs of all patients, though their prevalence differed between sites (Figures S6C and S6D). These findings raise the possibility that programs required for LN metastasis are dynamic and hence undetected in comparisons of primary tumors and LNs. Indeed, prior studies have also failed to detect genetic or transcriptional distinctions between tumors and locoregional metastases (Colella et al., 2008).
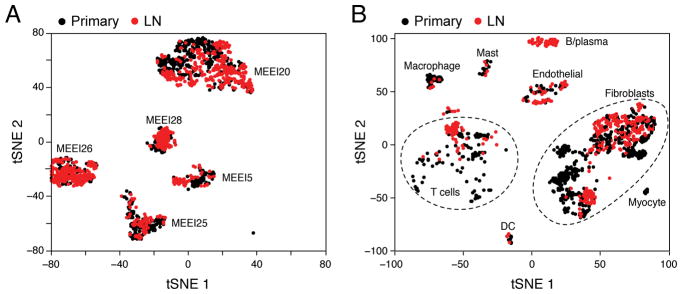
Figure 5. Intra-tumoral HNSCC heterogeneity recapitulated in nodal metastases.
(A) t-SNE plot of malignant cells (as in Figure 2) from five primary tumors (black) and their matched LNs (red). Malignant cells cluster by tumor rather than by site.
(B) t-SNE plot of non-malignant cells (as in Figure 2) from five primary tumors (black) and their matched LNs (red). Non-malignant cells are consistent across tumors but their representation and expression states vary between sites.
See Figure S6.
We also observed an overall concordance in the identity and representation of stromal and immune cells in LNs and matched primary tumors, albeit with some important distinctions. Although most clusters contained cells from both sites, myocytes were observed only in primary tumors and B/plasma cells were found only in LNs (Figure 5B). Fibroblast subsets were also differentially represented: LN fibroblasts were enriched for myofibroblasts and the CAF1 subtype (hypergeometric test; p<0.05), and preferentially expressed certain receptors and ligands (e.g. IL1R1, MMP11, SPARC) (Figures 5B, S2E and S6E). These differences support an altered signaling environment in the LN, but suggest that the TME remains largely stable upon locoregional metastasis.
These findings prompted us to examine the histology of LN specimens, using the markers described above. We found largely intact epithelial structures or ‘nests’ of malignant cells (Figures S6F and S6G) with p-EMT markers at their periphery, surrounded by CAFs and other TME components. These observations are consistent with a ‘collective migration’ model (Clark and Vignjevic, 2015; Lambert et al., 2017), where malignant and stromal cells move in clusters to spread lymphatogenously and form LN metastases. Alternatively, individual cells may disseminate and engraft at the same site (‘single-cell dissemination’), thereby recapitulating primary tumor heterogeneity within LN metastases.
HNSCC subtypes refined by deconvolution of bulk expression data
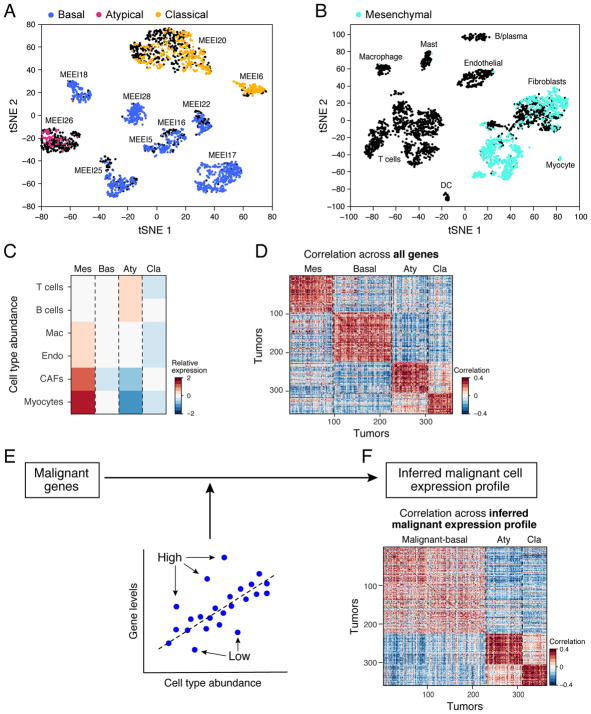
We next considered the generality and prognostic significance of the malignant and stromal expression programs identified from our scRNA-seq data. A recent TCGA study analyzed expression profiles for hundreds of HNSCC tumors, and classified them into four subtypes: basal, mesenchymal, classical, and atypical (Cancer Genome Atlas, 2015). Although the TCGA profiles were acquired from bulk tumors, we reasoned that expression programs of individual cellular components might enable us to extract additional insights. In particular, we asked whether molecular subtypes defined from these bulk data reflect differences in malignant programs, malignant cell composition, and/or TME composition.
We first determined the TCGA expression subtypes of our ten HNSCC tumors. We scored malignant cells from each tumor for their correspondence to subtype expression signatures. Strikingly, each tumor clearly mapped to just one of three subtypes: basal (n=7), classical (n=2), or atypical (n=1) (Figure 6A). None of the malignant cells mapped to the mesenchymal subtype, even though it is the second most frequent subtype among oral cavity tumors. However, when we expanded our analysis to include stromal and immune cells, we found that hundreds of CAFs, myofibroblasts, and myocytes mapped to the mesenchymal subtype (Figure 6B). This finding raised the possibility that the mesenchymal TCGA subtype reflects high stromal representation in bulk samples, rather than a distinct malignant cell program. Indeed, analysis of TCGA samples confirmed that mesenchymal subtype tumors highly expressed genes specific to CAFs and myocytes (Figure 6C). Furthermore, when we examined histology sections for HNSCC tumors from TCGA, we confirmed that mesenchymal tumors had roughly 2.7-fold more fibroblasts than basal tumors (t-test, p<0.0001; Figures S7A–D).
Figure 6. HNSCC subtypes revised by deconvolution of expression profiles from hundreds of tumors.
(A) t-SNE plot of malignant cells from ten tumors (as in Figure 2). Each cluster of cells corresponds to a different tumor. Cells are colored according to the TCGA expression subtype that they match. Black indicates no match. Each tumor can be clearly assigned to one of three subtypes: basal, atypical, or classical.
(B) t-SNE plot of non-malignant cells (as in Figure 2) from ten tumors. Each cluster of cells corresponds to a different cell type. Cells are colored according to the TCGA expression subtype that they match. Black indicates no match. Fibroblasts and myocytes highly express signature genes of the mesenchymal subtype, which likely reflects tumor profiles with high stromal representation.
(C) For each TCGA subtype (columns), heatmap shows relative expression of gene signatures for non-malignant cell types (rows), which were used as estimates of cell type abundances. Tumors classified as mesenchymal highly expressed genes specific to CAFs and myocytes, while atypical tumors were enriched for T- and B-cells.
(D) Heatmap depicts pairwise correlations between TCGA expression profiles ordered by their subtype annotations. This analysis included all genes and recovered all four subtypes.
(E) Schematic of linear regression used to subtract the influence of non-malignant cell frequency from bulk TCGA expression profiles, and thereby infer malignant cell-specific expression profiles.
(F) Heatmap depicts pairwise correlations between TCGA expression profiles ordered by their subtype annotations. This analysis was based on the inferred malignant cell-specific expression profiles in (E). Classical and atypical subtypes are maintained. However, basal and mesenchymal subtypes collapse to a single subtype, which we term ‘malignant-basal.’
See Figure S7.
To investigate the influence of TME composition on TCGA classifications further, we devised a computational approach to subtract the effect of non-malignant cells from TCGA profiles. We restricted the analysis to genes expressed by malignant cells. Since most of these genes were also expressed by non-malignant cells, we normalized their expression to remove the expected contribution of non-malignant cells. To this end, we used cell type-specific gene signatures to estimate the relative abundance of each cell type in each tumor and then, for each gene, we inferred a linear relationship between its bulk expression across tumors and the relative abundance of each cell type using multiple linear regression (Figure 6E). By using the residual of this regression model, we removed the influence of cell type frequencies, including malignant cell frequency (i.e. purity), and inferred a malignant cell-specific intrinsic expression profile for each TCGA tumor.
Remarkably, while standard analysis of TCGA data recovered all four subtypes (Figure 6D), analysis of inferred malignant cell-specific expression eliminated the mesenchymal subtype, while maintaining the other three subtypes (Figure 6F). Tumors previously classified as mesenchymal were found to be part of the previously described basal subtype (now referred to as ‘malignant-basal’). We validated that TCGA mesenchymal scores reflect genes primarily expressed by CAFs and do not correlate with the malignant cell-specific p-EMT program (Figure S4B–D). We therefore suggest that HNSCC tumors may be refined into three subtypes of malignant cells (malignant-basal, classical and atypical), with the previously described mesenchymal subtype reflecting malignant-basal tumors with a large stromal component. The combined malignant-basal subtype would be particularly prevalent, comprising >70% of oral cavity tumors in TCGA, consistent with the classification of seven out of ten tumors in our cohort.
p-EMT predicts metastasis and adverse pathological features
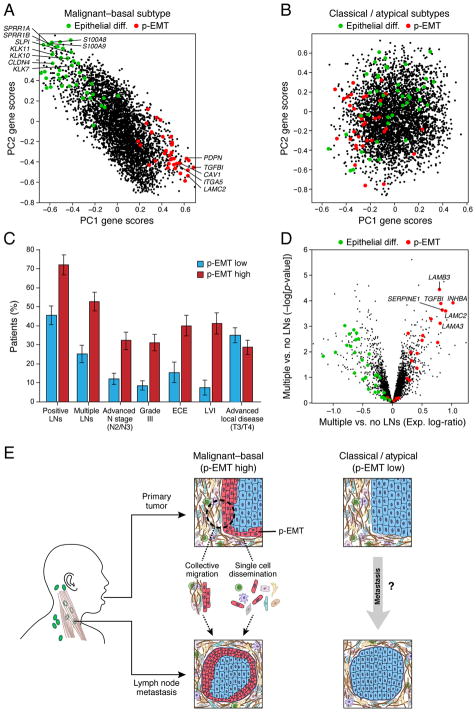
Incorporation of TCGA data gave us an opportunity to examine the prevalence and significance of the p-EMT program across a larger cohort. In our smaller cohort, the p-EMT program was evident in cells from seven of ten tumors (Figure S3B), which exactly correspond to the seven tumors that mapped to the malignant-basal subtype (Figure 6A). Consistent with our smaller cohort, p-EMT levels were highest in malignant-basal tumors in TCGA (Figure S7E). Furthermore, principal component analysis of malignant-basal TCGA tumors, but not atypical and classical tumors, revealed that the first two components were associated with expression of p-EMT genes and were inversely correlated with epithelial differentiation genes (Figures 7A, 7B, S7F and S7G). Remarkably, p-EMT programs defined from these unbiased analyses of bulk expression data were highly consistent with those defined by our scRNA-seq analyses (Figure 7A). They independently confirmed the absence of classical EMT TFs, except for SNAIL2 (Figure S7L), and further support an in vivo p-EMT state in human tumors. Thus, by controlling for confounding effects of TME composition, we demonstrate that differences in p-EMT program expression represents a predominant source of inter-tumoral variability in HNSCC tumors.
Figure 7. p-EMT predicts nodal metastasis and adverse pathologic features.
(A) PC1 and PC2 gene scores based on PCA of inferred malignant cell-specific profiles from all malignant-basal TCGA tumors (n=225). p-EMT genes (red) and epithelial differentiation genes (green) underlie variance among malignant-basal tumors.
(B) PC1 and PC2 gene scores based on PCA of inferred malignant cell-specific profiles from all classical and atypical TCGA tumors (n=156). p-EMT (red) and epithelial differentiation (green) genes are weakly associated with variance in these tumors.
(C) Plot depicts percentage of p-EMT high and p-EMT low malignant-basal tumors associated with each clinical feature. Higher p-EMT scores were associated with positive LNs, advanced nodal stage, high grade, extracapsular extension (ECE), and lymphovascular invasion (LVI) (hypergeometric test, p<0.05). Advanced local disease (T3/T4) as determined by T-stage did not correlate with p-EMT score.
(D) Volcano plot depicts gene expression differences between malignant-basal TCGA tumors with multiple LNs versus those without positive LNs. p-EMT genes (red) have increased expression, while epithelial differentiation genes (green) have decreased expression in metastatic tumors.
(E) Model of the in vivo p-EMT program associated with invasion and metastasis in malignant-basal HNSCC tumors.
See Figure S7.
Lymphatogenous spread of HNSCC tumors to form LN metastases is a major source of disease burden and mortality. Accordingly, resection of oral cavity tumors is typically accompanied by neck dissection to remove the first echelon of draining LNs, a procedure associated with patient morbidity. Tumors with poor prognostic features, such as extracapsular extension or lymphovascular invasion, also receive adjuvant therapy. We therefore tested whether the in vivo p-EMT signature might predict unfavorable pathological features or disease outcome in malignant-basal tumors.
We found that high p-EMT scores were associated with the existence and number of LN metastases and with higher nodal stage (hypergeometric test; p<0.05; Figure 7C). We also found an association with higher tumor grade, offering an explanation for the aggressiveness of poorly differentiated tumors. High p-EMT scores were similarly associated with adverse pathological characteristics, including extracapsular extension and lymphovascular invasion (Figure 7C), for which reliable biomarkers are lacking. Interestingly, p-EMT was not associated with primary tumor size (Figure 7C), suggesting a direct association with invasion and metastasis but not with tumor growth. Overall, p-EMT genes were among the top correlated genes with these clinical features, while other programs such as cell cycle or hypoxia did not correlate nearly as strongly (Figures 7D and S7H). In contrast, the epithelial differentiation program was negatively associated with metastasis (Figure S7H), consistent with our prior observation of an inverse correlation between p-EMT and epithelial differentiation. Importantly, the p-EMT program is a stronger predictor of nodal metastasis and local invasion (Figure S7I) than either the TCGA mesenchymal program or conventional EMT signatures, both of which primarily reflect CAF frequency (Figures S4A and S7I) (Cancer Genome Atlas, 2015; Tan et al., 2014). Current clinical practice relies on imperfect predictors of nodal metastasis, such as tumor thickness and size, resulting in a high rate (~80%) of unnecessary neck dissections (Monroe and Gross, 2012). The p-EMT score could help predict nodal metastasis and thus spare patient morbidity associated with unnecessary neck dissections (Figure S7J).
DISCUSSION
Intra-tumoral heterogeneity represents a major challenge in oncology. Among emerging technologies, scRNA-seq has facilitated the identification of developmental hierarchies, drug resistance programs, and patterns of immune infiltration relevant to tumor biology, diagnosis, and therapy (Kim et al., 2016; Li et al., 2017; Patel et al., 2014; Tirosh et al., 2016a; Tirosh et al., 2016b; Venteicher et al., 2017). Here, we applied the approach to characterize primary HNSCC tumors and matched LN metastases. Our analysis highlights a complex cellular ecosystem with active cross-talk between malignant and non-malignant cells, and an in vivo p-EMT program associated with metastasis (Figure 7E). Our study represents an important step towards understanding intra-tumoral expression heterogeneity in epithelial tumors, which encompass most solid malignancies.
Among our key findings is the identification of a p-EMT program in malignant cells in vivo. This program involves upregulation of certain mesenchymal genes and moderation of epithelial programs. Although reminiscent of an EMT-like process, the program lacks classical TFs thought to drive EMT, with exception of SNAIL2 (Nieto et al., 2016; Thiery et al., 2009; Ye and Weinberg, 2015). SNAIL2 levels do not correlate with the p-EMT program across individual cells in a tumor, but do correlate with the p-EMT program across tumors, both in our small cohort and in TCGA tumors (Figures S7K and S7L), hinting at post-transcriptional regulation. Prior studies have linked SNAIL2 to EMT-like changes required for wound healing (Savagner et al., 2005), raising the possibility that such physiologic responses are co-opted by invasive tumor cells.
Given the absence of classical regulatory programs, the retention of epithelial markers, and the likely transience of this expression state, we speculate that the p-EMT program reflects a ‘metastable’ state that recapitulates certain aspects of EMT, but may be fundamentally different from those defined in vitro (Lundgren et al., 2009; Nieto et al., 2016). Indeed, although we describe an isolated EMT-like program, the molecular description of EMT is currently being re-evaluated with increasing evidence for a continuum of states. It has also been hypothesized that a dynamic, partial EMT state confers invasive properties without losing tumor initiation capacity (Lambert et al., 2017). It remains unclear whether a full EMT state exists in HNSCC, or if the spectrum extends only to p-EMT. Regardless, our unbiased definition of an in vivo partial EMT-like program in patients should guide future studies of this process as it relates to human cancers and metastases.
Several observations suggest that the p-EMT program may promote local invasion and LN metastasis. First, IHC analyses clearly showed that the program localizes to the leading edge of primary tumors, potentially enabling the collective migration of cohorts of cells (Figure 7E) (Clark and Vignjevic, 2015; Lambert et al., 2017). Interestingly, p-EMT cells are in close proximity to CAFs in the surrounding TME, consistent with ligand-receptor analyses supporting regulatory cross-talk between these populations. Second, p-EMThigh HNSCC cells have increased invasive potential in vitro. Third, deconvolution of bulk expression profiles for hundreds of HNSCC tumors identified the p-EMT program as a leading source of variability between patients that is strongly predictive of nodal metastases, lymphovascular invasion, and extranodal extension. Importantly, although CAF abundance did not independently predict nodal metastasis and invasion, tumors with both high CAF scores and high p-EMT scores had a particularly high propensity for metastasis, consistent with a cooperative effect (Figure S7I). This may reflect a role for paracrine signaling between CAFs and malignant cells in promoting nodal disease.
At the same time, other observations temper our conclusions. First, an important caveat of our study is that only 10 tumors were deeply characterized. Analysis of more tumors may reveal additional stromal, immune and malignant cell states, potentially including malignant cells that have further progressed towards a mesenchymal state. Second, the p-EMT program is largely absent from classical and atypical HNSCC tumors, which nonetheless metastasize at similar rates. Thus, p-EMT may be relevant in some subtypes but not others, potentially explaining discord regarding the importance of EMT in tumor biology (Nieto et al., 2016; Thiery et al., 2009; Ye and Weinberg, 2015). Third, although our data imply that the p-EMT state is responsive to CAF signals, the program might simply be a function of increased TME interactions due to disrupted tumor borders, and thus a correlate but not a cause of metastasis. Further study is needed to define the precise mechanisms by which p-EMT and corresponding stromal interactions drive HNSCC metastasis.
Subtype classification schemes have been applied to several tumor types based on ‘bulk’ analyses, which cannot effectively parse intra-tumoral heterogeneity. Here, knowledge of the expression states of malignant, stromal, and immune cell types in HNSCC tumors enabled us to deconvolve bulk TCGA data and infer malignant cell-specific expression profiles. This analysis suggested that the mesenchymal subtype reflects the TME, namely the fraction of CAFs and myocytes within a tumor. Indeed, no malignant cells mapped to the mesenchymal subtype described by TCGA. Thus, the mesenchymal subtype may reflect stromal composition and should be re-evaluated in future studies. In contrast, we find strong support for the other three HNSCC subtypes (classical, atypical, basal). Malignant cells from each of our tumors map exclusively to one of those subtypes. These subtypes also remain stable when controlling for TME. Nonetheless, the potential of stromal components to offer orthogonal prognostic insight (Figure S7I) suggests that future classification systems may ultimately need to integrate both malignant and non-malignant components in a tumor.
In summary, our work provides important insights into HNSCC biology and an atlas of malignant, stromal, and immune cells that should prove relevant to other epithelial malignancies. Our computational approach for inferring malignant cell-specific profiles from bulk expression data refined HNSCC subtypes, and offers a general strategy to extract information from many other cancer datasets. Finally, our definition of a p-EMT program helps relate a large body of EMT data to the in vivo biology of a human tumor. Although further studies are needed, the association of this p-EMT program to unfavorable clinical features may guide future diagnostic strategies and treatment algorithms.
STAR METHODS
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal mouse CD45-vioblue, clone 5B1 | Miltenyi Biotec | Cat#130-092-880, RRID:AB_1103220 |
| Monoclonal mouse CD90-PE, clone 5E10, lot #4343763 | BD Biosciences | Cat#555596, RRID:AB_395970 |
| Monoclonal mouse CD31-PE-cy7, clone WM59, lot #4357750 | BD Biosciences | Cat#563651 |
| Monoclonal mouse CD3-PE-cy7, clone UCHT1, lot #E09903-1631 | ThermoFisher | Cat#25-0038-42 |
| Calcein AM | ThermoFisher | Cat#C3100MP |
| TO-PRO-3 iodide | ThermoFisher | Cat#T3605 |
| Monoclonal mouse p63, clone 4A4, lot #031915, 040416 | Biocare Medical | Cat#CM 163 A/B, RRID:AB_10582730 |
| Monoclonal mouse LAMC2, clone CL2980, lot #CL2980 | Novus Biologicals | Cat#NBP2-42388 |
| Polyclonal rabbit Beta Ig-h3/TGFBI, lot #QC14319-41943 | Novus Biologicals | Cat#NBP1-60049, RRID:AB_11005227 |
| Polyclonal rabbit CLDN4, lot #AA43131 | Novus Biologicals | Cat#NB100-91712, RRID:AB_1216500 |
| Monoclonal mouse MMP-10, clone 110304, lot #DRA0215031 | R&D Systems | Cat#MAB910, RRID:AB_2144566 |
| Polyclonal goat p63, lot #KFX0115111 | R&D Systems | Cat#AF1916, RRID:AB_2207174 |
| Polyclonal sheep PDPN, lot #XXO0115071 | R&D Systems | Cat#AF3670, RRID:AB_2162070 |
| Polyclonal rabbit LAMB3, lot #A74251 | Sigma-Aldrich | Cat#HPA008069, RRID:AB_1079228 |
| Polyclonal rabbit ITGA5, lot #B74062 | Sigma-Aldrich | Cat#HPA002642, RRID:AB_1078469 |
| Polyclonal rabbit SPRR1B, lot #SA100223AI | Sigma-Aldrich | Cat#SAB1301567 |
| Polyclonal rabbit FAP, lot #R84355 | Sigma-Aldrich | Cat#HPA059739 |
| Monoclonal mouse CXADR-PE, clone RmcB, lot #2766468 | EMD Millipore | Cat#FCMAB418PE, RRID:AB_10807695 |
| Polyclonal rabbit TGFBI, lot #75709 | LifeSpan Biosciences | Cat#LS-C325695 |
| Monoclonal mouse p16, clone E6H2 | Roche Tissue Diagnostics | Cat#725-4713 |
| RNAscope Probe HPV-HR18 | Advanced Cell Diagnostics | Cat#312591 |
| R-PE Rabbit IgG Labeling Kit | ThermoFisher | Cat#Z25355 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| See Table S1 for a list of patients included in the study. | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| A-83-01 | Tocris Bioscience | Cat#2939 |
| DMH-1 | Tocris Bioscience | Cat#4126 |
| CHIR99021 | Tocris Bioscience | Cat#4423 |
| Y-27632 | Selleck Chemicals | Cat#S1049 |
| Recombinant TGFβ1 | R&D Systems | Cat#240-B-010 |
| Recombinant TGFβ3 | R&D Systems | Cat#243-B3-010 |
| Critical Commercial Assays | ||
| Human Tumor Dissociation Kit | Miltenyi Biotec | Cat#130-095-929 |
| CellTiter-Glo | Promega | Cat#G7572 |
| BioCoat Matrigel Invasion Chambers | Corning | Cat#354480 |
| RNeasy Micro Kit | Qiagen | Cat#74004 |
| QIAamp DNA Blood Mini Kit | Qiagen | Cat#51106 |
| pENTR/D-TOPO Cloning Kit | ThermoFisher | Cat#K240020 |
| Gateway LR Clonase Enzyme Mix | ThermoFisher | Cat#11791019 |
| FuGENE HD Transfection Reagent | Promega | Cat#E2312 |
| PCR Supermix | ThermoFisher | Cat#10572014 |
| Deposited Data | ||
| Raw data | dbGAP | phs001474.v1.p1 |
| Processed data | GEO | GSE103322 |
| Experimental Models: Cell Lines | ||
| Cal27 | Ohio State University, James Rocco Lab | RRID:CVCL_1107 |
| SCC9 | Ohio State University, James Rocco Lab | RRID:CVCL_1685 |
| SCC4 | Ohio State University, James Rocco Lab | RRID:CVCL_1684 |
| SCC25 | Ohio State University, James Rocco Lab | RRID:CVCL_1682 |
| JHU-006 | Ohio State University, James Rocco Lab | RRID:CVCL_5985 |
| HEK293T | MGH, Bradley Bernstein Lab | RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| TGFBI forward: 5′-CAC CAT GGC GCT CTT CGT GCGG-3′ | IDT | Ref#150615285 |
| TGFBI reverse: 5′-CTA ATG CTT CAT CCT CTC-3′ | IDT | Ref#150615286 |
| TGFBI sgRNA1 forward: 5′-CAC CGA GCT GGT AGGGCG ACT TGG C-3′ | IDT | Ref#150619894 |
| TGFBI sgRNA1 reverse: 5′-AAA CGC CAA GTC GCCCTA CCA GCT C-3′ | IDT | Ref#150619895 |
| TGFBI sgRNA2 forward: 5′-CAC CGC GAC TTG GCG GGA CCC GCC A-3′ | IDT | Ref#150619896 |
| TGFBI sgRNA2 reverse: 5′-AAA CTG GCG GGT CCC GCC AAG TCG C-3′ | IDT | Ref#150619897 |
| TGFBI sgRNA3 forward: 5′-CAC CGC ATG CTC ACT ATC AAC GGG A-3′ | IDT | Ref#150619898 |
| TGFBI sgRNA3 reverse: 5′-AAA CTC CCG TTG ATA GTG AGC ATG C-3′ | IDT | Ref#150619899 |
| TGFBI NGS forward (sgRNA 1 and 2): 5′-TCC ATG GCG CTC TTC GTG-3′ | IDT | Ref#160658478 |
| TGFBI NGS reverse (sgRNA 1 and 2): 5′-GAC TAC CTG ACC TTC CGC AG-3′ | IDT | Ref#160658479 |
| TGFBI NGS forward (sgRNA3): 5′-GTG GAC CCT GAC TTG ACC TG-3′ | IDT | Ref#160658480 |
| TGFBI NGS reverse (sgRNA3): 5′-GTA GTG GAT CAC CCC GTT GG-3′ | IDT | Ref#160658481 |
| Recombinant DNA | ||
| pDNR-Dual-TGFBI | Harvard Plasmid Consortium | Cat#HsCD00003120 |
| pMAL | MGH, Bradley Bernstein Lab | van Galen et al. (2014) |
| pMAL-Luc | MGH, Bradley Bernstein Lab | van Galen et al. (2014) |
| pMAX-GFP | MGH, Bradley Bernstein Lab | van Galen et al. (2014) |
| lentiCRISPRv2 | Addgene | 52961 |
| Non-targeting control plasmid | Broad Institute | BRDN0001478216 |
| Software and Algorithms | ||
| FlowJo version 10.2 | TreeStar | https://www.flowjo.com/solutions/flowjo |
| NIS-Elements Advanced Research version 3.10 | Nikon | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research |
| GraphPad Prism version 4.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| MatLab version 2014b | MathWorks | https://www.mathworks.com/products/matlab.html |
| MatLab scripts for analyses | Trinity Cancer Transcriptome Analysis Toolkit | https://github.com/NCIP/Trinity_CTAT/wiki |
| Other | ||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents may be directed to, and will be fulfilled by, the Lead Contact Bradley Bernstein (bernstein.bradley@mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Tumor Specimens
Patients at the Massachusetts Eye and Ear Infirmary (MEEI) were consented preoperatively to take part in the study following Institutional Review Board approval (Protocol #11-024H). Age and gender of human subjects providing samples are summarized in Table S1 and listed as follows: MEEI5 69/F; MEEI6 88/F; MEEI7 71/F; MEEI8 82/F; MEEI9 77/F; MEEI10 76/M; MEEI12 80/M; MEEI13 52/F; MEEI16 63/F; MEEI17 59/M; MEEI18 41/M; MEEI20 53/M; MEE22 77/M; MEEI23 56/M; MEEI24 78/F; MEEI25 76/F; MEEI26 51/M; MEEI28 58/M. Fresh biopsies of oral cavity head and neck squamous cell carcinoma (HNSCC) were collected at the time of surgical resection, either from the primary tumor or lymph node (LN) dissection. A small fragment was snap frozen for bulk whole exome sequencing and the remainder of the provided tissue was processed for single-cell RNA-seq (scRNA-seq).
Cell Lines
Oral cavity HNSCC cell lines (Cal-27, SCC9, SCC4, SCC25, and JHU-006; all derived from male patients) were generously provided by Dr. James Rocco and colleagues after confirmation by short tandem repeat (STR) analysis (data not shown). They were cultured as follows: JHU-006 cells were grown in RPMI 1640 media (ThermoFisher Scientific), while others cells were grown in 3:1 Ham’s F12 (ThermoFisher Scientific):DMEM (ThermoFisher Scientific). 10% fetal bovine serum (FBS; Peak Serum, Fort Collins, CO) and 1X penicillin-streptomycin-glutamine (PSG; ThermoFisher Scientific) were added to all growth media.
METHOD DETAILS
Tumor Dissociation
Fresh biopsy samples of oral cavity HNSCC were minced, washed with phosphate buffered saline (PBS; ThermoFisher Scientific, Waltham, MA), and dissociated using a Human Tumor Dissociation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) per manufacturer guidelines. Viability was confirmed to be >90% in all samples using trypan blue (ThermoFisher Scientific) exclusion. Cell suspensions were filtered using a 70 μm filter (ThermoFisher Scientific), and dissociated cells were pelleted and re-suspended in PBS with 1% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO). Cells were stained with CD45-vioblue (Miltenyi Biotec), along with either the combination of CD90-PE (BD Biosciences, Franklin Lakes, NJ) and CD31-PE-cy7 (BD Biosciences) or CD3-PE-cy7 (ThermoFisher Scientific), then washed with cold PBS, and re-suspended for flow cytometry analyses.
Sorting of Patient Samples
Cells were stained for viability with 1 μM calcein AM (ThermoFisher Scientific) and 0.33 μM TO-PRO-3 iodide (ThermoFisher Scientific) immediately prior to sorting. Fluorescence-activated cell sorting (FACS) was performed on FACSAria Fusion Special Order System (BD Biosciences) using 488 nm (calcein AM, 530/30 filter), 640 nm (TO-PRO-3, 670/14 filter), 405 nm (Vioblue, 450/50 filter), 561 nm (PE, 586/15 filter; PE-Cy7, 780/60 filter) lasers. Standard forward scatter height versus area criteria were used to discard doublets and capture singlets. Viable cells were identified as calceinhigh and TO-PROlow and additional gates were used to enrich or deplete specific cell types in each plate. For each tumor, plates were sorted containing CD45-cells (to deplete immune cells), CD45-/CD90-/CD31-cells (to further deplete fibroblasts and endothelium and enrich for malignant cells), CD45+ cells (to enrich for immune cells), and CD45+/CD3+ cells (to enrich specifically for T-cells). Single cells were sorted into 96-well plates containing TCL buffer (Qiagen, Hilden, Germany) with 1% β-mercaptoethanol. Plates were briefly centrifuged, snap frozen, and stored at −80 °C before cDNA synthesis and library construction. For each tumor sample, at least one CD45- and one CD45+ plate was sequenced.
cDNA Synthesis and Library Construction
Libraries for isolated single cells were generated based on the SMART-Seq2 protocol (Picelli et al., 2014) with the following modifications: RNA was purified using Agencourt RNAClean XP beads (Beckman Coulter, Brea, CA), prior to reverse transcription with Superscript II (ThermoFisher Scientific) or Maxima (ThermoFisher Scientific) reverse transcriptase and whole transcriptome amplification using KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). Full length cDNA libraries were tagmented using the Nextera XT Library Prep Kit (Illumina, San Diego, CA). 384 samples were pooled and sequenced as paired-end 38 base reads on a NextSeq 500 instrument (Illumina).
Whole Exome and Targeted Sequencing
Snap frozen fresh biopsy and matched whole blood samples were processed by the Genomics Platform at the Broad Institute. Whole exome sequencing was performed per standard protocols using Illumina technology (Illumina). Briefly, library construction was performed as previously described (Fisher et al., 2011). Subsequently, hybridization and capture were performed using the Rapid Capture Exome Kit (Illumina) per manufacturer protocol. After post-capture enrichment, library pools were quantified using an automated qPCR assay on the Agilent Bravo (Agilent Technologies, Santa Clara, CA). Cluster amplification of denatured templates was performed per manufacturer’s protocol using HiSeq 4000 cluster chemistry and HiSeq 4000 flowcells (Illumina). Flowcells were sequenced using v1 Sequencing-by-Synthesis chemistry for HiSeq 4000 flowcells. The flowcells were then analyzed using RTA v.1.18.64 or later (Illumina). In addition, SnAPShot next generation sequencing v2 assay was performed on FFPE samples at the MGH Center for Integrated Diagnostics per standard protocols as previously described (Zheng et al., 2014). Sequencing was performed on an Illumina NextSeq (Illumina). Novoalign (Novocraft Technologies, Selangor, Malaysia) was used to align reads to the hg19 human genome reference. Single nucleotide and indel variants were detected using MuTect1 (Cibulskis et al., 2013), LoFreq (Wilm et al., 2012), and GATK (DePristo et al., 2011; McKenna et al., 2010; Van der Auwera et al., 2013). Exons from 91 gene targets were sequenced.
RNA-seq of Cell Lines
For scRNA-seq, cells were harvested, stained for viability, and sorted into 96-well plates, as described above. cDNA synthesis, library construction, and sequencing were also performed as described. For bulk RNA, RNA was isolated from 1,000 pooled cells using RNEasy Micro Kit (Qiagen).
Flow Cytometry and Sorting of Cell Lines
Sorting of SCC9 cells was performed using TGFBI antibody (LifeSpan Biosciences, Seattle, WA) conjugated to PE using the R-PE IgG labeling kit (ThermoFisher Scientific) per manufacturer specifications. Cells were sorted as described above. For stained samples, cells were considered marker-positive if marker signal was at least as high as the top ~2% of cells in the unstained control. For repopulation experiments, 105 TGFBIhigh, TGFBIlow, and bulk sorted cells were plated and propagated. Cells were harvested after 4 hours, 24 hours, 4 days, and 7 days, stained with TGFBI-PE as described, and re-analyzed by FACS. Cells harvested at 4 hours were not re-stained prior to FACS analysis. Final analysis was performed in FlowJo version 10.2 (TreeStar, Ashland, OR). In addition, single cells in each condition at the 7 day time point were sorted into 96-well plates for scRNA-seq.
Modification of Culture Conditions
For hypoxia cultures, SCC9 cells were grown for seven days in a Galaxy 48R CO2 incubator (Eppendorf, Hamburg, Germany), with 2% O2, 5% CO2. Cells were then harvested and FACS sorted for scRNA-seq. For co-culture experiments, a tumor biopsy from MEEI18 was used to derive CAFs by the Broad Institute Cancer Cell Line Factory. Briefly, the tissue was washed with PBS (ThermoFisher Scientific) and minced using a scalpel. It was digested in 5 mL media with 1 mL 10X collagenase-hyaluronidase (StemCell Technologies, Vancouver, Canada) and 1 mL dispase (StemCell Technologies) for one hour at 37°C. Cells were then centrifuged at 1000 rpm for 5 minutes, followed by RBC lysis with a 5 minute incubation in ACK lysis buffer (ThermoFisher Scientific), followed by 3 minutes in 1 mL media with 1:6 DNase I (StemCell Technologies). Cells were then washed and plated for propagation in ACL4 media (RPMI with L-glutamine (ThermoFisher Scientific) with 5% FBS (Sigma-Aldrich), 0.5% BSA (Rockland Immunochemicals, Limerick, PA), 10 mM HEPES (Sigma-Aldrich), 0.5 mM sodium pyruvate (Sigma-Aldrich), 0.02 mg/mL insulin (Sigma-Aldrich), 0.01 mg/mL transferrin (Sigma-Aldrich), 25 nM sodium selenite (Sigma-Aldrich), 50 nM hydrocortisone (Sigma-Aldrich), and 1 ng/mL epidermal growth factor (Sigma-Aldrich)). Growth of a pure population of fibroblasts was confirmed by a PCR-based targeted sequencing assay using the TruSeq Custom Amplicon platform (Illumina). These tumor-derived fibroblasts were initially plated at a 1:3 ratio with SCC9 cells, and cells were harvested after 48 hours when the ratio of tumor-derived fibroblasts to SCC9 cells was approximately 1:1.
TGFβ Treatment and TGFBI Overexpression
For drug treatment experiments, SCC9 cells were grown in vehicle (4μM HCl with 1μg/mL BSA), TGFβ, or TGFβ-inhibitor. For TGFβ-treated cells, 10 ng/mL recombinant TGFβ1 (R&D Systems, Minneapolis, MN) or TGFβ3 (R&D systems) was applied. Cells in the TGFβ-inhibitor condition were either grown in 3:1 F12:DMEM (ThermoFisher Scientific) with 1μM A-83-01 (Tocris Bioscience, Bristol, UK) or small airway basal medium (Lonza, Basel, Switzerland) with four inhibitors of the TGFβ pathway: 1 μM DMH-1, 1 μM A-83-01, 1 μM CHIR99021 (Tocris Bioscience), and 10 μM Y-27632 (Selleck Chemicals, Houston, TX). For scRNA-seq, cells in each condition were harvested 4 hours after treatment. For bulk RNA-seq, cells were harvested 2, 4, or 6 days after treatment and titrated for analysis. For matrigel invasion assay and cell proliferation assays, cells were maintained in the given conditions for the duration of the experiment.
For TGFBI overexpression, TGFBI was PCR-amplified from pDNR-Dual-TGFBI (Harvard Plasmid Consortium, Cambridge, MA) using the following primers (Integrated DNA Technologies, Coralville, IA): For: 5′-CAC CAT GGC GCT CTT CGT GCG G-3′ and Rev: 5′-CTA ATG CTT CAT CCT CTC-3′. The PCR product was then cloned into pMAL (van Galen et al., 2014) using the pENTR/D-TOPO Cloning Kit (ThermoFisher Scientific) and the Gateway LR Clonase protocol (ThermoFisher Scientific). SCC9 cells at 50–70% confluence were transfected with pMAL-TGFBI or pMAL-Luc (van Galen et al., 2014) using the FuGENE HD transfection reagent (Promega, Madison, WI) per manufacturer protocol. Transfection with pMAX-GFP (van Galen et al., 2014) in parallel conditions confirmed adequate transfection efficiency. Cells were harvested 24 hours after transfection.
TGFBI Knockout Using CRISPR-Cas9
CRISPR sgRNAs were subcloned into lentiCRISPRv2 (Addgene, Cambridge, MA) using primers listed in the Key Resources Table. The target sequences were: sgRNA1 (exon 1 CDS, antisense): 5′-AGC TGG TAG GGC GAC TTG GC-3′; sgRNA2 (exon 1 CDS, antisense): 5′-CGA CTT GGC GGG ACC CGC CA-3′; and sgRNA3 (exon 8 CDS, sense): 5′-CAT GCT CAC TAT CAA CGG GA-3′. A non-targeting control (“mock”) plasmid (BRDN0001478216, Broad Genetic Perturbation Platform, Broad Institute, Cambridge MA) was used for comparison. CRISPR plasmids were co-transfected into 293T cells with GAG/POL and VSVG plasmids, per the Addgene third generation lentiviral system, using the FuGENE HD transfection reagent (Promega) per manufacturer’s protocol. At 36 hours post-transfection, the supernatant was collected and concentrated using Lenti-X Concentrator (Clontech), per manufacturer’s protocol. SCC9 cells at 70% confluence (approximately 2.5 × 104 cells) in 24-well plates were infected with concentrated virus for 36 hours, allowed to recover for multiple passages, and selected with 1 μg/mL puromycin (Life Technologies) for 48 hours, prior to harvesting for matrigel and sequencing assays. Genomic DNA was isolated from 3 × 106 cells using QIAamp DNA Blood Mini Kit (Qiagen). A ~200 bp fragment surrounding the CRISPR cut site of each sample was PCR amplified (PCR Supermix, ThermoFisher Scientific) using TGFBI NGS primers listed in the Key Resources Table. Efficient genome editing was confirmed with next generation sequencing of PCR products at the Massachusetts General Hospital (MGH) Center for Computational & Integrative Biology (CCIB) DNA Core per standard core protocols. Briefly, this entailed Illumina adapter ligation, low-cycle PCR amplification, and sequencing on the Illumina MiSeq (Illumina). Results were analyzed using the CRISPResso software pipeline (Pinello et al., 2016).
Matrigel Invasion Assay
Matrigel invasion assay was performed as previously described (Puram et al., 2012). Preformed matrigel invasion chambers (Corning, Corning, NY) were prepared per manufacturer protocol. Serum-containing media was placed below the invasion chambers and 2.5 × 104 cells suspended in 500 μL serum-free media were placed above the invasion chambers and incubated for 24 hours. Cells on the lower surface of the membrane were fixed with methanol, stained with crystal violet, and counted in a blinded manner. Cells in serum-containing media were used as a negative control.
Cell Proliferation Assay
CellTiter-Glo (CTG) proliferation assay were performed per manufacturer protocol. Cells were plated in 96-well plates in 6–9 replicates per condition at 1,000 cells per well. Cells were lysed on days 2, 4, and 6 by adding CTG reagent (Promega), and point luminescence was measured via the BioTek Synergy HTX Platereader (BioTek, Winooski, VT). For all experiments, a proportional sampling of cells were also lysed at 1 hour after initial plating to ensure that equal numbers were plated across conditions. For cells lysed on day 6, fresh media was added on day 3. CTG luminescence values for individual wells were normalized by subtracting background luminescence (mean luminescence values for wells containing PBS, with CTG reagent added), adjusting for 2μM adenosine triphosphate (ATP) luminescence measured on the same 96-well plate, and normalizing by numbers of plated cells in each condition (as measured by T0 luminescence).
Staining of Tissue Sections
Sectioning and immunohistochemical (IHC) staining of formalin fixed, paraffin-embedded (FFPE) HNSCC specimens was performed by the MGH Histopathology Core per standard protocols. All sections were 5 μm thick. Briefly, antigen retrieval was performed in a decloaker (Biocare Medical) using citrate buffer at pH 6.0. Sections were deparaffinized through xylenes and graded ethanol. Primary antibodies were visualized with HRP- or AP-linked secondary antibodies, followed by diaminobenzidine (DAB; Dako, Glostrup, Denmark) or AP-red (Dako) chromogens, respectively. Sections were counterstained with hematoxylin (ThermoFisher Scientific). Human papillomavirus (HPV) in situ hybridization (ISH) was performed per Advanced Cell Diagnostics RNAscope DAB ISH protocol (Advanced Cell Diagnostics, Newark, CA), with dewaxing followed by a 95-minute target retrieval step, incubation with the RNAscope enzyme, and a 6-hour hybridization. Stained sections were visualized using a Nikon Eclipse 90i microscope with a Nikon DS-Fi1 high definition color camera and NIS-Elements Advanced Research version 3.10 software (Nikon, Melville, NY). Images were captured with a 20X objective and were reviewed by a dedicated head and neck pathologist (W.C.F.).
TCGA Stromal Quantification
Digital hematoxylin and eosin stained slides for TCGA tumors were downloaded and entire sections were examined in a blinded manner. Working with a dedicated head and neck pathologist (W.C.F.), the stromal content of each basal and mesenchymal tumor was quantified by percent and scored as 0 (<10% stromal content), 1+ (10% to <20%), 2+ (20% to <30%), 3+ (30% to <50%), or 4+ (≥50%).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed with GraphPad Prism version 7. (GraphPad Software, La Jolla, CA) or MatLab version 2014b (MathWorks, Natick, MA). Parameters such as sample size, the number of replicates, the number of independent experiments, measures of center, dispersion, and precision (mean ± SD or SEM), and statistical significance are reported in Figures and Figure Legends. Results were considered statistically significant when p < 0.05, or a lower threshold when indicated, by the appropriate test (ANOVA, t-test, Pearson correlation). The Student’s t-test, permutation test, and hypergeometric test were utilized for comparisons in experiments with two sample groups. In experiments with more than two sample groups, analysis of variance (ANOVA) was performed followed by Bonferroni’s post-hoc test.
Single-Cell RNA-seq Data Processing
Expression levels were quantified as Ei,j=log2(TPMi,j/10+1), where TPMi,j refers to transcript-per-million for gene i in sample j, as calculated by RSEM (Li and Dewey, 2011). TPM values are then divided by 10 since we estimate the complexity of single-cell libraries to be on the order of 100,000 transcripts and would like to avoid counting each transcript ~10 times, as would be the case with TPM, which may inflate the difference between the expression level of a gene in cells in which the gene is detected and those in which it is not detected. This modification has a minimal influence on the expression values (Spearman correlation of 1, Pearson correlation of 0.98), but decreases the difference between the expression values of undetected genes (i.e. zero) and that of detected genes (data not shown), thereby reducing the impact of dropouts on downstream analysis. We note that the SMART-Seq2 protocol cannot incorporate unique molecular identifiers (UMI) and therefore we cannot directly identify duplicate reads.
For each cell, we quantified two quality measures: (i) the number of genes for which at least one read was mapped, which is indicative of library complexity and (ii) the average expression level (E) of a curated list of housekeeping genes (Tirosh et al., 2016a), which is meant to verify that genes which are expected to be expressed highly, regardless of cell type, are indeed detected as highly expressed. Scatter plot analyses of all profiled cells separated low and high quality cells based on the these two measures (data not shown), and we therefore conservatively excluded all cells with either fewer than 2,000 detected genes or an average housekeeping expression level (E) below 2.5, as done in previous studies (Patel et al., 2014; Tirosh et al., 2016a). For cells passing these quality controls, the median number of reads were 1.34 million per cell, with a 52.2% transcriptome mapping rate and 3,880 detected genes.
We used the remaining cells (k=5,902) to identify genes that are expressed at high or intermediate levels by calculating the aggregate expression of each gene i across the k cells, as Ea(i)=log2(average(TPM(i)1...k)+1), and excluded genes with Ea<4. For the remaining cells and genes, we defined relative expression by centering the expression levels, Eri,j=Ei,j-average[Ei,1...k]. The relative expression levels, across the remaining subset of cells and genes, were used for downstream analysis. Although normalization approaches can potentially introduce bias into initial clustering, relative expression levels, as defined above and as defined with an alternative normalization method (Bacher et al., 2017) were highly similar. The use of alternative normalization had a limited influence on downstream results such as the distribution of p-EMT scores (data not shown).
To test for batch effects, we performed preliminary clustering of all cells using t-SNE with perplexity of 30 followed by density clustering (DBscan with parameters epsilon=5 and MinPoints=15). The resulting clusters showed limited impact of sequencing batches but an apparent batch effect linked to the enzyme used for reverse transcription (Superscript II or Maxima; data not shown). Since these batch effects have a different impact on the transcriptomes of distinct cell types, we corrected the effect in two steps. First, of the 27 clusters identified in our preliminary clustering described below (see Classification to Malignant and Non-malignant Cells and Figure S1D), we identified seven pairs of clusters that differed by the enzyme used but otherwise were highly similar (as defined by an average Pearson correlation above 0.9); each of these pairs of clusters were then merged, thereby reducing the impact of enzyme usage on cluster assignment. We then normalized the data within each cluster to correct for within-cluster differences that may be linked to enzyme usage. In each cluster, we calculated, for each gene, the average expression among cells processed with Superscript II, the average expression among cells processed with Maxima, and the difference between those. We then subtracted the difference from all cells processed with Maxima in order to correct for the average differences between the two subsets of cells, and make all data comparable to that generated by Superscript II.
Annotation of t-SNE clusters (as in Figures 2A and 2C) by the reverse transcription enzyme revealed that all non-malignant clusters and most malignant clusters contained cells processed with both enzymes (data not shown), suggesting that the choice of enzymes has a minimal effect on the final clustering pattern. Five malignant clusters (each corresponding to all malignant cells from a specific tumor) included cells processed only with Superscript II or only with Maxima. Four of these clusters included only cells processed by Superscript II; since the normalization was done to make all data comparable to Superscript II (by only correcting the Maxima-generated data) these clusters should remain comparable to all other clusters. One malignant cluster contained only cells processed by Maxima, corresponding to all malignant cells of MEEI28, which could theoretically introduce variability between MEEI28 and other malignant clusters; however, this tumor had few differentially expressed genes compared to other tumors (Figure 2D), indicating that batch effects are unlikely to explain the differences between tumors. Importantly, variability of the p-EMT and epithelial differentiation programs was not influenced by the enzyme used for reverse transcription (data not shown).
Epithelial Classification
We defined a set of potential epithelial markers consisting of all cytokeratins, EPCAM, and SFN. We excluded potential markers that were lowly expressed (Ea<4) or not co-regulated with the other markers across all single cells (Pearson R<0.4 with the average of all other markers). The average expression (E) of the 14 remaining genes was used to quantify an epithelial score, which was bimodally distributed (Figure 1C). Epithelial and non-epithelial cells were defined as those with epithelial scores above 3 and below 1.5, respectively, and the remaining cells (with intermediate scores) were unresolved.
CNV Estimation
Initial CNVs (CNV0) were estimated by sorting the analyzed genes by their chromosomal location and applying a moving average to the relative expression values, with a sliding window of 100 genes within each chromosome, as previously described (Patel et al., 2014; Tirosh et al., 2016a). To avoid considerable impact of any particular gene on the moving average, we limited the relative expression values to [−3,3] by replacing all values above 3 by a ceiling of 3, and replacing values below −3 by a floor of −3. This was performed only in the context of CNV estimation. We scored each cell for the extent of CNV signal, defined as the mean of squares of CNV0 values across the genome, and for the correlation between the CNV0 profile of each cell with the average CNV0 profile of all cells from the corresponding tumor. Putative malignant cells were then defined as those with CNV signal above 0.05 and CNV correlation above 0.5, putative non-malignant cells as those below the two cutoffs, and unresolved cells as those above only one of the thresholds. This initial analysis was based on the average CNV0 of all cells as a reference, which is biased due to the inclusion of many malignant cells. We thus redefined CNV estimations, the CNV signal, and CNV correlations values using the average patterns of non-malignant cells as a reference. Non-malignant cells were separated into distinct clusters based on t-SNE as described below. For each cluster we defined a baseline reflecting the average CNV0 estimates of all cells in that cluster, and based on these distinct baselines we defined the maximal (BaseMax) and minimal (BaseMin) baseline at each window. The final CNV estimate of cell i at position j was defined as:
Classification to Malignant and Non-malignant Cells
Epithelial and CNV-based classifications were highly concordant and enabled robust assignment of single cells as malignant or non-malignant. To further support these classifications, we reasoned that global similarity of gene expression programs should also distinguish between malignant and non-malignant cells. We examined 27 clusters as defined by the preliminary clustering described above. Most clusters contained exclusively malignant or non-malignant cells by the above two criteria. Five clusters of smaller sizes were associated primarily with cells that had unresolved or inconsistent assignments by the above two criteria. These clusters were also associated with low complexity (number of genes detected in each cell) and low expression of housekeeping genes, leading us to suspect that they reflect low-quality data. Exclusion of these 324 cells was therefore useful both in order to maintain confidence in malignant classifications and to remove cells of low quality for which the global expression profile and associated clustering may be highly affected by their low data quality.
Identification of Differentially Expressed Genes
To identify differentially expressed genes between different clusters, including comparisons of non-malignant clusters and of malignant clusters, we combined three criteria: (i) an average fold-change of 2, (ii) a t-test p-value below 10−10, and (iii) a permutation test p-value below 0.001. The latter criterion was defined by shuffling the assignments of cells to clusters 10,000 times and counting the fraction of times where an equal or larger difference was obtained between the average expression of each cluster and that of the remaining clusters. The cutoff in the second criterion ensures the control for multiple testing (a stringent Bonferroni correction would result in a corrected p-value of 6.5 × 10−6, as there are at most 10 × 6,465 tests in the family of hypotheses for differential expression).
Classifying Non-malignant Cells
t-SNE analysis of all non-malignant cells using perplexity of 30 was followed by DBscan clustering (with parameters 5 and 15) to identify eight major clusters. Clustering using this approach was highly consistent with an alternative approach (Figure S1E) (Bacher et al., 2017). Furthermore, additional t-SNE analyses with multiple perplexity parameters (15, 20, 25, 30 and 35) and six instances for each perplexity parameter confirmed the robustness of the clustering patterns (data not shown). For each original cluster, we quantified its robustness in each alternative t-SNE instance by the fraction of cells for which the five nearest neighbors (in the alternative t-SNE) are all assigned to the same cluster as the cells being examined. This analysis demonstrated an average rate (across the 30 alternative t-SNE analyses) of consistent clustering larger than 99.6% for each of the clusters. Inspection of the top differentially expressed genes revealed classical cell type markers; for each cluster, we thus defined a set of marker genes, which were both identified as differentially expressed and previously associated with a specific cell type. The average expression profiles of those gene-sets were indeed highly specific to the corresponding clusters (Figure S1F), supporting the cell type classifications.
To further identify subtypes we focused on the two cell types with the largest numbers of cells: T-cells and fibroblasts. We used refined DBscan clustering of the t-SNE analysis (with parameters Epsilon=3, and MinPoints=5) to separate each of those clusters to sub-clusters, and further examined the results with multiple t-SNE analyses to evaluate the robustness of cluster assignments (data not shown).
The T-cell cluster was subdivided into four subtypes, which were annotated based on the differential expression of T cell markers (Figure S2A). This clustering was not strict as variability among T cells was continuous, yet the four clusters were used to represent the main patterns of variability that we observed among T cells (exhausted, CD4, CD8, Tregs).
For fibroblasts, we first observed two robust sub-clusters (myofibroblasts and CAFs, each with more than 98% consistent clustering as defined above) and a third intermediate sub-cluster which was less robust (89% consistent clustering, data not shown). In subsequent analysis, we explored further the diversity of fibroblasts using a focused PCA (Figure S2D). This analysis was restricted to fibroblasts and to genes that are preferentially expressed by fibroblasts (defined as Ea of fibroblast higher than Ea of all other non-malignant cells combined). It recapitulated the three sub-clusters defined above, but also demonstrated that CAFs may be further separated into two subtypes (CAF1 and CAF2) that differ in the expression of many ligands, receptors, and other fibroblast-related genes (Figure S2E).
Expression Programs of Intra-tumoral Heterogeneity
For each of the 10 tumors, non-negative matrix factorization (as implemented by the Matlab nnmf function, with the number of factors set to 10) was used to identify variable expression programs. NNMF was applied to the relative expression values (Er), by transforming all negative values to zero. Notably, undetected genes include many drop-out events (genes that are expressed but are not detected in particular cells due to the incomplete transcriptome coverage), which introduce challenges for normalization of single-cell RNA-seq; since NNMF avoids the exact normalized values of undetected genes (as they are all zero), it may be beneficial in analysis of single-cell RNA-seq (data not shown). We retained only programs for which the standard deviation in cell scores within the respective tumor was larger than 0.8, which resulted in a total of 60 programs across the 10 tumors. The 60 programs were compared by hierarchical clustering (data not shown), using one minus the Pearson correlation coefficient over all gene scores as a distance metric. Six clusters of programs were identified manually (Figure 3B) and used to define meta-signatures. For each cluster, NNMF gene scores were log2-transformed and then averaged across the programs in the cluster, and genes were ranked by their average scores (see Table S6 for the top 50 genes in each cluster). The top 30 genes for each cluster were defined as the meta-signature that was used to define cell scores (see Table S7); each of those genes had average scores above 1 and a t-test p-value below 0.05, based on their scores across the individual programs in the cluster. Since the number of programs in a cluster was small this analysis was not powered to correct for multiple testing and thus we refer to an uncorrected p-value and selected the top ranked genes. However, while confidence is difficult to establish for individual genes in each meta-program, each gene-set defined as a meta-program is highly significant in its co-variation in tumors. For each of the meta-programs, and within each of the tumors included in those meta-programs (2–8 tumors for each meta-program), the average Pearson correlation between all pairs of genes included in the gene-set (calculated across single malignant cells from the respective tumor) was higher than that obtained for 10,000 control gene-sets, which were selected to reproduce the overall distribution of expression levels of the meta-program genes (see also Defining Cell and Sample Scores).
To show the robustness of the NNMF-derived programs with regards to the number of NNMF factors in our dataset, we repeated the NNMF analysis with the number of factors between 5 and 15 (data not shown). We then compared the resulting NNMF programs to the meta-programs defined in our original analysis, with a threshold of global Pearson correlation (across all genes) of 0.2. This threshold is highly significant as it was never observed among 10,000 permutation analyses, in which we permuted the centered expression data of each cell and repeated the analysis. Each of the six meta-programs was identified with each of the NNMF parameters.
Defining Cell and Sample Scores
We used cell scores in order to evaluate the degree to which individual cells express a certain pre-defined expression program. These are initially based on the average expression of the genes from the pre-defined program in the respective cell: Given an input set of genes (Gj), we define a score, SCj(i), for each cell i, as the average relative expression (Er) of the genes in Gj. However, such initial scores may be confounded by cell complexity, as cells with higher complexity have more genes detected (i.e. less zeros) and consequently would be expected to have higher cell scores for any gene-set. To control for this effect we also add a control gene-set (Gjcont); we calculate a similar cell score with the control gene-set and subtract it from the initial cell scores: SCj(i)=average[Er(Gj,i)] – average[Er(Gjcont,i)]. The control gene-set is selected in a way that ensures similar properties (distribution of expression levels) to that of the input gene-set to properly control for the effect of complexity. First, all analyzed genes are binned into 25 bins of equal size based on their aggregate expression levels (Ea). Next, for each gene in the given gene-set, we randomly select 100 genes from the same expression bin. In this way, the control gene-set has a comparable distribution of expression levels to that of the considered gene-set, and is 100-fold larger, such that its average expression is analogous to averaging over 100 randomly-selected gene-sets of the same size as the considered gene-set. A similar approach was used to define bulk sample scores from TCGA.
Flow Cytometry and Sorting of Cell Lines
We performed n=3 independent experiments for TGFBI staining. For stained samples, cells were considered marker-positive if marker signal was at least as high as the top ~2% of cells in the unstained control.
Matrigel Invasion Assay
We performed n=3 independent experiments per condition, and n=4–6 replicates per independent experiment. Invaded cells in each well were counted in a blinded manner across four distinct high powered fields and averaged. Error was calculated as SEM for a representative experiment.
Cell Proliferation Assay
We performed n=3–4 independent experiments per condition, and n=6–9 replicates per independent experiment. CTG luminescence values for individual wells were normalized by subtracting background luminescence (mean luminescence values for wells containing PBS, with CTG reagent added), adjusting for 2μM adenosine triphosphate (ATP) luminescence measured on the same 96-well plate, and normalizing by numbers of plated cells in each condition (as measured by T0 luminescence). Error was calculated as SEM for a representative experiment.
Putative Interactions Between Cell Types
We identified putative interactions between any pair of cell types based on expression of a receptor by one cell type and expression of an interacting ligand by the other cell type: whenever a ligand transcript is “expressed” by cell type A and the interacting receptor transcript is “expressed” by cell type B, we define it as a potential interaction between A and B. If the malignant cells express the receptor or the ligand, then the corresponding interaction was defined as incoming or outgoing, respectively. This analysis required two additional definitions. First, the set of potential receptor-ligand interactions were obtained from Ramilowski et al. (Nature Communications, 2015). Second, a ligand or receptor transcript was defined as “expressed” by a given cell type if its average expression in that cell type was above our threshold of 4 (in values of log2(TPM+1)).
TCGA Subtype Analysis
Bulk RNA-seq data of HNSCC tumors (rnaseqv2-RSEM_genes_normalized) was downloaded from the Broad Firehose website (https://gdac.broadinstitute.org/), along with additional tumor and clinical annotations. Expression data was log2-transformed, filtered to include only the top 10,000 genes (based on average expression), centered for each gene, and compared between subtypes. We identified all genes preferentially expressed in each of the four subtypes (fold-change >2 and p<0.01 by t-test, when comparing a given subtype to each of the other three subtypes) and scored single cells by the four subtype gene-sets (Figures 6A and 6B). To further examine the classification of TCGA samples, we first calculated the average Pearson correlation of each sample with all samples classified by TCGA into a given subtype; samples with an average correlation above 0.1 to one (and only one) subtype were retained for further analysis (Figures 6C–F), while samples with lower correlations for all four subtypes or higher correlation to more than one subtype were excluded.
Inferring Cancer-cell Specific Expression
We first excluded all genes that are not expressed by the malignant cells (i.e., are only expressed by the TME) based on the single-cell data. We retained cells with Ea above 3 (as calculated only over the malignant cells). While this step reduces the influence of TME on bulk expression profiles, it is not sufficient to control for the effect of TME because most genes expressed by malignant cells are also expressed at comparable levels by additional cell types in the TME. We thus aimed to remove this influence using regression analysis. For each of the cell types (t) (both TME and malignant cells) we used the average expression of cell type-specific genes to estimate the relative abundance of the cell type (Frt) across all bulk tumors. These estimates were then used for a multiple linear regression seeking to approximate Ex(i,g), the (log-transformed and centered) expression level of gene g in bulk tumor i, by the sum of Frt(i), the estimated relative cell type frequencies of tumor i, multiplied by gene-specific and cell type-specific scaling factors Xt(g):
Tg includes all the cell types for which the average expression of gene g is lower than that of the malignant cells by at most 2-fold; note that this definition includes also the malignant cell as a cell type, which enables the regression to account for purity. This regression defines the scaling factors Xt(g) that minimize the sum of squares of the residuals, R(i,g), which reflect the component of expression level that is not accounted by the expression of cell types Tg based on the assumption of linear relationship between cell type abundances and total expression level; we define the residuals as the inferred cancer-cell specific expression.
p-EMT Stratification of TCGA samples
Since p-EMT and epithelial differentiation scores were a prominent source of variability in malignant-basal tumors, but not in classical and atypical, we classified only those tumors into p-EMT high and p-EMT low. We defined sample scores (see Defining Cell and Sample Scores) for all malignant–basal tumors based on the inferred cancer-cell specific expression of the p-EMT and epithelial differentiation (Epi. Diff. 2) signatures; only the subset of genes from these signatures which were included in the inferred cancer-cell specific expression were used for these scores. We then ranked the tumors based on their p-EMT score minus the epithelial differentiation, and defined the highest 40% as p-EMT high and the lowest 40% as p-EMT low, while excluding the remaining 20% of tumors with intermediate scores.
Prognostic analysis of p-EMT and CAF scores
To evaluate the effect of p-EMT on seven clinical features (Figure 7C), we compared the fractions of patients with that feature between p-EMT high and p-EMT low tumors, and evaluated the significance of enrichments with a hypergeometric test. To further evaluate the effect of p-EMT while also taking CAF frequency (which is highly consistent with TCGA mesenchymal scores) into account, we used a binomial logistic regression model as implemented by MATLAB fitglm function, with binomial distribution and included interactions. These models fit a logistic regression of two effects (p-EMT scores and CAF frequency scores) and their interactions, in order to predict the clinical features, with a separate model for each feature. The p-values from these models are shown in the bottom panel of Figure S7I.
DATA AND SOFTWARE AVAILABILITY
Raw expression and WES data is available through dbGAP (https://www.ncbi.nlm.nih.gov/gap) with accession number phs001474.v1.p1. Processed expression data is available through the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE103322. Matlab scripts for analyses are available through the Trinity Cancer Transcriptome Analysis Toolkit (https://github.com/NCIP/Trinity_CTAT/wiki).
Supplementary Material
Figure S1. Cells are classified as malignant and non-malignant based on CNVs and epithelial marker expression, Related to Figures 1 and 2.
(A) Histograms show distribution of cells ordered by numbers of reads (Left; median 1.34 million reads), percent of reads mapped to the transcriptome (Middle; median 52.2%), and number of unique genes detected (Right; median 3,880 detected genes).
(B) Heatmap shows large-scale CNVs for individual cells (rows) from 18 tumors, inferred based on the average expression of 100 genes surrounding each chromosomal position (columns). Red: Amplifications; Blue: Deletions.
(C) Large-scale CNVs of seven samples (rows) from three patients as defined by whole exome sequencing analysis.
(D) Stacked bar plots of 27 clusters show percent of malignant (blue) and non-malignant (red) cells, as classified by one (light color) or two (dark color) independent methods: epithelial marker scoring and CNVs. 22 of 27 clusters contain >95% malignant or non-malignant cells; cells in the remaining five clusters were excluded from further analysis.
(E) t-SNE plot of non-malignant cells (as shown in Figure 2A) colored by their assignment to 14 clusters by SC3 (Bacher et al., 2017) with default parameters, demonstrating high consistency between SC3 clusters and tSNE coordinates.
(F) t-SNE plot of non-malignant cells from 10 tumors (same as Figure 2A) with cells colored based on the average expression of sets of marker genes for particular cell types (marker genes and associated cell types are indicated next to each plot). Zero expression level (for all markers of a given cell type) is indicated with small circles, and positive expression is indicated by larger circles, with higher levels indicated by shades of red.
Figure S2. Expression heterogeneity of stromal and immune cells in the HNSCC ecosystem, Related to Figure 2.
(A) (Top) Zoomed in t-SNE plot of T-cells with four distinct clusters identified. (Bottom) Heat map of differentially expressed genes (rows) facilitates annotation of the four clusters (columns) as naïve-like, regulatory, cytotoxic, and exhausted.
(B) Bar plot shows percent of exhausted CD8+ T-cells in six tumors. Asterisks indicate a significant deviation from the mean (hypergeometric test, p<0.01).
(C) (Top) Zoomed in t-SNE plot of fibroblasts with two distinct clusters and a set of intermediates identified. (Bottom) Heat map of differentially expressed genes (rows) facilitates annotation of the clusters (columns) as myofibroblasts, activated CAFs, and intermediate (resting) fibroblasts lacking coherent expression of genes consistent with either myofibroblasts or CAFs.
(D) PC1 and PC2 from a principal component analysis of all fibroblasts, colored based on their assignments to the three clusters as in (D), demonstrates that PC2 further separates the CAF cluster into two subpopulations (CAF1 and CAF2, defined as CAFs with PC2>0, and PC2<0, respectively).
(E) Heatmap of differentially expressed genes (rows) between the CAF1 and CAF2 subpopulations. Selected genes are indicated by name.
(F) Heatmap shows distribution of relative CNVs (columns) for upregulated genes from 10 tumors (rows). Relative CNVs are calculated as the CNV value in the respective tumor minus the average CNVs of all other tumors.
(G) Bar plot shows percentage of upregulated genes (blue) and other genes (red) with relative CNV>0.15 in each tumor, demonstrating a significant enrichment of upregulated genes with high CNVs in all cases (hypergeometric test with Bonferroni correction, p<0.05).
(H) t-SNE plots of malignant (Left; same as Figure 2C) and non-malignant (Right; same as Figure 2A) cells colored by number of unique genes detected. These plots show that clustering is not driven by the detected number of genes. Additional analyses with clusters annotated by batch demonstrate clusters are not determined by batch effects (data not shown).
(I) (Top) Heatmap shows absolute expression of housekeeping (positive) genes (top rows) and immune marker (negative) genes (bottom rows) in single cells (columns) from MEEI25 (same as Figure 3A). (Middle) Heatmap shows absolute expression of genes defining distinct meta-programs (rows) identified by NNMF in single cells (columns) from MEEI25. (Bottom) Bar plot shows number of detected genes in single cells (columns) from MEEI25, with cells ordered as in top and middle panels. Variability in the number of genes detected is not linked to the expression programs identified.
Figure S3. Defining the p-EMT program in HNSCC tumors and cell lines, Related to Figure 3.
(A) Each panel (from top to bottom) shows the meta-signature scores (top section of panel) and a heat map with expression of the top 10 genes for that meta-signature (bottom section of panel) for each of the six coherent expression programs in malignant cells. Cells from ten HNSCC tumors are included and sorted (left to right) first by tumor, within a tumor by sample (primary followed by LN, when applicable), and within a sample by the corresponding meta-signature score (black line).
(B) Each panel (from top to bottom) shows violin plots that depict scores for one of the six meta-signatures in (A) for malignant cells from ten tumors. Violin plots in the second panel depict p-EMT scores, revealing distinct cohorts of p-EMT low (blue) and p-EMT high (red) tumors. Tumors in all panels are ordered identically.
(C–F) Line graphs show smoothed expression (moving average with a window of 100 cells) for selected genes (as labeled); cells from ten HNSCC tumors were included and rank ordered by p-EMT program expression. The selected genes include six of the top p-EMT genes (C), eight epithelial genes negatively correlated with p-EMT scores (D), six epithelial genes not correlated with p-EMT scores (E), and canonical EMT transcription factors (TFs) (F).
(G) Heatmap depicts pairwise Pearson correlations of global expression profiles of malignant cells from ten tumors and five oral cavity HNSCC cell lines. Correlations were calculated across all genes with average expression (Ea) above four in at least one of the tumors or cell lines and after centering the expression levels of genes across all samples included. Clustering indicates that cell lines are more similar to one another than to primary tumor samples and also illustrates the distinction between tumor samples of different subtypes.
(H) Heatmaps show pairwise correlations of expression profiles from individual cells in five oral cavity HNSCC cell lines, ordered by hierarchical clustering. SCC9 includes a subpopulation of cells with an expression profile reminiscent of the p-EMT program, while SCC25 has a subpopulation with an expression profile similar to the stress program. Selected genes preferentially expressed within these subpopulations are highlighted, with markers used for sorting experiments (TGFBI, CXADR) in bold.
Figure S4. Distinguishing the p-EMT program in HNSCC tumors from previously described EMT programs and modeling p-EMT in vitro, Related to Figure 3.
(A) Correlation plot demonstrates pairwise Pearson correlations between EMT and p-EMT programs, including signatures from previous work, as well as this work. Previously described TCGA-Mesenchymal genes (“Mes”), EMT signatures from tumors (“Tumor”), and cell lines (“Culture”) strongly correlate with the expression program of CAFs. These programs weakly correlate with the p-EMT program (“Orig.”) described in this study. Focusing on malignant-specific p-EMT genes (“Malig.”) and p-EMT genes identified after deconvolution (“Decon.”) reveals a more limited correlation of p-EMT with TCGA-Mes and previous EMT signatures, indicating this program is distinct from prior EMT descriptions.
(B) Scatter plot demonstrates three cohorts of TCGA tumors, with (1) high TCGA-mes/intermediate p-EMT, (2) high p-EMT, and (3) low p-EMT scores.
(C) Heatmap demonstrates relative expression of TCGA-Mes, CAF, and p-EMT genes (rows) in TCGA tumors (columns) from the cohorts described in (B), with the eight malignant-specific p-EMT genes (“Malig.”) shown at the bottom.
(D) Bar plots show average expression of each of the gene sets described in (C) in CAFs, malignant cells, and all other immune and stromal cell types detected in our cohort. The p-EMT signature is highly specific to malignant cells, while the TCGA-mes signature is associated with CAFs.
(E) Line graphs show percentage of cycling malignant cells within a sliding window of 20 cells, rank ordered by p-EMT scores. Seven p-EMT high tumors are included; in each tumor, a p-value is shown (permutation test), corresponding to the enrichment of cycling cells among the 30% of cells with lowest p-EMT scores in that tumor. Low p-EMT is significantly enriched with cycling cells among the three tumors with the highest p-EMT scores (MEEI16, MEEI17, and MEEI25).
(F) Bar plot depicts relative invasiveness of SCC9 cells transfected with TGFBI or vector in matrigel invasion assays (error bars reflect SEM; t-test, p<0.005, n=3).
(G) Bar plot shows relative proliferation of SCC9 treated as in (F) (error bars reflect SEM; ANOVA, p<0.0001, n=4).
(H) (Top left) Fluorescence-activated cell sorting plot identifies p-EMThigh and p-EMTlow SCC9 cells isolated based on TGFBI expression. (Top right) Histogram (offset) reveals the distribution of TGFBI expression across cells from the respective isolates (p-EMThigh and p-EMTlow; separated by dashed line) immediately after sorting. (Bottom) Histograms (offset) reveal the distribution of TGFBI expression across cells from the respective isolates (p-EMThigh and p-EMTlow; separated by dashed line) after 4 hours, 24 hours, 4 days, and 7 days in culture. The p-EMThigh and p-EMTlow populations remained distinct 4 hours and 24 hours after sorting (representative experiment; t-test, p<0.0001, n=3).
Figure S5. p-EMT program is localized at the leading edge, distinct from the epithelial differentiation program at the core, Related to Figure 4.
(A–C) Immunohistochemical staining of representative tumors (MEEI5, MEEI16, MEEI17, MEEI25, MEEI28) for p-EMT (LAMC2, MMP10, TGFBI) with the malignant cell-specific marker p63. Scale bar = 100 μM. The leading edges of tumors co-stain with p63 and p-EMT markers. Additional staining with the marker p-EMT marker ITGA5 further validated localization of p-EMT at the leading edge (data not shown).
(D) Immunohistochemical staining of representative tumors (MEEI17, MEEI28) for multiple p-EMT markers (LAMC2, TGFBI). p-EMT markers co-localize at the leading edge.
(E–G) Immunohistochemical staining of representative p-EMT low tumors (MEEI20, MEEI26) for p-EMT (PDPN, LAMB3, LAMC2) with the malignant cell-specific marker p63. p-EMT low tumors show minimal staining for p-EMT markers at the leading edge. Additional staining with the marker ITGA5 confirmed minimal staining for the p-EMT program in these tumors (data not shown).
(H and I) Immunohistochemical staining of representative tumors (MEEI16, MEEI17) for epithelial differentiation (SPRR1B, CLDN4) and the malignant cell-specific marker p63.
(J and K) Immunohistochemical staining of representative tumor (MEEI17) for p-EMT (LAMC2, PDPN) and epithelial differentiation (CLDN4). Markers demonstrate distinct spatial localization of p-EMT and epithelial differentiation programs, at the leading edge and core, respectively.
(L) Bar plot shows statistical significance (minus log10 of p-value defined by hypergeometric test) of number of observed outgoing interactions between ten listed cell types and malignant cells. Bars above the x-axis indicate a greater number of interactions than expected, while bars below the x-axis indicate fewer interactions than expected.
(M) Immunohistochemical staining of representative tumors (MEEI16, MEEI18) for p-EMT and CAFs (FAP) with the malignant cell-specific marker p63. FAP staining is present both at the leading edge of tumors nests and in the stroma, highlighting activated CAFs.
(N) Bar plot depicts relative proliferation of SCC9 cells treated with vehicle, TGFβ, or TGFβ pathway inhibitors (error bars reflect SEM; ANOVA, p<0.0001, n=4).
(O) Histograms show percent of sequencing reads with insertions or deletions (indels) of specified size in mock infected SCC9 cells (Top left) and SCC9 TGFBI CRISPR knockout cells (other panels). Each of the TGFBI-targeting sgRNAs resulted in >98.8% of reads containing indels, indicating efficient knockout of TGFBI.
(P) Bar plot depicts relative invasiveness of mock infected SCC9 cells or SCC9 TGFBI CRISPR knockout cells after treatment with vehicle or TGFβ in matrigel invasion assays (error bars reflect SEM; ANOVA, p<0.0001, n=3).
(Q) Violin plot depicts hypoxia program scoring of SCC9 cells grown in normoxic or hypoxic conditions. Hypoxic conditions are associated with significantly increased hypoxia score (t-test, p<0.05).
(R) Violin plot depicts scoring of SCC9 cells for p-EMT scores after growth in standard conditions (control), hypoxic conditions, or in co-culture with CAFs derived from MEEI18. p-EMT expression is not significantly changed across these conditions.
Figure S6. Variability in the p-EMT program and cancer-associated fibroblasts across tumor subsites (primary and lymph node), Related to Figure 5.
(A) Comparison of point mutations between primary and LN samples in three individual tumors (MEEI26, MEEI20, and MEEI25 from top to bottom) as detected by whole exome sequencing. In each tumor, we examined all mutations identified in at least one of the samples (primary or LN) and assigned it one of three values in each sample: “detected” (black), “not detected” (white), or unresolved due to “low coverage.” A single mutant read was sufficient to define a mutation as “detected,” but zero mutant reads were defined as “not detected” only if the probability of detecting zero mutant reads in that sample was below 0.05 (as defined by binomial test, given the number of reads covering that base and assuming the same frequency of the mutant reads as in the sample(s) where it is detected). Mutations were then ordered by their identification across the samples and assigned to four classes: shared among primary and LN, specific to primary, specific to LN, and unresolved. Note that for MEEI26 two LN samples are included corresponding to the left (ipsilateral) and right (contralateral) LNs, denoted as LNL and LNR, respectively.
(B) Heatmap of differentially expressed genes between primary and LN samples across multiple patients. For each of the five patients with matched primary and LN samples, we identified significant differentially expressed genes (defined by p<0.001 and fold-change>2). All genes defined as upregulated in at least two patients (left panel) or downregulated in at least two patients (right panel) are shown. Red: upregulated; Blue: downregulated. Darker shades indicate significant differential expression, while lighter shades denote borderline differential expression (p<0.05 and fold-change>1.5).
(C) Violin plot depicts p-EMT score of malignant cells from five primary tumors and matched LN.
(D) Scatter plot shows the average (x-axis) and the variability (y-axis) of p-EMT scores across individual malignant cells within each sample; five primary tumors (black) and matched LNs (red) are included and matched samples are connected with lines. p-EMT high tumors display both higher average and higher variability of p-EMT scores.
(E) Fibroblasts from primary (black) and LN (red) samples, scored by the relative expression of gene-sets distinguishing CAFs from myofibroblasts (x-axis) and those distinguishing the CAF1 and CAF2 subsets (y-axis), demonstrating that LN CAFs are biased towards the CAF1 subset (hypergeometric test, p<0.05).
(F and G) Immunohistochemical staining of representative LN metastases (MEEI25, MEEI28) for p-EMT (PDPN, LAMB3) with the malignant-cell specific marker p63.
Figure S7. p-EMT program is negatively correlated with epithelial differentiation and may predict nodal metastasis, Related to Figures 6 and 7.
(A) Hematoxylin-eosin (H&E) stained sections from representative mesenchymal (Left) and basal (Right) TCGA tumors demonstrate substantially more stromal infiltrate in mesenchymal than basal tumors. Scale bar = 400 μM.
(B) (Left) Bar plot shows significantly higher percent of stromal infiltrate in mesenchymal tumors compared to basal tumors (t-test, p<0.0001; n=203 tumors). (Right) Bar plot shows number of tumors with H&E stromal scores ranging from 0 (lowest) to 4 (highest) for mesenchymal and basal subtype TCGA tumors.
(C and D) Scatter plots demonstrate a correlation between H&E stromal score (indicated by dot color) with CAF and TCGA mesenchymal scores (C), but not p-EMT scores (D).
(E) Line graph shows distribution of p-EMT scores across TCGA tumors of each subtype.
(F) Scatter plot shows scoring of TCGA basal and mesenchymal tumors for epithelial differentiation and p-EMT which are significantly negatively correlated in this subset of tumors (Pearson correlation, p<0.05); black lines indicate linear regression.
(G) Scatter plot shows scoring of TCGA classical and atypical tumors for epithelial differentiation and p-EMT, which are not significantly correlated in this subset of tumors; black lines indicate linear regression.
(H) Bar plot shows direction and statistical significance (p-value based on a t-test) of the association between each of six coherent meta-signatures and the presence of multiple versus no metastatic LNs in TCGA malignant-basal tumors. The p-EMT and epithelial differentiation programs, which were inversely correlated in expression studies, had opposite associations with metastasis. The other programs show no significant association with LN metastases.
(I) (Top) Bar plot shows the percent of patients with adverse clinical features (positive LNs, multiple LNs, advanced N stage, grade III, extranodal extension, lymphovascular invasion, and advanced local disease) in cohorts with high and low p-EMT scores stratified by high and low CAF scores. (Bottom) Heatmap shows the statistical significance of p-EMT and CAF effects on adverse clinical features based on a binomial logistic regression with two predictive variables (p-EMT and variable scores) and an interaction effect. Only the p-EMT effect is predictive of clinical features associated with metastasis and invasion (positive LNs, multiple LNs, advanced nodal stage, extracapsular extension, and lymphovascular invasion) (Bottom, first row). In contrast, the CAF effect has no significant predictive value for features associated with metastasis, but instead, predicts high grade disease and advanced local disease (T3/T4) (Bottom, second row). The p-EMT and CAF effects did act cooperatively to influence the risk of nodal metastasis (Bottom, third row), consistent with a putative ligand-receptor interaction between CAFs and p-EMT cells.
(J) Percent of patients from TCGA for which neck dissection was justified using varying thresholds of p-EMT scores and stratified by tumor (T) stage. Justified neck dissection refers to patients with initial clinical diagnosis of lymph node-negative (cN0) for which neck dissection revealed a positive metastatic lymph node (pN1-N3); the percentage of justified neck dissections was calculated out of all patients with clinical node-negative disease that underwent neck dissection. A higher p-EMT threshold is associated with a higher rate of justified neck dissection, regardless of T-stage (permutation test, p<0.05).
(K) Correlations of genes with the p-EMT program within (x-axis) and across (y-axis) tumors in our cohort of ten patients. Within-tumor correlations were calculated separately in each tumor and averaged; across-tumor correlations were calculated between the average levels of genes and those of the p-EMT program across all malignant cells in each tumor. Selected genes are indicated.
(L) Scatter plot shows the correlations of genes with p-EMT (x-axis) and epithelial differentiation (y-axis) programs based on inferred malignant cell-specific profiles from TCGA malignant-basal tumors. Genes of the p-EMT (red) and epithelial differentiation (green) programs as well as EMT TFs (black) are indicated, demonstrating a high p-EMT correlation with SNAIL2 but not of other EMT TFs.
Table S1. Patients and samples included in dataset, Related to Figure 1.
Table S2. Clinical and pathologic features of deeply sequenced samples, Related to Figure 1. p16 immunohistochemistry and HPV in situ hybridization were negative for all samples.
Table S3. Mutations and copy number variations detected in profiled primary tumors, Related to Figure 1. Common mutations evaluated by whole exome sequencing of a subset of samples and SNaPshot next generation sequencing assay of all samples include the top 5 mutations in TCGA HNSCC tumors, as well as mutations in TERT promoter. CNVs evaluated include top 4 abnormalities noted in TCGA HNSCC tumors.
Table S4. Mutations detected by whole exome sequencing, Related to Figure 1. Mutations are sorted by patient number, within patient by primary tumor followed by lymph node, and within sample by location within the genome.
Table S5. Differentially expressed genes between CAF subsets, Related to Figure 2. Genes are sorted from most to least significant.
Table S6. Expression programs detected by NNMF in each of 10 patients, Related to Figure 3. Clusters are ordered as in Figure 3B, and within each cluster the genes are ordered from most to least significant. For each cluster, headers also indicate the patient from which it was derived and an inferred annotation. See also online tables.
Table S7. Six meta-signatures, each derived from multiple related NNMF programs, Related to Figure 3. Genes in each program are ordered from most to least significant.
Acknowledgments
We thank P. van Galen and members of the Bernstein and Regev laboratories for critical review of the manuscript. We also thank H. Ravichandran, P. Della Pelle, N. Hayes, and K. Hoadley for technical assistance. This research was supported by an American Academy of Otolaryngology Resident Grant (S.V.P.), a New England Otolaryngology Society Resident Grant (S.V.P.), the Human Frontiers Science Program (I.T.), a NRSA T32 Training Grant (A.S.P.), the Massachusetts Eye and Ear Infirmary, the National Cancer Institute, the NIH Common Fund, the Broad Institute, the Klarman Cell Observatory, the Starr Foundation, the Ludwig Center, and an NIH Shared Instrumentation grant for cytometry. B.E.B. is the Bernard and Mildred Kayden Endowed Research Institute Chair and an American Cancer Society Research Professor. A.R. is a Howard Hughes Medical Institute Investigator.
Footnotes
AUTHOR CONTRIBUTIONS
S.V.P., I.T., A.S.P, A.P.P., S.G., and C.R. designed and performed experiments. C.L.L. and R.M. provided guidance for FACS analyses, K.Y. assisted with WES, and W.C.F. led all histology. E.A.M., K.S.E, D.G.G., M.A.V., O.R., and J.W.R. provided input on experimental and study design. S.V.P., I.T., and A.S.P. wrote the manuscript with input from A.R. and B.E.B.. D.T.L., A.R., and B.E.B. supervised the project.
References
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Chu LF, Leng N, Gasch AP, Thomson JA, Stewart RM, Newton M, Kendziorski C. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017;14:584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Vignjevic DM. Modes of cancer cell invasion and the role of the microenvironment. Curr Opin Cell Biol. 2015;36:13–22. doi: 10.1016/j.ceb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Colella S, Richards KL, Bachinski LL, Baggerly KA, Tsavachidis S, Lang JC, Schuller DE, Krahe R. Molecular signatures of metastasis in head and neck cancer. Head Neck. 2008;30:1273–1283. doi: 10.1002/hed.20871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Kim KT, Lee HW, Lee HO, Song HJ, Jeong da E, Shin S, Kim H, Shin Y, Nam DH, Jeong BC, et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. doi: 10.1186/s13059-016-0945-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics (England) 2011:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJ, Kong SL, Chua C, Hon LK, Tan WS, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017 doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Nordenskjold B, Landberg G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. Br J Cancer. 2009;101:1769–1781. doi: 10.1038/sj.bjc.6605369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madar S, Goldstein I, Rotter V. ‘Cancer associated fibroblasts’--more than meets the eye. Trends Mol Med. 2013;19:447–453. doi: 10.1016/j.molmed.2013.05.004. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe MM, Gross ND. Evidence-based practice: management of the clinical node-negative neck in early-stage oral cavity squamous cell carcinoma. Otolaryngol Clin North Am. 2012;45:1181–1193. doi: 10.1016/j.otc.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Mechanisms of TGFbeta-Induced Epithelial-Mesenchymal Transition. J Clin Med. 2016:5. doi: 10.3390/jcm5070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Liu SJ, Di Lullo E, Malatesta M, Pollen AA, Nowakowski TJ, Kohanbash G, Aghi M, Kriegstein AR, Lim DA, et al. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12:889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25:1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, Yuan GC. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Rocco JW. Molecular Aspects of Head and Neck Cancer Therapy. Hematol Oncol Clin North Am. 2015;29:971–992. doi: 10.1016/j.hoc.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Yeung CM, Jahani-Asl A, Lin C, de la Iglesia N, Konopka G, Jackson-Grusby L, Bonni A. STAT3-iNOS Signaling Mediates EGFRvIII-Induced Glial Proliferation and Transformation. J Neurosci. 2012;32:7806–7818. doi: 10.1523/JNEUROSCI.3243-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri D, Rosato B, Nanni M, Magenta A, Belleudi F, Torrisi MR. Expression of the FGFR2 mesenchymal splicing variant in epithelial cells drives epithelial-mesenchymal transition. Oncotarget. 2016;7:5440–5460. doi: 10.18632/oncotarget.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey DC, Weymouth N, Shi Z. Smooth muscle alpha actin (Acta2) and myofibroblast function during hepatic wound healing. PLoS One. 2013;8:e77166. doi: 10.1371/journal.pone.0077166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P, Kusewitt DF, Carver EA, Magnino F, Choi C, Gridley T, Hudson LG. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J Cell Physiol. 2005;202:858–866. doi: 10.1002/jcp.20188. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TZ, Miow QH, Miki Y, Noda T, Mori S, Huang RY, Thiery JP. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol Med. 2014;6:1279–1293. doi: 10.15252/emmm.201404208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016a;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016b;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk D, Nainys J, Sharma R, Kathail P, Carr AJ, Moon KR, Mazutis L, Wolf G, Krishnaswamy S, Pe’er D. MAGIC: A diffusion-based imputation method reveals gene-gene interactions in single-cell RNA-sequencing data. BioRxiv 2017 Pre-print. [Google Scholar]
- van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, Xie S, Laurenti E, Hermans K, Eppert K, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- Venteicher AS, Tirosh I, Hebert C, Yizhak KCN, Filbin MG, Hoverstadt V, Escalante LE, Saw ML, Rodman C, et al. Decoupling genetics, lineages and tumor micro-environment in IDH-mutant gliomas by single-cell RNA-seq. Science. 2017:355. doi: 10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. Coming full circle-from endless complexity to simplicity and back again. Cell. 2014;157:267–271. doi: 10.1016/j.cell.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Li P, Song H, Song F, Qu Y, Ma X, Shi R, Wu J. CXCL12/CXCR4 Axis Upregulates Twist to Induce EMT in Human Glioblastoma. Mol Neurobiol. 2016;53:3948–3953. doi: 10.1007/s12035-015-9340-x. [DOI] [PubMed] [Google Scholar]
- Ye X, Weinberg RA. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, Chen J, Robinson HE, Shim HS, Chmielecki J, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cells are classified as malignant and non-malignant based on CNVs and epithelial marker expression, Related to Figures 1 and 2.
(A) Histograms show distribution of cells ordered by numbers of reads (Left; median 1.34 million reads), percent of reads mapped to the transcriptome (Middle; median 52.2%), and number of unique genes detected (Right; median 3,880 detected genes).
(B) Heatmap shows large-scale CNVs for individual cells (rows) from 18 tumors, inferred based on the average expression of 100 genes surrounding each chromosomal position (columns). Red: Amplifications; Blue: Deletions.
(C) Large-scale CNVs of seven samples (rows) from three patients as defined by whole exome sequencing analysis.
(D) Stacked bar plots of 27 clusters show percent of malignant (blue) and non-malignant (red) cells, as classified by one (light color) or two (dark color) independent methods: epithelial marker scoring and CNVs. 22 of 27 clusters contain >95% malignant or non-malignant cells; cells in the remaining five clusters were excluded from further analysis.
(E) t-SNE plot of non-malignant cells (as shown in Figure 2A) colored by their assignment to 14 clusters by SC3 (Bacher et al., 2017) with default parameters, demonstrating high consistency between SC3 clusters and tSNE coordinates.
(F) t-SNE plot of non-malignant cells from 10 tumors (same as Figure 2A) with cells colored based on the average expression of sets of marker genes for particular cell types (marker genes and associated cell types are indicated next to each plot). Zero expression level (for all markers of a given cell type) is indicated with small circles, and positive expression is indicated by larger circles, with higher levels indicated by shades of red.
Figure S2. Expression heterogeneity of stromal and immune cells in the HNSCC ecosystem, Related to Figure 2.
(A) (Top) Zoomed in t-SNE plot of T-cells with four distinct clusters identified. (Bottom) Heat map of differentially expressed genes (rows) facilitates annotation of the four clusters (columns) as naïve-like, regulatory, cytotoxic, and exhausted.
(B) Bar plot shows percent of exhausted CD8+ T-cells in six tumors. Asterisks indicate a significant deviation from the mean (hypergeometric test, p<0.01).
(C) (Top) Zoomed in t-SNE plot of fibroblasts with two distinct clusters and a set of intermediates identified. (Bottom) Heat map of differentially expressed genes (rows) facilitates annotation of the clusters (columns) as myofibroblasts, activated CAFs, and intermediate (resting) fibroblasts lacking coherent expression of genes consistent with either myofibroblasts or CAFs.
(D) PC1 and PC2 from a principal component analysis of all fibroblasts, colored based on their assignments to the three clusters as in (D), demonstrates that PC2 further separates the CAF cluster into two subpopulations (CAF1 and CAF2, defined as CAFs with PC2>0, and PC2<0, respectively).
(E) Heatmap of differentially expressed genes (rows) between the CAF1 and CAF2 subpopulations. Selected genes are indicated by name.
(F) Heatmap shows distribution of relative CNVs (columns) for upregulated genes from 10 tumors (rows). Relative CNVs are calculated as the CNV value in the respective tumor minus the average CNVs of all other tumors.
(G) Bar plot shows percentage of upregulated genes (blue) and other genes (red) with relative CNV>0.15 in each tumor, demonstrating a significant enrichment of upregulated genes with high CNVs in all cases (hypergeometric test with Bonferroni correction, p<0.05).
(H) t-SNE plots of malignant (Left; same as Figure 2C) and non-malignant (Right; same as Figure 2A) cells colored by number of unique genes detected. These plots show that clustering is not driven by the detected number of genes. Additional analyses with clusters annotated by batch demonstrate clusters are not determined by batch effects (data not shown).
(I) (Top) Heatmap shows absolute expression of housekeeping (positive) genes (top rows) and immune marker (negative) genes (bottom rows) in single cells (columns) from MEEI25 (same as Figure 3A). (Middle) Heatmap shows absolute expression of genes defining distinct meta-programs (rows) identified by NNMF in single cells (columns) from MEEI25. (Bottom) Bar plot shows number of detected genes in single cells (columns) from MEEI25, with cells ordered as in top and middle panels. Variability in the number of genes detected is not linked to the expression programs identified.
Figure S3. Defining the p-EMT program in HNSCC tumors and cell lines, Related to Figure 3.
(A) Each panel (from top to bottom) shows the meta-signature scores (top section of panel) and a heat map with expression of the top 10 genes for that meta-signature (bottom section of panel) for each of the six coherent expression programs in malignant cells. Cells from ten HNSCC tumors are included and sorted (left to right) first by tumor, within a tumor by sample (primary followed by LN, when applicable), and within a sample by the corresponding meta-signature score (black line).
(B) Each panel (from top to bottom) shows violin plots that depict scores for one of the six meta-signatures in (A) for malignant cells from ten tumors. Violin plots in the second panel depict p-EMT scores, revealing distinct cohorts of p-EMT low (blue) and p-EMT high (red) tumors. Tumors in all panels are ordered identically.
(C–F) Line graphs show smoothed expression (moving average with a window of 100 cells) for selected genes (as labeled); cells from ten HNSCC tumors were included and rank ordered by p-EMT program expression. The selected genes include six of the top p-EMT genes (C), eight epithelial genes negatively correlated with p-EMT scores (D), six epithelial genes not correlated with p-EMT scores (E), and canonical EMT transcription factors (TFs) (F).
(G) Heatmap depicts pairwise Pearson correlations of global expression profiles of malignant cells from ten tumors and five oral cavity HNSCC cell lines. Correlations were calculated across all genes with average expression (Ea) above four in at least one of the tumors or cell lines and after centering the expression levels of genes across all samples included. Clustering indicates that cell lines are more similar to one another than to primary tumor samples and also illustrates the distinction between tumor samples of different subtypes.
(H) Heatmaps show pairwise correlations of expression profiles from individual cells in five oral cavity HNSCC cell lines, ordered by hierarchical clustering. SCC9 includes a subpopulation of cells with an expression profile reminiscent of the p-EMT program, while SCC25 has a subpopulation with an expression profile similar to the stress program. Selected genes preferentially expressed within these subpopulations are highlighted, with markers used for sorting experiments (TGFBI, CXADR) in bold.
Figure S4. Distinguishing the p-EMT program in HNSCC tumors from previously described EMT programs and modeling p-EMT in vitro, Related to Figure 3.
(A) Correlation plot demonstrates pairwise Pearson correlations between EMT and p-EMT programs, including signatures from previous work, as well as this work. Previously described TCGA-Mesenchymal genes (“Mes”), EMT signatures from tumors (“Tumor”), and cell lines (“Culture”) strongly correlate with the expression program of CAFs. These programs weakly correlate with the p-EMT program (“Orig.”) described in this study. Focusing on malignant-specific p-EMT genes (“Malig.”) and p-EMT genes identified after deconvolution (“Decon.”) reveals a more limited correlation of p-EMT with TCGA-Mes and previous EMT signatures, indicating this program is distinct from prior EMT descriptions.
(B) Scatter plot demonstrates three cohorts of TCGA tumors, with (1) high TCGA-mes/intermediate p-EMT, (2) high p-EMT, and (3) low p-EMT scores.
(C) Heatmap demonstrates relative expression of TCGA-Mes, CAF, and p-EMT genes (rows) in TCGA tumors (columns) from the cohorts described in (B), with the eight malignant-specific p-EMT genes (“Malig.”) shown at the bottom.
(D) Bar plots show average expression of each of the gene sets described in (C) in CAFs, malignant cells, and all other immune and stromal cell types detected in our cohort. The p-EMT signature is highly specific to malignant cells, while the TCGA-mes signature is associated with CAFs.
(E) Line graphs show percentage of cycling malignant cells within a sliding window of 20 cells, rank ordered by p-EMT scores. Seven p-EMT high tumors are included; in each tumor, a p-value is shown (permutation test), corresponding to the enrichment of cycling cells among the 30% of cells with lowest p-EMT scores in that tumor. Low p-EMT is significantly enriched with cycling cells among the three tumors with the highest p-EMT scores (MEEI16, MEEI17, and MEEI25).
(F) Bar plot depicts relative invasiveness of SCC9 cells transfected with TGFBI or vector in matrigel invasion assays (error bars reflect SEM; t-test, p<0.005, n=3).
(G) Bar plot shows relative proliferation of SCC9 treated as in (F) (error bars reflect SEM; ANOVA, p<0.0001, n=4).
(H) (Top left) Fluorescence-activated cell sorting plot identifies p-EMThigh and p-EMTlow SCC9 cells isolated based on TGFBI expression. (Top right) Histogram (offset) reveals the distribution of TGFBI expression across cells from the respective isolates (p-EMThigh and p-EMTlow; separated by dashed line) immediately after sorting. (Bottom) Histograms (offset) reveal the distribution of TGFBI expression across cells from the respective isolates (p-EMThigh and p-EMTlow; separated by dashed line) after 4 hours, 24 hours, 4 days, and 7 days in culture. The p-EMThigh and p-EMTlow populations remained distinct 4 hours and 24 hours after sorting (representative experiment; t-test, p<0.0001, n=3).
Figure S5. p-EMT program is localized at the leading edge, distinct from the epithelial differentiation program at the core, Related to Figure 4.
(A–C) Immunohistochemical staining of representative tumors (MEEI5, MEEI16, MEEI17, MEEI25, MEEI28) for p-EMT (LAMC2, MMP10, TGFBI) with the malignant cell-specific marker p63. Scale bar = 100 μM. The leading edges of tumors co-stain with p63 and p-EMT markers. Additional staining with the marker p-EMT marker ITGA5 further validated localization of p-EMT at the leading edge (data not shown).
(D) Immunohistochemical staining of representative tumors (MEEI17, MEEI28) for multiple p-EMT markers (LAMC2, TGFBI). p-EMT markers co-localize at the leading edge.
(E–G) Immunohistochemical staining of representative p-EMT low tumors (MEEI20, MEEI26) for p-EMT (PDPN, LAMB3, LAMC2) with the malignant cell-specific marker p63. p-EMT low tumors show minimal staining for p-EMT markers at the leading edge. Additional staining with the marker ITGA5 confirmed minimal staining for the p-EMT program in these tumors (data not shown).
(H and I) Immunohistochemical staining of representative tumors (MEEI16, MEEI17) for epithelial differentiation (SPRR1B, CLDN4) and the malignant cell-specific marker p63.
(J and K) Immunohistochemical staining of representative tumor (MEEI17) for p-EMT (LAMC2, PDPN) and epithelial differentiation (CLDN4). Markers demonstrate distinct spatial localization of p-EMT and epithelial differentiation programs, at the leading edge and core, respectively.
(L) Bar plot shows statistical significance (minus log10 of p-value defined by hypergeometric test) of number of observed outgoing interactions between ten listed cell types and malignant cells. Bars above the x-axis indicate a greater number of interactions than expected, while bars below the x-axis indicate fewer interactions than expected.
(M) Immunohistochemical staining of representative tumors (MEEI16, MEEI18) for p-EMT and CAFs (FAP) with the malignant cell-specific marker p63. FAP staining is present both at the leading edge of tumors nests and in the stroma, highlighting activated CAFs.
(N) Bar plot depicts relative proliferation of SCC9 cells treated with vehicle, TGFβ, or TGFβ pathway inhibitors (error bars reflect SEM; ANOVA, p<0.0001, n=4).
(O) Histograms show percent of sequencing reads with insertions or deletions (indels) of specified size in mock infected SCC9 cells (Top left) and SCC9 TGFBI CRISPR knockout cells (other panels). Each of the TGFBI-targeting sgRNAs resulted in >98.8% of reads containing indels, indicating efficient knockout of TGFBI.
(P) Bar plot depicts relative invasiveness of mock infected SCC9 cells or SCC9 TGFBI CRISPR knockout cells after treatment with vehicle or TGFβ in matrigel invasion assays (error bars reflect SEM; ANOVA, p<0.0001, n=3).
(Q) Violin plot depicts hypoxia program scoring of SCC9 cells grown in normoxic or hypoxic conditions. Hypoxic conditions are associated with significantly increased hypoxia score (t-test, p<0.05).
(R) Violin plot depicts scoring of SCC9 cells for p-EMT scores after growth in standard conditions (control), hypoxic conditions, or in co-culture with CAFs derived from MEEI18. p-EMT expression is not significantly changed across these conditions.
Figure S6. Variability in the p-EMT program and cancer-associated fibroblasts across tumor subsites (primary and lymph node), Related to Figure 5.
(A) Comparison of point mutations between primary and LN samples in three individual tumors (MEEI26, MEEI20, and MEEI25 from top to bottom) as detected by whole exome sequencing. In each tumor, we examined all mutations identified in at least one of the samples (primary or LN) and assigned it one of three values in each sample: “detected” (black), “not detected” (white), or unresolved due to “low coverage.” A single mutant read was sufficient to define a mutation as “detected,” but zero mutant reads were defined as “not detected” only if the probability of detecting zero mutant reads in that sample was below 0.05 (as defined by binomial test, given the number of reads covering that base and assuming the same frequency of the mutant reads as in the sample(s) where it is detected). Mutations were then ordered by their identification across the samples and assigned to four classes: shared among primary and LN, specific to primary, specific to LN, and unresolved. Note that for MEEI26 two LN samples are included corresponding to the left (ipsilateral) and right (contralateral) LNs, denoted as LNL and LNR, respectively.
(B) Heatmap of differentially expressed genes between primary and LN samples across multiple patients. For each of the five patients with matched primary and LN samples, we identified significant differentially expressed genes (defined by p<0.001 and fold-change>2). All genes defined as upregulated in at least two patients (left panel) or downregulated in at least two patients (right panel) are shown. Red: upregulated; Blue: downregulated. Darker shades indicate significant differential expression, while lighter shades denote borderline differential expression (p<0.05 and fold-change>1.5).
(C) Violin plot depicts p-EMT score of malignant cells from five primary tumors and matched LN.
(D) Scatter plot shows the average (x-axis) and the variability (y-axis) of p-EMT scores across individual malignant cells within each sample; five primary tumors (black) and matched LNs (red) are included and matched samples are connected with lines. p-EMT high tumors display both higher average and higher variability of p-EMT scores.
(E) Fibroblasts from primary (black) and LN (red) samples, scored by the relative expression of gene-sets distinguishing CAFs from myofibroblasts (x-axis) and those distinguishing the CAF1 and CAF2 subsets (y-axis), demonstrating that LN CAFs are biased towards the CAF1 subset (hypergeometric test, p<0.05).
(F and G) Immunohistochemical staining of representative LN metastases (MEEI25, MEEI28) for p-EMT (PDPN, LAMB3) with the malignant-cell specific marker p63.
Figure S7. p-EMT program is negatively correlated with epithelial differentiation and may predict nodal metastasis, Related to Figures 6 and 7.
(A) Hematoxylin-eosin (H&E) stained sections from representative mesenchymal (Left) and basal (Right) TCGA tumors demonstrate substantially more stromal infiltrate in mesenchymal than basal tumors. Scale bar = 400 μM.
(B) (Left) Bar plot shows significantly higher percent of stromal infiltrate in mesenchymal tumors compared to basal tumors (t-test, p<0.0001; n=203 tumors). (Right) Bar plot shows number of tumors with H&E stromal scores ranging from 0 (lowest) to 4 (highest) for mesenchymal and basal subtype TCGA tumors.
(C and D) Scatter plots demonstrate a correlation between H&E stromal score (indicated by dot color) with CAF and TCGA mesenchymal scores (C), but not p-EMT scores (D).
(E) Line graph shows distribution of p-EMT scores across TCGA tumors of each subtype.
(F) Scatter plot shows scoring of TCGA basal and mesenchymal tumors for epithelial differentiation and p-EMT which are significantly negatively correlated in this subset of tumors (Pearson correlation, p<0.05); black lines indicate linear regression.
(G) Scatter plot shows scoring of TCGA classical and atypical tumors for epithelial differentiation and p-EMT, which are not significantly correlated in this subset of tumors; black lines indicate linear regression.
(H) Bar plot shows direction and statistical significance (p-value based on a t-test) of the association between each of six coherent meta-signatures and the presence of multiple versus no metastatic LNs in TCGA malignant-basal tumors. The p-EMT and epithelial differentiation programs, which were inversely correlated in expression studies, had opposite associations with metastasis. The other programs show no significant association with LN metastases.
(I) (Top) Bar plot shows the percent of patients with adverse clinical features (positive LNs, multiple LNs, advanced N stage, grade III, extranodal extension, lymphovascular invasion, and advanced local disease) in cohorts with high and low p-EMT scores stratified by high and low CAF scores. (Bottom) Heatmap shows the statistical significance of p-EMT and CAF effects on adverse clinical features based on a binomial logistic regression with two predictive variables (p-EMT and variable scores) and an interaction effect. Only the p-EMT effect is predictive of clinical features associated with metastasis and invasion (positive LNs, multiple LNs, advanced nodal stage, extracapsular extension, and lymphovascular invasion) (Bottom, first row). In contrast, the CAF effect has no significant predictive value for features associated with metastasis, but instead, predicts high grade disease and advanced local disease (T3/T4) (Bottom, second row). The p-EMT and CAF effects did act cooperatively to influence the risk of nodal metastasis (Bottom, third row), consistent with a putative ligand-receptor interaction between CAFs and p-EMT cells.
(J) Percent of patients from TCGA for which neck dissection was justified using varying thresholds of p-EMT scores and stratified by tumor (T) stage. Justified neck dissection refers to patients with initial clinical diagnosis of lymph node-negative (cN0) for which neck dissection revealed a positive metastatic lymph node (pN1-N3); the percentage of justified neck dissections was calculated out of all patients with clinical node-negative disease that underwent neck dissection. A higher p-EMT threshold is associated with a higher rate of justified neck dissection, regardless of T-stage (permutation test, p<0.05).
(K) Correlations of genes with the p-EMT program within (x-axis) and across (y-axis) tumors in our cohort of ten patients. Within-tumor correlations were calculated separately in each tumor and averaged; across-tumor correlations were calculated between the average levels of genes and those of the p-EMT program across all malignant cells in each tumor. Selected genes are indicated.
(L) Scatter plot shows the correlations of genes with p-EMT (x-axis) and epithelial differentiation (y-axis) programs based on inferred malignant cell-specific profiles from TCGA malignant-basal tumors. Genes of the p-EMT (red) and epithelial differentiation (green) programs as well as EMT TFs (black) are indicated, demonstrating a high p-EMT correlation with SNAIL2 but not of other EMT TFs.
Table S1. Patients and samples included in dataset, Related to Figure 1.
Table S2. Clinical and pathologic features of deeply sequenced samples, Related to Figure 1. p16 immunohistochemistry and HPV in situ hybridization were negative for all samples.
Table S3. Mutations and copy number variations detected in profiled primary tumors, Related to Figure 1. Common mutations evaluated by whole exome sequencing of a subset of samples and SNaPshot next generation sequencing assay of all samples include the top 5 mutations in TCGA HNSCC tumors, as well as mutations in TERT promoter. CNVs evaluated include top 4 abnormalities noted in TCGA HNSCC tumors.
Table S4. Mutations detected by whole exome sequencing, Related to Figure 1. Mutations are sorted by patient number, within patient by primary tumor followed by lymph node, and within sample by location within the genome.
Table S5. Differentially expressed genes between CAF subsets, Related to Figure 2. Genes are sorted from most to least significant.
Table S6. Expression programs detected by NNMF in each of 10 patients, Related to Figure 3. Clusters are ordered as in Figure 3B, and within each cluster the genes are ordered from most to least significant. For each cluster, headers also indicate the patient from which it was derived and an inferred annotation. See also online tables.
Table S7. Six meta-signatures, each derived from multiple related NNMF programs, Related to Figure 3. Genes in each program are ordered from most to least significant.