Abstract
Actin networks are adaptive materials enabling dynamic and static functions of living cells. A central element for tuning their underlying structural and mechanical properties is the ability to reversibly connect, i.e., transiently crosslink, filaments within the networks. Natural crosslinkers, however, vary across many parameters. Therefore, systematically studying the impact of their fundamental properties like size and binding strength is unfeasible since their structural parameters cannot be independently tuned. Herein, this problem is circumvented by employing a modular strategy to construct purely synthetic actin crosslinkers from DNA and peptides. These crosslinkers mimic both intuitive and noncanonical mechanical properties of their natural counterparts. By isolating binding affinity as the primary control parameter, effects on structural and dynamic behaviors of actin networks are characterized. A concentration-dependent triphasic behavior arises from both strong and weak crosslinkers due to emergent structural polymorphism. Beyond a certain threshold, strong binding leads to a nonmonotonic elastic pulse, which is a consequence of self-destruction of the mechanical structure of the underlying network. The modular design also facilitates an orthogonal regulatory mechanism based on enzymatic cleaving. This approach can be used to guide the rational design of further biomimetic components for programmable modulation of the properties of biomaterials and cells.
Keywords: biomimetic materials, biopolymers, crosslinkers, DNA nanotechnology, molecular design
Living cells exhibit an elaborate mixture of both dynamic and stabile material behaviors that enable them to rapidly adapt to complex environments.[1] The filamentous, cytoskeletal biopolymer actin is at the heart of this ability, simultaneously forming load-bearing networks, mechanosensing protrusions, and motility-inducing ratchets.[1–3] Actin-associated proteins that form transient, physical crosslinks between filaments have been implicated in nearly every actin-dependent process and their role in directing structural morphology, mechanical stasis, and active behaviors has been well documented.[1] In contrast to permanent chemical crosslinks in classical polymer systems, these biological crosslinks have a far broader parameter space defined by the molecular details of their binding and connective regions. Factors such as on/off-rates, size, flexibility, orientation, and biochemical switches allow these components to exert control over the material properties of both networks and cells. This goes beyond simply imposing local affine responses against external deformations. Overall, these factors render actin-based structures highly adaptive.[2,4–7]
The resulting abundance of structural phases in soft, polymeric materials has been captured in a theoretical framework built upon the classical Onsager approach describing isotropic to nematic phase transitions in rigid rods, where transient crosslinks are modeled as distance-dependent interrod attractive potentials.[8] In contrast to the established triphasic (isotropic, nematic, and coexistence) character for noninteracting rods, the addition of nonpermanent crosslinks revealed a highly complex, polyphasic behavior dependent upon the energetic preference for parallel or perpendicular orientation between linked filaments.[8] A complex mixture of states more closely resembling both experimental observations and biological structures has been predicted in analytical and simulation-based studies. Here, stress fiber-like bundles and regular, cubatic networks coexist with the previously known isotropic and nematic phases.[8–10]
Still, a critical discrepancy remains; this coexistence of phases favored by parallel and perpendicular binding preferences should only occur in an unphysiologically narrow range.[8] However, the simultaneous appearance of bundles and regular networks are ubiquitous in mixtures of biopolymers such as actin and the naturally occurring transient crosslinker α-actinin, possibly due to the low binding affinity of the binding interaction.[8,11–13] It is likely that the molecular and physical details of the biological constructs enable the structural polymorphism of actin that is of crucial importance to the mechanical behaviors of cells. However, a critical limitation arises when attempting to systematically investigate the details of such biological constructs in order to develop biomimetic or bioinspired materials: using different native biological crosslinkers to explore the phase space of one parameter (e.g., using α-actinin and fascin to vary binding strength) is unavoidably coupled with unwanted variations of several other parameters.[14,15] Genetic mutations can be used to perturb crosslinker properties such as binding kinetics or size;[16–18] however, their empirical nature and complexity inhibits systematic studies where different key parameters can be modified in a completely decoupled way.
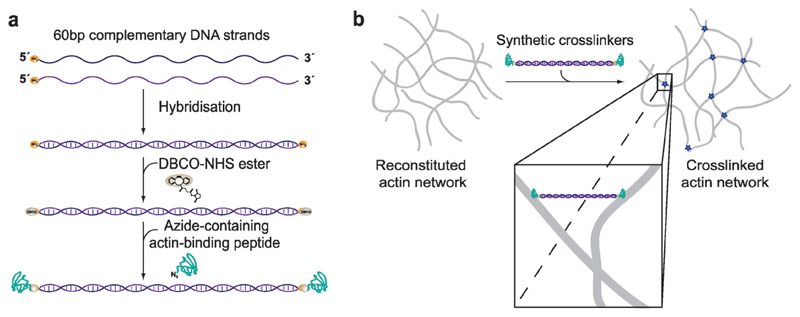
Here, we resolve this natural limitation by introducing synthetic actin crosslinkers based on a well-defined DNA template. The core building block is a segment of double-stranded DNA (60 bases ≙ length of 20 nm), which can be covalently attached to actin binding peptides via copper-free click chemistry (Figure 1a; see the Experimental Section).[19] This approach allows us to conserve the basic geometry of the crosslinkers while independently altering the affinity of their binding domains to actin filaments, in order to examine their impact on structural and bulk mechanical properties. We used this plug and play platform to conjugate either the linear, 17-residue actin binding peptide commercially known as LifeAct (KD (F-Actin) ≈ 2.3 ± 0.9 × 10−6 M)[20] or the bicyclic heptapeptide phalloidin (KD (F-Actin) ≈ 9 ± 2 × 10−9 M)[21] to both ends of the DNA template to mimic weak or strong binding, respectively (see also Figure S1 in the Supporting Information).[20,21] For simplicity, we henceforth denote the weak LifeAct-based and strong phalloidin-based crosslinking constructs as wLX and sPX. To quantitatively evaluate emergent effects of these two crosslinkers, we employed dynamic shear rheology to test a broad range of crosslinker concentrations (ratio R = ccrosslinker/cactin) in viscoelastic actin filament networks (see the Experimental Section). These biomimetic crosslinkers reconstruct many of the trivial morphological and rheological behaviors consistent with actin networks that are crosslinked by native proteins. Moreover, we found that imposing a strong binding energy leads to the time-dependent build-up and subsequent decay of prestress within the network, a striking signature of soft glassy materials previously seen in actin networks tightly bundled by fascin.[14,22] Finally, we investigated a regulatory mechanism based on the enzymatic severing of the core template in order to function as an orthogonal “off-switch” for the crosslinking activity.
Figure 1. Constructing synthetic actin crosslinkers.
a) A copper-free click chemistry approach[19] was used to covalently attach two actin-binding peptides to double-stranded DNA. After hybridization of the complementary strands, dibenzocyclooctyne-NHS ester covalently reacted with primary amine groups on each end of the double-stranded DNA spacer. Subsequently, an azide-containing actin binding peptide formed a covalent bond with the previously attached DBCO group. b) Since the two binding domains were connected by double-stranded DNA, the construct can physically crosslink actin filaments within the system.
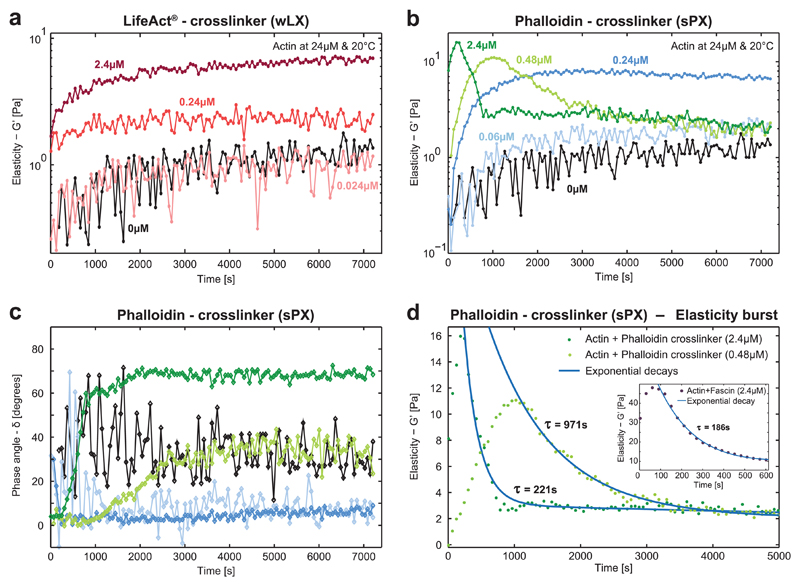
The elasticity (G′) of polymerizing actin solutions increased monotonically over time, which is the expected signature of elongating actin filaments arranging into an isotropic, entangled network. Once the steady state of actin polymerization was reached, the elasticity remained stable (Figure 2a). These qualitative characteristics were conserved when including the weakly binding wLX in the polymerizing actin networks, although they were superimposed by additional crosslink-dependent effects. The overall elasticity displayed a notable sixfold increase compared to pure actin for higher wLX concentrations of 2.4 × 10−6 m (R = 0.1). This stiffening illustrates the effect of physically, albeit transiently connecting individual filaments, which has been reported previously for the weakly binding natural crosslinker α-actinin.[14,23] For the high-affinity sPX, a similar behavior was observed for comparatively lower concentrations. In this case, a sixfold increase of G′ has been measured at a molar ratio of R = 0.01, or ≈10% of the concentration of wLX required to reach a similar magnitude of elasticity (Figure 2b—blue curves). In comparison to pure, viscoelastic actin networks, which display a phase angle δ around 40° (with δ being defined as the arctangent of the ratio G″/G′, where G″ and G′ are the frequency-dependent viscous and elastic shear moduli, respectively, with 0° being a purely elastic (solid-like) response and 90° being a purely viscous (i.e., Newtonian fluid) response), the phase angle for both types of crosslinked networks was shifted to lower values, indicating their increasing elastic response (Figure 2c—blue curves). Once again, equivalent behavior can be found in the natural analog, where these signatures of gelation have been reported for similarly low ratios of the high-affinity natural crosslinker human fascin.[14] For these cases, the time evolution of the network mechanics is mainly dominated by the ongoing filament and network formation.
Figure 2. Time evolution of the elasticity during actin polymerization in the presence of synthetic actin crosslinkers.
a) Mechanical properties of actin networks (24 × 10−6 m) enriched with different concentrations (R = 0, 0.001, 0.01, 0.1) of the wLX were determined via dynamic shear rheology. Measurements always started with monomeric actin and the monotonically increasing G′ of pure actin (black) illustrates the formation of filaments and their arrangements into entangled networks. Upon addition of the crosslinker, the elasticity increased in a concentration-dependent manner similar to the natural crosslinker α-actinin (corresponding crosslinker concentrations are given in different shades of red). b) Strong phalloidin crosslinkers induced entirely different mechanical fingerprints comparable to findings reported for the strong natural crosslinker fascin.[14,22] Networks are initially monotonically stiffened similar to wLX, but at much lower concentrations (R = 0, 0.0025, 0.01, 0.02, 0.1). When reaching a concentration threshold, the time evolution of G′ became nonmonotonic. First, the elasticity drastically increased until reaching a peak, which was subsequently followed by an exponential decay—a time evolution that is referred to as the “elasticity burst.”[14] c) The corresponding phase angles illustrate that low crosslinker concentrations (blue curves) induced a predominantly elastic response. High crosslinker densities initially induced a predominant elastic response (green curves), but in the time evolution the systems became increasingly viscous. d) The higher sPX concentration (dark green) induced the elasticity burst earlier with a higher magnitude of G′ with a fast subsequent relaxation indicated by the shorter decay time. Inset: Similar characteristic fingerprints have been observed in actin networks with a high concentration of the natural crosslinker fascin.[14]
Most systems reached a steady state at ≈2000 s after initializing polymerization (Figure 2a,b—blue curves, as well as Figure S5 in the Supporting Information) and remained stable until the actin started to denature after ≈1 d (data not shown). Actin networks enriched with the highest wLX concentration used, however, did not reach this steady state after 2000 s presumably due to the high number of crosslinkers retarding the underlying glassy dynamics, which drive the system toward equilibrium.
However, this mechanical fingerprint drastically changed at higher sPX concentrations. When reaching a threshold of R values between 0.01 and 0.02, the time evolution of G′ became nonmonotonic, marked by an initial “burst” that relaxed to a viscous dominated state (Figure 2b—green curves). The nature of this elastic pulse was further revealed through the concurrent evolution of the phase angle δ over time (Figure 2c—green curves). Before the peak of G′ was reached, δ remained stably low and the system exhibited a predominantly elastic response. In the subsequent regime of decaying G′, δ monotonically increased before reaching a stable plateau, corresponding to the emergence of an increasingly viscoelastic response. Interestingly, for the highest sPX concentration (Figure 2c—dark green curve) δ reached significantly higher values than for solutions of purely entangled actin filaments (Figure 2c—black curve), and the system predominantly behaved as a viscous fluid.
This dynamic pulse behavior closely resembles previously reported findings as well as our own measurements for the natural, strongly binding actin crosslinker fascin (Figure S7, Supporting Information).[14,22] Simultaneous polymerization and strong crosslinking have been shown to generate prestressed structures that are bent beyond their equilibrium configuration[22], causing the forming actin networks to experience a corresponding emergence and increase of internal stress.[22,24] This build up occurs because the typical filament elongation rates (≈10 μM−1 s−1)[25] far exceed the off-rates of the actin-binding domains (0.12 s−1 for fascin[26] and 0.00026 s−1 for phalloidin[21]). The off-rate is the primary kinetic mechanism responsible for relaxation of sterically trapped nonequilibrium configurations. The subsequent relaxation is likely induced by glassy dynamics aiming to release the stress toward equilibrium[22] and the breaking of filaments subjected to significant bending forces.[27] The stochastic unbinding of a crosslink from a filament is followed by its subsequent rebinding in a more relaxed configuration. Consequently, the network contains less internal stress driving the decay.[11] In turn, this decay gradually decreases the unbinding probability with increasing network age.[11,22] This suggested deceleration in the change of the network’s elasticity is supported by the exponentially decaying relaxation behavior shown in Figure 2b,d.
However, the increasingly dominant fluidization observed from the phase angle at the high-sPX limit cannot be solely explained by relaxation of prestress, but likely also reflects mesoscale structural evolution within the network. We hypothesize that short, oligomeric actin filaments formed at the onset of polymerization are almost instantaneously crosslinked to other such protofilaments in their entropically favored, parallel bundled configuration. These bundles grow due to actin polymerization, while simultaneously depleting the surrounding monomer pool. These simultaneous processes can even be enhanced by the concentration-dependent tendency of crosslinkers to suppress actin depolymerization kinetics.[28] Initially, this suppression leads to the observed burst of G′ as the rapidly elongating bundles merge due to high crosslink concentrations. The growing structures become percolated throughout the network and are sterically trapped in prestressed configurations.[22,24] Eventually, the pool of actin monomers is depleted to its steady-state concentration, thus halting further elongation of the constituent filaments. Beyond a critical turning point, bundles become increasingly disconnected from the rest of the network since the energetic preference of strong crosslinkers promotes the binding of parallel pairs of filaments.[9,10] This is possibly enhanced by the breakage of filaments under bending and torsional stress.[27] The resulting depercolation of the system resulted in the observed drop in G′ as well as the simultaneous increase in the loss modulus G″ (and accordingly δ) shown in Figure 2c. Previously published Monte Carlo simulations support this possible transition and suggested that bundle-dominated networks contain a larger number of crosslinkers forming bridges between parallel filaments within a single bundle, and fewer contributing to overall network percolation as the degree of bundling becomes more pronounced.[10]
We hypothesize that the peak of the elasticity burst is reached faster for the highest sPX concentration (R = 0.1) due to enhanced depletion of the monomer pool[28] and trapping of prestressed structures during polymerization. The characteristic decay time τ significantly increased for lower sPX concentrations and this nonmonotonic effect disappeared altogether for sPX concentrations below R = 0.01 (Figure 2d). The measured decay times are comparable to a previous study on fascin employing the same crosslinker/actin concentrations as in our investigations here (Figure 2d, inset).[14] In contrast, the lower actin concentrations utilized by Lieleg et al. resulted in a much longer decay time τ for fascin-induced elasticity bursts (Table S3, Supporting Information),[22] likely due to combined effects of (a) smaller bundles inducing a lower collective restoring force[29,30] and (b) fewer bundles driving depercolation during the relaxation process. Similar to actin/fascin networks,[14,22] G′ in the final steady-state is lower in cases where an elasticity burst occurred compared to the cases where no burst was observed. Since we were using the same basic geometries for our different synthetic crosslinkers, the induced elasticity burst appears to be a general feature of crosslinkers with low off-rates and is seemingly independent of other parameters such as their length or structure.
This pulse-like nature of the initial elasticity burst effectively acts as a reporter signal of the material’s molecular constituents. Rather than imposing inherent, static material properties like strain hardening or fixed shear moduli, this burst effectively acts as a rudimentary biohybrid signal processor, where both the amplitude and width/decay time of the pulse are determined by the biopolymer and crosslinker densities (Table S3, Supporting Information).
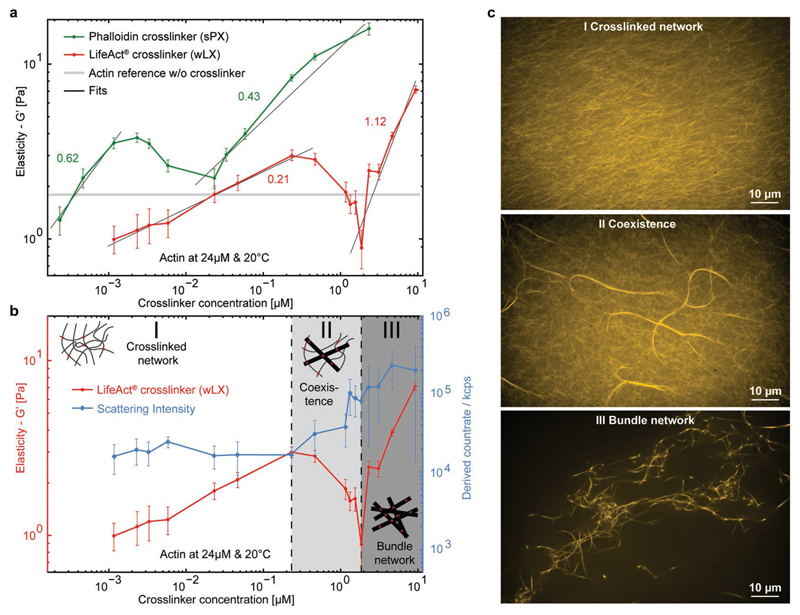
These findings already suggest that the formation of different morphologies such as networks and/or bundles leads to nontrivial mechanical characteristics that highly depend on the crosslinker type and concentration. For both sPX and wLX, we found a qualitatively similar, nonmonotonic, concentration-dependent behavior for the network elasticity (Figure 3a). For the lowest respective concentrations, G′ was slightly lower than compared to purely entangled actin networks. In this regime, each network was not percolated, but crosslinkers already induced local heterogeneities that triggered a global softening.[5,31] With increasing concentrations, the networks were progressively more percolated leading to a rising elasticity as illustrated by the positive slopes in Figure 3a. However, comparing these slopes for the different crosslinker binding affinities reveals that sPX stiffened the networks much more efficiently than wLX, displaying a power law exponent of 0.62 compared to 0.21 for the weaker case.
Figure 3. Synthetic crosslinkers nonmonotonically influence the elasticity of actin networks.
a) Elastic moduli of actin networks were investigated for different concentrations of wLX (red) and sPX (green) crosslinkers. Both initially stiffened the networks with increasing concentration before reaching a peak, which was followed by a decrease of G′ to a minimum with a subsequent monotonic increase (power-law fits (black line) and exponents are shown next to the corresponding G′ increase). This nonmonotonic behavior can be attributed to different structural morphologies, which were captured by b) light scattering (blue; error bars indicate standard deviations of the mean values) and c) fluorescence microscopy.
Interestingly, both cases exhibited a local maximum of G′, which subsequently decreased to a minimum and rose again with a different exponential behavior for the highest concentrations. Although the onset of the different regimes varied, the qualitative curve progression for both binding strengths resembles one another in appearance. Generally, this nonmonotonic fingerprint can be attributed to different structural morphologies induced at the corresponding crosslinker concentrations.[5] In the following explanation, the observed triphasic behavior is described for networks containing wLX; however, we expect that the characteristics will be generally valid for other synthetic and natural actin crosslinkers, as also indicated by prior findings.[5,15]
In the isotropic regime (I) occurring at low wLX concentrations (R ≤ 0.01), actin filaments were arranged in a weakly crosslinked network. With increasing wLX concentrations, networks remained mostly isotropic, as illustrated by the constant light scattering intensity (Figure 3b—blue curve) and visually shown in Figure 3c. Above a threshold concentration, the networks entered a coexistence regime (II), where higher wLX densities increasingly favored alignment of filaments. This was marked by the coexistence of bundles within an isotropic background network and a concentration-dependent decrease of G′. This new structural arrangement was captured by the sudden change of the light scattering profile at the onset of the elasticity decrease (Figure 3b) and was directly observable with fluorescence microscopy (Figure 3c). Within this regime (0.01 < R ≤ 0.08), bundles can be considered as local anisotropies. Thus, the effective actin concentration in the percolated background network was reduced, which weakened the overall structure and resulted in the observed decrease of G′. A previous study based on Monte Carlo simulations emphasized that bundled configurations incorporate more crosslinkers than their isotropic counterparts, suggesting that this increasing anisotropy comes at the cost of overall network percolation.[10] This combination of both reducing the effective actin concentration and concurrently reducing percolation in the background network accounts for the drastic decrease of G′ to values even below those found for much smaller crosslinker concentrations.[10] Finally, at high wLX ratios (R > 0.8), the elastic response was dominated by a network of percolated bundles (III). Here, higher crosslink densities continue to favor an increasing prevalence of bundled structures (Figure 3c), which is also illustrated by the rising light scattering intensity. Accordingly, the elasticity increased monotonically. This qualitative structural trend can also be seen in networks containing sPX, although light scattering analysis was limited due to the time-dependent evolution of structural anisotropies. However, clear differences became apparent when inspecting the sample with fluorescence microscopy (see Figure S8 and Movies S1–S3 in the Supporting Information). In addition to the apparent visual differences, the fluctuations of the formed bundles decreased with increasing sPX concentration as a direct result of their thicker, more tightly packed structure.[30,32]
This structural polymorphism has been previously observed for natural crosslinkers[5,33] and is supported by both coarse-grained molecular dynamics[8,34] and Monte Carlo[9,10] simulations. In the study by Borukhov et al., modeling crosslinkers as an effective rod–rod attraction within a variant of the classical Onsager theory revealed a rich phase space including networks, bundles, and their coexistence.[8] Interestingly, the measured slopes for increasing G′ vary differently between regimes I and III for the two crosslinkers. The stronger actin-binding affinity of sPX induced a roughly threefold steeper increase in regime I than the comparatively weak interaction of wLX. This was likely caused by the longer binding times and thus more efficient percolation of the network. However, the trend was inverted in regime III: wLX induced a roughly 2.6-fold higher concentration-dependent increase in G′ than sPX. Since unbinding kinetics of phalloidin are much slower compared to LifeAct, we expect that sPX have an energetic preference to be stored in multi-filament bundles. Thus, fewer crosslinkers contribute to the percolation of the system, as is consistent with previous simulations.[10] Effectively, this tendency implies that strong-binding crosslinkers generally have a lower “percolation efficiency” in the presence of bundles than their weakly binding counterparts, accounting for the steeper concentration-dependent increase of G′ for wLX in regime III.
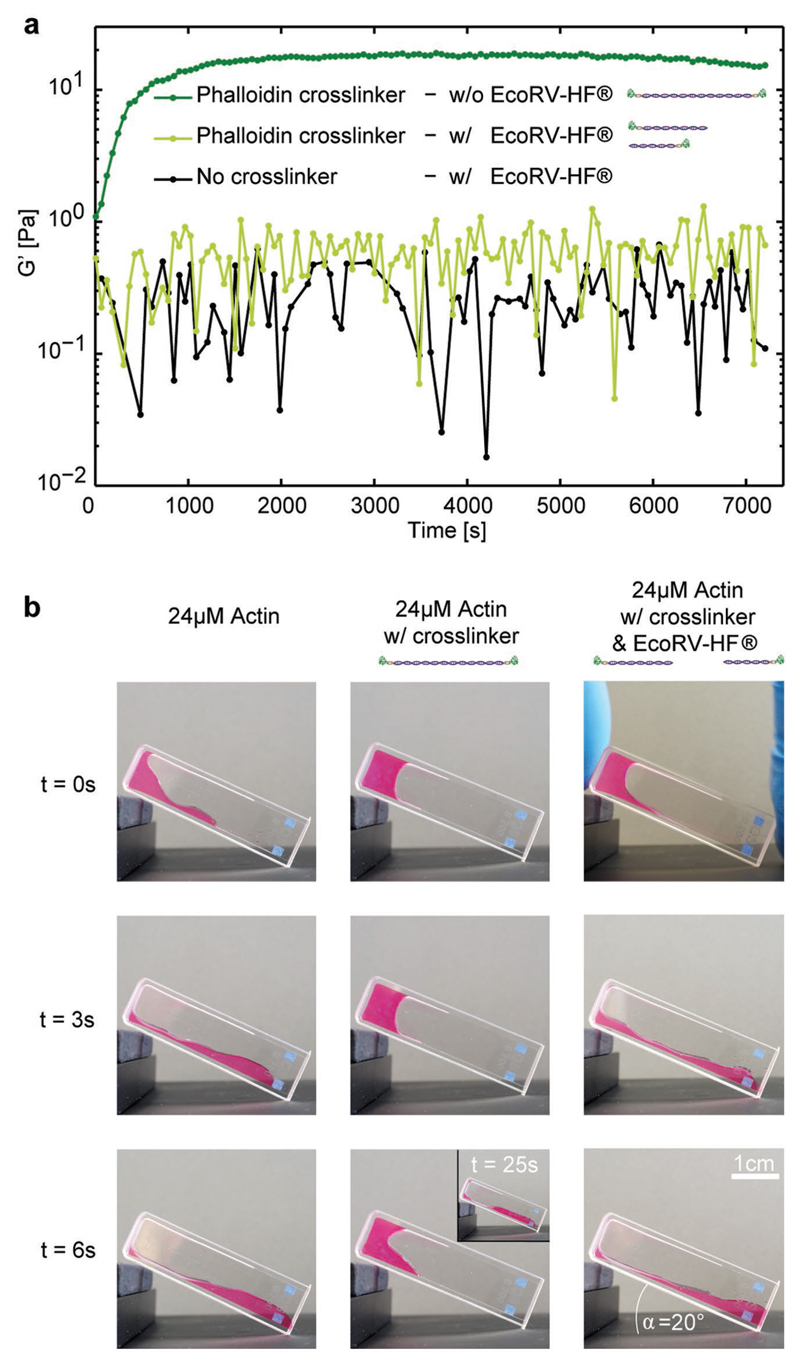
In addition to mimicking natural mechanical signatures, our modular, DNA-based design also enables the integration of rudimentary regulatory mechanisms in the form of orthogonal state switches. Here, the sequence for the DNA connector included a recognition site for the EcoRV-HF restriction enzyme so that the crosslinker could be cleaved into two segments, thereby effectively “breaking” the link between filaments (see the Experimental Section and Figure S2, Supporting Information). Incubating crosslinkers with EcoRV-HF prior to measurement resulted in a drastic decrease of the elastic modulus to values close to the actin-only control (Figure 4). Marginal differences are well within the sample-to-sample variation, likely resulting from small numbers of undigested crosslinkers (Figure S2B, Supporting Information).
Figure 4. Reversibility of synthetic actin crosslinkers.
a) The time-dependent elasticity of actin polymerization was investigated in the presence of intact sPX (dark green, c = 0.24 × 10−6 m (R = 0.01), i.e., below the threshold of the elasticity burst) and compared to EcoRV-HF-cleaved sPX (light green, c = 0.24 × 10−6 m (R = 0.01)). Cleaving the crosslinkers reversed the stiffening effect illustrating the possibility to switch between different mechanical states. The cleavage was additionally verified via native polyacrylamide gel electrophoresis (Figure S2, Supporting Information) and has also b) a drastic effect on the macroscopic behavior of actin as shown in an inclined cuvette monitoring the bulk properties over time. Left row: Pure actin was polymerized at 24 × 10−6 m. Middle row: Actin was polymerized at 24 × 10−6 m with 9.6 × 10−6 m sPX. Right row: Actin was polymerized at 24 × 10−6 m, including 9.6 × 10−6 m sPX and 300 units EcoRV-HF.
The mechanical impact of crosslink-induced stiffening and enzyme-based regulation can also be seen in macroscopic gels. A purely entangled, viscoelastic solution of actin filaments flowed down a gradient within seconds (Figure 4b, left column) while the addition of crosslinks amplified the elastic behavior, slowing the flow by more than fivefold (Figure 4b, center). Enzymatically severing the crosslinks restored the dominance of the fluid-like contribution, once again inducing flow effectively indistinguishable from the actin-only solution (Figure 4b, right).
The effects of the synthetic crosslinkers described here are striking examples of how soft, biopolymer-based materials can be fundamentally altered by rationally designed molecular components. To our knowledge, these constructs are the first example of purely synthetic actin crosslinkers and facilitated a systematic study into the global impact of their binding affinity when it is decoupled from other structural parameters. Varying the interaction strength and concentration of the crosslinkers enabled us to control the actin network elasticity by over two orders of magnitude. Remarkably, the two different synthetic crosslinkers closely reconstruct the mechanical fingerprints of analogous natural crosslinkers such as α-actinin and fascin. Similar to fascin, high concentrations of the strongly binding, phalloidin-based crosslinker sPX induced a nonmonotonic time evolution and out-of-equilibrium dynamics (elastic pulse) during the formation of the actin networks.[22,24] Since the natural strong crosslinker fascin and its synthetic analog here are only similar in their high binding affinity to actin, this special time evolution seems to predominantly depend on binding strength and concentration. While this relaxation reported here is effectively an irreversible transition due to the loss of mechanical structure, the molecular constituents in the system—namely, actin proteins and DNA-peptide crosslinkers—are themselves likely unharmed by the nonchemical nature of the decay. Therefore, this mechanism points toward a possible strategy for engineering soft, mechanical transducers, based on the recyclable, self-destruction of network’s underlying mesostructure.
Additionally, different concentrations of both crosslinkers induced three distinct regimes for structural morphology and mechanical behavior, which have also been reported for natural crosslinkers[5] and hypothesized in analytical and simulation approaches.[8–10,34] Ultimately, the crosslinker/filament ratio and strength of the actin-binding domains impact the degree of bundling and percolation within the system resulting in the observed nonmonotonic concentration dependency of G′.
Beyond simply reconstructing behaviors arising from natural actin crosslinking proteins, this programmable toolbox gives an entry point for fabricating noncanonical modulators of biohybrid materials that are not constrained by typical biological function. This approach is not only limited to alterations of structural or physical parameters such as the length of the crosslinker, heterogeneous affinities of the binding sites on either end, or the number of available binding sites, but also enables the integration of additional components for stimulus-response mechanisms. While we showed this effect in a very simple form through the sequence-specific enzymatic severing of the crosslinker, more complex components such as aptamer-based logic gating, light or pH-triggered conformation switches, complex transcription circuits, or more can be implemented to convert a specific stimulus into a distinct structural or mechanical signature.[35–38] Such techniques potentially serve as a foundation for strategies to design signal-responsive, programmable, smart biomaterials for cell-based or dynamic sensing applications or as intracellular tools to synthetically model the impact of crosslink mutation in disease pathology.[17]
Experimental Section
Actin Preparation
Globular actin was prepared from rabbit muscle as described previously.[39] Actin was polymerized by the addition of 8.75 μL 20x F-buffer at a final concentration of 24 × 10−6 m and a volume of 175 μL.
Synthetic Actin Crosslinkers
Lyophilized oligonucleotides (Table S1, Supporting Information; biomers.net GmbH, Germany) were resuspended in Millipore water and concentrations were spectrophotometrically determined by a NanoDrop 1000 (Thermo Fisher Scientific Inc., USA). Complementary oligonucleotides were hybridized in 100 × 10−3 m KH2PO4 pH 7.2 at a final concentration of 25 × 10−3 m in a thermocycler (denaturation for 10 min at 95 °C; complementary base pairing for 15 min at 71.6 °C; quick drop to 4 °C; TProfessional Standard PCR Thermocycler, Core Life Sciences Inc., USA). Dibenzocyclooctyne-N-hydroxysuccinimidyl ester (DBCO-NHS ester) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 × 10−3 m, added in a 100-fold molar excess to previously hybridized DNA, and incubated over night at room temperature. DBCO-functionalized dsDNA was purified via ethanol precipitation. To introduce an azide group, amino-phalloidin (((R)-4-hydroxy-4-methyl-Orn7)-Phalloidin; Bachem, Germany) was prefunctionalized with azidopropionic acid sulfo-NHS ester (Jena Bioscience, Germany). Therefore, azidopropionic acid sulfo-NHS ester was dissolved in DMSO at a concentration of 10 × 10−3 m, mixed in equal molar amounts with amino-phalloidin in 100 × 10−3 m KH2PO4 pH 7.2, and incubated over night at room temperature. No further purifications were performed. Azide-containing peptides (Tab. S2; LifeAct, Peptide Specialty Laboratories GmbH, Germany; prefunctionalized Azide-Phalloidin) were added in a 50–100-fold molar excess to DBCO-DNA and incubated over night at room temperature. To remove excess peptides, samples were Amicon filtered (Amicon Ultra-4, PLGC Ultracel-PL Membrane, 10 kDa, Merck Millipore, Germany) in 1x Dulbecco’s phosphate-buffered saline (DPBS, w/o calcium, w/o magnesium; Thermo Fisher Scientific Inc., USA). The successful synthesis and purity of the samples was verified with a native polyacrylamide gel (PAGE) (Figure S1, Supporting Information). Synthetic crosslinkers were stored at −20 °C with no detectable degradation. They were added at different concentrations ranging from 0.24 × 10−9 m to 9.6 × 10−6 m (R = 0.0001 to 0.4).
Since it is known that phalloidin itself influences actin filaments by doubling their persistence length,[40] it was verified that corresponding effects were insignificant compared to crosslinking effects (Figure S3, Supporting Information).[44–47] This comparison illustrates that the physical connection between the binding domains is the crucial property necessary to induce the observed dynamics and the rich structural/mechanical phase space.
EcoRV-HF-Digestion of sPX
To investigate the necessity of two actin binding domains on one crosslinker for the stiffening of reconstituted actin networks, bulk rheology measurements were performed on actin with functional, undigested, as well as EcoRV-HF-digested sPX (incubated for 1 h at 37 °C). Crosslinker DNA was designed with an EcoRV digestion site almost in the middle of the double strand. A digestion resulted in a 32 bp and a 28 bp fragment. Prior to rheology measurements, 1624 ng sPX (final concentration of 0.24 × 10−6 m in 175 μL final sample volume; R = 0.01) were incubated with 5 μL of EcoRV-HF (20 000 units per milliliter, New England Biolabs Inc., USA) and 1.75 μL of 10x CutSmart buffer in a final volume of 20 μL for 1 h at 37 °C. No inactivation of the enzyme was performed. Corresponding PAGE analysis showed a digestion rate of at least 80% after 1 h of incubation (Figure S2, Supporting Information). Additionally, double stranded control DNA was incubated without an EcoRV digestion site. No detectable digestion of control DNA was observed due to sequence specificity of the enzyme (data not shown). After EcoRV-HF-incubation, the digested sample was directly added to actin, 10x G-buffer and 20x KME-buffer to a final volume of 175 μL. Thereby, actin polymerization was induced and rheological measurements were performed.
Rheology
175 μL of sample was loaded to the dynamic shear rheometer (ARES, TA Instruments, USA) equipped with a cone (diameter 25 mm, 0.04 rad). The network between cone and plate was surrounded with a 2.5 mL 1x F-buffer bath similarly as described previously[41,42] to avoid direct contact of the sample with air. The sample chamber was sealed with a cap equipped with wet sponges to suppress evaporation. Measurements were performed at 20 °C and followed the sequence: (i) The time evolution of the polymerization was monitored for 2 h (one data point per minute; γ = 5%; f = 1 Hz), which was subsequently followed by a (ii) short f sweep (γ = 5%; f = 0.01–30 Hz; five data points per decade), (iii) long f sweep (γ = 5%; f = 0.001–30 Hz; 21 data points per decade), (iv) short f sweep, (v) γ sweep (f = 1 Hz; γ = 0.0125–100%; 20 data points per decade), (vi) short f sweep, and (vii) γ sweep. The sequence has been designed to test the robustness of the system over time and the frequency of 1 Hz has been chosen to display the values for G0.
Light Scattering
Static light scattering (Malvern Instruments Ltd., Zetasizer Nano ZSP, UK) was used to observe the dependence of actin morphology[14,43] on synthetic crosslinker concentrations. The final actin concentration was 24 × 10−6 m. The crosslinker concentration ranged from 0 × 10−6 m to 9.6 × 10−6 m (R = 0–0.4). The scattering of the sample was measured every minute for 1.5 h and scattering intensities were arithmetically averaged after 30 min of equilibration.
Spinning Disk Confocal Microscopy
For visualization, monomeric actin was mixed at a molar ratio of 3:1 with phalloidin tetramethylrhodamine isothiocyanate (phalloidin-TRITC) purchased from Sigma-Aldrich. Synthetic crosslinkers with two LifeAct binding domains (wLX) were added to yield a final R = 0.333, R = 0.01, and R = 0.001 or with two phalloidin binding domains (sPX) to yield a final R = 1, R = 0.1, and R = 0.01, respectively. Polymerization was initialized by increasing the salt concentration to 1x F-buffer conditions after mixing all components with a final actin concentration of 3 × 10−6 m. Immediately after starting the polymerization process, the premixed solution was deposited into a sample chamber as described previously.[11] Measurements were performed on a spinning disc confocal microscope (inverted Axio Observer.Z1/Yokogawa CSU-X1A 5000 (Carl Zeiss Microscopy GmbH, Germany), 100x oil immersion objective (Plan-Apochromat 100x/1.40 Oil DIC M27)) and recorded with a Hamamatsu camera at an exposure time of 50 ms.
Macroscopic Behavior of Actin in an Inclined Cuvette
Three different samples were prepared similar to as described above and 150 μL of final sample solutions was pipetted into a small cuvette (first sample: 24 × 10−6 m actin; second sample: 24 × 10−6 m actin with synthetic crosslinkers featuring two phalloidin binding domains (sPX) at R = 0.4; third sample: 24 × 10−6 m actin with sPX at R = 0.4, which were incubated for 1 h at 37 °C with 300 units of EcoRV-HF (New England Biolabs Inc., USA)) prior to the experiments. A small amount of free Phalloidin-TRITC was added to dye the solution for a better contrast and visibility. After 40 min resting time, the cuvette with the respective sample was placed on an inclined plane with a 20° angle. Images were recorded every second using a commercially available digital camera.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
J.S.L. and J.S. contributed equally to this work. The authors would like to thank Klaus Kroy, Manlio Tassieri, and Aftab Taiyab for fruitful discussions as well as Sebastian Schmidt, Patricia Warne, and Emilia Wisotzki for proofreading this manuscript. The authors gratefully acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG-1116/17-1), the European Research Council (ERC-741350) and the Fraunhofer Attract project 601 683.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adma.201706092.
Contributor Information
Dr. Jessica S. Lorenz, Fraunhofer Institute for Cell Therapy and Immunology (IZI) 04103 Leipzig, Germany
Martin Sajfutdinow, Fraunhofer Institute for Cell Therapy and Immunology (IZI) 04103 Leipzig, Germany.
Carsten Schuldt, Fraunhofer Institute for Cell Therapy and Immunology (IZI) 04103 Leipzig, Germany; Peter Debye Institute for Soft Matter Physics Leipzig University, 04103 Leipzig, Germany.
Prof Josef A. Käs, Peter Debye Institute for Soft Matter Physics Leipzig University, 04103 Leipzig, Germany.
Dr David M. Smith, Fraunhofer Institute for Cell Therapy and Immunology (IZI) 04103 Leipzig, Germany.
References
- [1].Huber F, Schnauss J, Ronicke S, Rauch P, Muller K, Futterer C, Kas J. Adv Phys. 2013;62:1. doi: 10.1080/00018732.2013.771509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wachsstock DH, Schwarz WH, Pollard TD. Biophys J. 1994;66:801. doi: 10.1016/s0006-3495(94)80856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Physiol Rev. 2014;94:235. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- [4].Lieleg O, Schmoller KM, Claessens MMAE, Bausch AR. Biophys J. 2009;96:4725. doi: 10.1016/j.bpj.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lieleg O, Claessens MMAE, Bausch AR. Soft Matter. 2010;6:218. [Google Scholar]
- [6].Gardel ML, Shin JH, MacKintosh FC, Mahadevan L, Matsudaira P, Weitz DA. Science. 2004;304:1301. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- [7].Wu J, Cai L-H, Weitz DA. Adv Mater. 2017;29:1702616. doi: 10.1002/adma.201702616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Borukhov I, Bruinsma RF, Gelbart WM, Liu AJ. Proc Natl Acad Sci USA. 2005;102:3673. doi: 10.1073/pnas.0404140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chelakkot R, Lipowsky R, Gruhn T. Soft Matter. 2009;5:1504. [Google Scholar]
- [10].Chelakkot R, Gruhn T. Soft Matter. 2012;8:11746. [Google Scholar]
- [11].Strehle D, Schnauss J, Heussinger C, Alvarado J, Bathe M, Kas J, Gentry B. Eur Biophys J. 2011;40:93. doi: 10.1007/s00249-010-0621-z. [DOI] [PubMed] [Google Scholar]
- [12].Limozin L, Sackmann E. Phys Rev Lett. 2002;89:168103. doi: 10.1103/PhysRevLett.89.168103. [DOI] [PubMed] [Google Scholar]
- [13].Meyer RK, Aebi U. J Cell Biol. 1990;110:2013. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tseng Y, Fedorov E, McCaffery JM, Almo SC, Wirtz D. J Mol Biol. 2001;310:351. doi: 10.1006/jmbi.2001.4716. [DOI] [PubMed] [Google Scholar]
- [15].Wachsstock DH, Schwartz WH, Pollard TD. Biophys J. 1993;65:205. doi: 10.1016/S0006-3495(93)81059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ward SMV, Weins A, Pollak MR, Weitz DA. Biophys J. 2008;95:4915. doi: 10.1529/biophysj.108.131722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ehrlicher AJ, Krishnan R, Guo M, Bidan CM, Weitz DA, Pollak MR. Proc Natl Acad Sci USA. 2015;112:6619. doi: 10.1073/pnas.1505652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wagner B, Tharmann R, Haase I, Fischer M, Bausch AR. Proc Natl Acad Sci USA. 2006;103:13974. doi: 10.1073/pnas.0510190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- [20].Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, et al. Nat Methods. 2008;5:605. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].De La Cruz EM, Pollard TD. Biochemistry. 1996;35:14054. doi: 10.1021/bi961047t. [DOI] [PubMed] [Google Scholar]
- [22].Lieleg O, Kayser J, Brambilla G, Cipelletti L, Bausch AR. Nat Mater. 2011;10:236. doi: 10.1038/nmat2939. [DOI] [PubMed] [Google Scholar]
- [23].Janmey PA, Hvidt S, Lamb J, Stossel TP. Nature. 1990;345:89. doi: 10.1038/345089a0. [DOI] [PubMed] [Google Scholar]
- [24].Schmoller KM, Lieleg O, Bausch AR. Soft Matter. 2008;4:2365. [Google Scholar]
- [25].Pollard TD. J Cell Biol. 1986;103:2747. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Aratyn YS, Schaus TE, Taylor EW, Borisy GG. Mol Biol Cell. 2007;18:3928. doi: 10.1091/mbc.E07-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Proc Natl Acad Sci USA. 1996;93:12937. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schmoller KM, Semmrich C, Bausch AR. J Struct Biol. 2011;173:350. doi: 10.1016/j.jsb.2010.09.003. [DOI] [PubMed] [Google Scholar]
- [29].Claessens MMAE, Bathe M, Frey E, Bausch AR. Nat Mater. 2006;5:748. doi: 10.1038/nmat1718. [DOI] [PubMed] [Google Scholar]
- [30].Heussinger C, Bathe M, Frey E. Phys Rev Lett. 2007;99:48101. doi: 10.1103/PhysRevLett.99.048101. [DOI] [PubMed] [Google Scholar]
- [31].Tempel M, Isenberg G, Sackmann E. Phys Rev E. 1996;54:1802. doi: 10.1103/physreve.54.1802. [DOI] [PubMed] [Google Scholar]
- [32].Strehle D, Mollenkopf P, Glaser M, Golde T, Schuldt C, Käs JA, Schnauß J. Molecules. 2017;22:1804. doi: 10.3390/molecules22101804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Falzone TT, Lenz M, Kovar DR, Gardel ML. Nat Commun. 2012;3:861. doi: 10.1038/ncomms1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nguyen LT, Yang W, Wang Q, Hirst LS. Soft Matter. 2009;5:2033. [Google Scholar]
- [35].Zhang DY, Seelig G. Nat Chem. 2011;3:103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- [36].Phan AT. Nucleic Acids Res. 2002;30:4618. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hamaguchi N, Ellington A, Stanton M. Anal Biochem. 2001;294:126. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- [38].Asanuma H, Ito T, Yoshida T, Liang X, Komiyama M. Angew Chem, Int Ed. 1999;38:2393. doi: 10.1002/(sici)1521-3773(19990816)38:16<2393::aid-anie2393>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- [39].Gentry B, Smith DM, Käs JA. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2009;79:31916. doi: 10.1103/PhysRevE.79.031916. [DOI] [PubMed] [Google Scholar]
- [40].Isambert H, Venier P, Maggs A, Fattoum A, Kassab R, Pantaloni D, Carlier M. J Biol Chem. 1995;270:11437. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- [41].van Oosten ASG, Vahabi M, Licup AJ, Sharma A, Galie PA, MacKintosh FC, Janmey PA. Sci Rep. 2016;6:19270. doi: 10.1038/srep19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vahabi M, Sharma A, Licup AJ, van Oosten ASG, Galie PA, Janmey PA, MacKintosh FC. Soft Matter. 2016;12:5050. doi: 10.1039/c6sm00606j. [DOI] [PubMed] [Google Scholar]
- [43].Tang JX, Janmey PA. J Biol Chem. 1996;271:8556. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- [44].Yin P, Hariadi RF, Sahu S, Choi HMT, Park SH, Labean TH, Reif JH. Science. 2008;321:824. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- [45].Schiffels D, Liedl T, Fygenson DK. ACS Nano. 2013;7:6700. doi: 10.1021/nn401362p. [DOI] [PubMed] [Google Scholar]
- [46].Schuldt C, Schnauß J, Händler T, Glaser M, Lorenz J, Golde T, Käs JA, Smith DM. Phys Rev Lett. 2016;117:197801. doi: 10.1103/PhysRevLett.117.197801. [DOI] [PubMed] [Google Scholar]
- [47].Glaser M, Schnauß J, Tschirner T, Schmidt BUS, Moebius-Winkler M, Käs JA, Smith DM. New J Phys. 2016;18:55001. [Google Scholar]
- [48].Schmoller KM, Lieleg O, Bausch AR. Phys Rev Lett. 2008;101:118102. doi: 10.1103/PhysRevLett.101.118102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.