Abstract
Background
Strong evidence supports the role of both genetic and environmental factors in pediatric-onset multiple sclerosis (POMS) etiology.
Objective
We comprehensively investigated the association between established MHC and non-MHC adult MS–associated variants and susceptibility to POMS.
Methods
Cases with onset <18 years (n=569) and controls (n=16,251) were included from the U.S. and Sweden. Adjusted logistic regression and meta–analyses were performed for individual risk variants and a weighted genetic risk score (wGRS) for non–MHC variants. Results were compared to adult MS cases (n=7,588).
Results
HLA–DRB1*15:01 was strongly associated with POMS (ORmeta= 2.95, p<2.0 × 10−16). Further, 28 of 104 non–MHC variants studied (23%) were associated (p<0.05); POMS cases carried, on average, a higher burden of these 28 variants compared to adults (ORavg=1.24 vs. 1.13, respectively), though the difference was not significant. The wGRS was strongly associated with POMS (ORmeta=2.77, 95% CI 2.33, 3.32, p<2.0 × 10−16) and higher, on average, when compared to adult cases. Additional class III risk variants in the MHC region associated with POMS were revealed after accounting for HLA–DRB1*15:01 and HLA–A*02.
Conclusion
Pediatric and adult MS share many genetic variants suggesting similar biological processes are present. MHC variants beyond HLA–DRB1*15:01 and HLA–A*02 are also associated with POMS.
Search terms: multiple sclerosis, pediatrics, genetics, epidemiology
INTRODUCTION
Multiple sclerosis (MS) disease onset occurs on average around 34 years of age, although approximately 5% of all MS patients have symptom onset before 18 years.1,2 Environmental factors established in adult–onset MS, such as remote infection with Epstein–Barr virus and exposure to cigarette smoking, have also been strongly implicated in pediatric–onset disease susceptibility.3 However, unlike adult–onset MS, pediatric cases almost exclusively have a relapsing–remitting course, and, in North America, are less likely to be of white/European ancestry.1,2
While limited evidence supports a role for similar genetic risk factors, such as HLA–DRB1, in pediatric MS susceptibility,4,5 to date, the 110 non–MHC adult MS susceptibility loci identified through a genome–wide association study,6 as well as the remainder of the MHC, have not been studied in pediatric–onset MS (POMS). Sorting out which adult MS risk variants are associated with onset in childhood will help determine whether the genetic contributions to the development of MS pathology occur irrespective of age. In addition, identifying variants with larger effects in POMS could point to critical biological pathways determining an earlier onset. The goal of this study was to: (1) determine whether established MS risk variants from GWAS in adults are also associated with POMS and have higher effect sizes; (2) to study HLA–DRB1*15, HLA–A*02 and the MHC region in POMS using high density SNP genotyping; (3) to directly compare results from pediatric–onset and adult cases for differences in overall genetic burden and effect size; and (4) to determine whether genetic risk variants are associated with age of onset in POMS cases.
METHODS
U.S. Participants
POMS cases were enrolled through pediatric MS centers established across the U.S. between January 2006 and December 2014 (Appendix 2). Consecutive patients with onset of MS or clinically isolated syndrome suggestive of early MS before age 18 seen at some of these pediatric MS clinics were offered participation as previously described.7 In addition, we utilized adult cases from Kaiser Permanente Northern California (KPNC) with reported age of first symptom onset <18 years.8 Age of onset of MS was determined as year of first self–reported clinical symptom, based on the following questions: “How old were you when you had your first clinical symptoms of MS?”, “What were your first clinical symptoms of MS?”. Year of disease onset was calculated using date of birth and was verified in the medical record when possible. KPNC MS cases with age of onset > 18 years were included for comparison in the study (n=1,103).8 Established diagnostic criteria were used for all pediatric (n=738)9 and adult–onset cases (n=1,103).10,11
Control individuals without a diagnosis of MS or related condition (optic neuritis, transverse myelitis, or demyelination disease) were recruited from Northern California,8 including individuals from the Genetic Epidemiology Research on Adult Health and Aging cohort who participated in the KPNC Research Program on Genes, Environment, and Health (RPGEH) (dbGaP phs000674.v2.p2), and controls recruited as part of a pediatric MS case–control study (total n=12,300).12
Swedish Participants
Data were collected from two population based case–control studies in Sweden of incident (Epidemiological Investigation of MS [EIMS]) and prevalent (Genes and Environment in MS [GEMS]) MS patients. The EIMS study (2005–2014) inclusion criteria were: age 16–70 years, diagnosed MS according to the to the McDonald criteria10,11 within two years, and ability to understand the Swedish language. GEMS study participants were identified from the Swedish National MS registry, fulfilled the McDonald criteria, and were recruited during 2009–2011. For both studies, controls were randomly chosen from the population register and matched to cases by sex, age at inclusion in the study, and region of residence. Two controls were matched to each case in the EIMS study and one control per case in the GEMS study. All participants in the EIMS study were distinct from those in the GEMS study. Details of the study design have been described elsewhere.13 Age of onset of MS was determined as year of first self–reported clinical symptom and confirmed when possible, as described above. Data for 175 POMS cases (reported age of onset <18 years), 6,485 adult–onset MS cases, and 5,376 controls were available for the current study.
Standard Protocol Approvals
Study protocols were approved by the Institutional Review Boards for Human Subjects at UCSF, Stony Brook, Children’s Hospital of Philadelphia, Texas Children’s Hospital, University of Colorado School of Medicine, University of Texas Southwestern, State University of New York Buffalo, Loma Linda University, Mayo Clinic, University of Alabama at Birmingham, Ann & Robert Lurie Children’s Hospital of Chicago, University of Utah, Boston Children’s Hospital, Massachusetts General Hospital, Washington University St. Louis, Children’s National Medical Center, Kaiser Permanente Division of Research, UC Berkeley, Regional Ethical Review Board in North Stockholm, and Karolinska Institutet. Informed consent or assent (children) was obtained for all study participants and their parents when appropriate.
Statistical Analyses
Genotyping and quality control procedures are outlined in the supplemental materials (Appendix 2). Characterization of HLA–DRB1*15 was assessed for each study participant; for the U.S. study, a tag SNP (rs3135388) was used.14 Whole genome data were used to test the previously established 110 MS GWAS SNP genotypes6 for association with POMS. Each SNP genotype was considered as zero, one, or two minor alleles (additive effect) for analysis. Further, a weighted genetic risk score (wGRS) that combines the weighted odds ratio (OR) from each of 110 non– MHC MS susceptibility loci was calculated for each pediatric–onset case and control by multiplying the number of risk alleles for each locus by the weight for that variant and then taking the sum across the 110 loci.15 The weight for each locus is the natural log of the OR for each allele. Three of 110 SNPs were missing in the U.S. study (rs201202118, rs201847125 and imm_5_141486748), and one SNP failed Hardy–Weinberg equilibrium, rs2744148. Three of 110 SNPs were missing from the Swedish data (rs2028597, rs4679081, and imm_5_141486748). Adequate SNP tagging proxies were not identified; therefore, these SNPs were not included in the individual SNP meta–analyses (n=6) or each cohort’s respective wGRS.
Logistic regression modeling was used adjusting for genetic ancestry and other covariates when noted for case–control analyses. 95% confidence intervals (CI), allelic OR (unless otherwise noted) and two–sided p–values were reported. For the MHC region, individual variants were analyzed spanning 6p22.1 to 6p21.3 (~29Mb to 33Mb on hg19 assembly). Meta–analyses were performed under a random effects model. Linear regression was performed in case–only analyses with age of onset as the outcome; values were transformed to the fifth power (x5) to normalize the negatively skewed distribution. All analyses were performed in PLINK v.1.07 or R v.3.2.2. Because the study of candidate SNPs and wGRS was strongly hypothesis–based, we used p<0.05 as a type I error rate for reporting significance. For the MHC region analyses (beyond HLA– DRB1*15:01 and HLA–A2), the number of independent MHC haplotype blocks was used to control for multiple comparisons and guide interpretation of results.
Role of Funding Source
Funding sources listed below (see Acknowledgements) had no involvement in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
RESULTS
Subject Characteristics
We restricted our study sample to white non–Hispanic individuals (European ancestry), the largest group in our dataset, to ensure a genetically homogenous sample and avoid the possibility of confounding by genetic ancestry (Supplementary Figure 1). After excluding population outliers, the final dataset was comprised of 394 POMS cases and 10,875 controls from the U.S., as well as 175 POMS cases and 5,376 controls from Sweden (total N=16,820). Demographic features and clinical characteristics of participants are shown in Table 1. Mean age at symptom onset for POMS cases was 14.05 years (+/− 3.30 years) in the U.S. study and 14.91 (+/− 2.67) in the Swedish study. We also included 1,103 and 6,485 adult–onset MS cases from the U.S. and Sweden, respectively (total N=7,588), for comparison of genetic associations observed in POMS (Supplementary Table 1).
Table 1.
Characteristics of POMS Case and Control Participants
| U.S. POMS Cases |
U.S. Controls |
Sweden POMS Cases |
Sweden Controls |
|
|---|---|---|---|---|
| N | 394 | 10,875 | 175 | 5,376 |
| Females:Males | 3·0:1 | 1·6:1 | 2·5:1 | 3·2:1 |
| Age of onset | 14·05 (3·30) | — | 14.91 (2·67) | — |
| wGRS | 12·50 (0·66) | 12·06 (0·68) | 12.73 (0·66) | 12·22 (0·69) |
| HLA–DRB1*15:01 | ||||
| 0 | 193 (49) | 7,973 (72) | 57 (33) | 3,806 (71) |
| 1–2 | 201 (51) | 2,902 (28) | 118 (67) | 1,570 (29) |
MS= multiple sclerosis; wGRS=weighted genetic risk score
Table values are mean (SD) for continuous variables or No. (%) for categorical variables.
Established Non–MHC MS Genetic Risk Variants
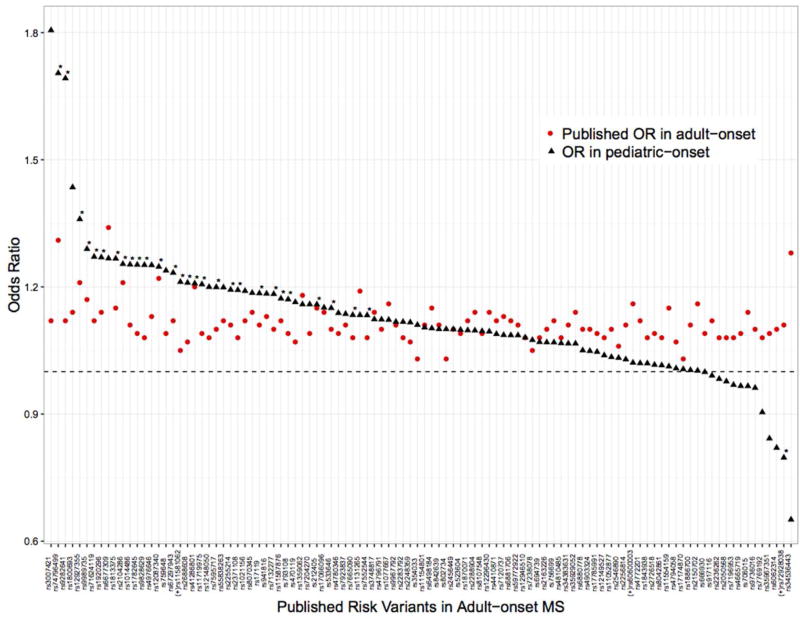
Results from the meta–analysis of MS variants identified in recent GWAS demonstrated that 28 of 104 established MS SNPs tested were significantly associated with pediatric–onset (Table 2; Supplementary Table 2). POMS cases carried, on average, a higher burden of the 28 variants compared to adult MS cases carrying the same polymorphisms (Figure 1) though the difference was not significant; the average OR of the 28 significant variants was 1.24 (95% CI 1.09, 1.41) in POMS cases compared to 1.13 (95% CI 1.08, 1.19) in adult MS cases, using estimates published by the International Multiple Sclerosis Genetics Consortium, with confidence intervals overlapping.6
Table 2.
Significant meta-analysis results for 104 non-MHC MS risk loci in POMS cases and controls (p<0 05)
| U.S. | Sweden | Meta-Analysisa | Published Estimate in Adults |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| CHR | SNP | GENE | A | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P |
| 1 | rs11581062 | SLC30A7 | G | 1·20 | 1·03, 1·39 | 0·02 | 1·25 | 0·99, 1·59 | 0·06 | 1·21 | 1·06, 1·38 | <0·01 | 1·12 | 1·07, 1·17 | <0·01 |
| 1 | rs12087340 | BCL10 | A | 1·32 | 1·05, 1·67 | 0·02 | 1·06 | 0·72, 1·55 | 0·78 | 1·24 | 1·02, 1·52 | 0·03 | 1·22 | 1·15, 1·29 | <0·01 |
| 1 | rs41286801 | EVI5 | A | 1·26 | 1·04, 1·51 | 0·02 | 1·10 | 0·82, 1·47 | 0·53 | 1·21 | 1·04, 1·41 | 0·01 | 1·20 | 1·15, 1·25 | <0·01 |
| 2 | rs7595717 | PLEK | A | 1·24 | 1·06, 1·44 | 0·01 | 1·12 | 0·88, 1·41 | 0·37 | 1·20 | 1·05, 1·36 | 0·01 | 1·10 | 1·06, 1·14 | <0·01 |
| 2 | rs9989735 | SP140 | C | 1·30 | 1·10, 1·54 | <0·01 | 1·27 | 0·98, 1·65 | 0·07 | 1·29 | 1·12, 1·49 | <0·01 | 1·17 | 1·12, 1·22 | <0·01 |
| 3 | rs9282641 | CD86 | G | 1·44 | 1·08, 1·93 | 0·01 | 2·21 | 1·33, 3·66 | <0·01 | 1·25 | 1·10, 1·43 | 0·01 | 1·12 | 1·05, 1·19 | <0·01 |
| 3 | rs11719975 | SATB1 | C | 1·18 | 1·01, 1·38 | 0·04 | 1·26 | 1·00, 1·58 | 0·05 | 1·21 | 1·06, 1·38 | <0·01 | 1·09 | 1·05, 1·13 | <0·01 |
| 3 | rs1014486 | IL12A | G | 1·14 | 0·99, 1·32 | 0·06 | 1·42 | 1·14, 1·76 | <0·01 | 1·25 | 1·01, 1·54 | 0·04 | 1·11 | 1·07, 1·14 | <0·01 |
| 3 | rs2255214 | CD86 | G | 1·20 | 1·04, 1·39 | 0·01 | 1·18 | 0·95, 1·46 | 0·13 | 1·19 | 1·06, 1·34 | <0·01 | 1·11 | 1·08, 1·15 | <0·01 |
| 3 | rs2371108 | E OMES | A | 1·23 | 1·06, 1·42 | 0·01 | 1·12 | 0·90, 1·39 | 0·31 | 1·19 | 1·06, 1·34 | <0·01 | 1·08 | 1·05, 1·12 | <0·01 |
| 3 | rs9828629 | F OXP1 | G | 1·27 | 1·09, 1·47 | <0·01 | 1·22 | 0·97, 1·53 | 0·09 | 1·70 | 1·14, 2·53 | <0·01 | 1·08 | 1·05, 1·12 | <0·01 |
| 3 | rs1920296 | IQCB1 | C | 1·26 | 1·08, 1·46 | <0·01 | 1·30 | 1·03, 1·65 | 0·03 | 1·27 | 1·12, 1·45 | <0·01 | 1·14 | 1·11, 1·18 | <0·01 |
| 3 | rs1813375 | CMC1 | A | 1·29 | 1·11, 1·48 | <0·01 | 1·23 | 0·99, 1·52 | 0·06 | 1·27 | 1·13, 1·42 | <0·01 | 1·15 | 1·12, 1·19 | <0·01 |
| 4 | rs7665090 | – | G | 1·11 | 0·96, 1·28 | 0·15 | 1·18 | 0·96, 1·47 | 0·12 | 1·14 | 1·01, 1·28 | 0·03 | 1·08 | 1·05, 1·12 | <0·01 |
| 5 | rs71624119 | ANKRD55 | G | 1·17 | 0·98, 1·40 | 0·08 | 1·47 | 1·10, 1·96 | 0·01 | 1·27 | 1·03, 1·57 | 0·03 | 1·12 | 1·08, 1·17 | <0·01 |
| 6 | rs67297943 | – | T | 1·25 | 1·02, 1·52 | 0·03 | 1·21 | 0·93, 1·58 | 0·15 | 1·23 | 1·05, 1·44 | 0·01 | 1·12 | 1·07, 1·16 | <0·01 |
| 6 | rs72928038 | BACH2 | A | 0·77 | 0·61, 0·97 | 0·03 | 0·85 | 0·62, 1·15 | 0·29 | 0·80 | 0·66, 0·96 | 0·02 | 1·11 | 1·07, 1·16 | <0·01 |
| 6 | rs17119 | JARID2 | A | 1·19 | 0·99, 1·44 | 0·07 | 1·17 | 0·87, 1·58 | 0·30 | 1·18 | 1·00, 1·39 | 0·05 | 1·11 | 1·06, 1·15 | <0·01 |
| 6 | rs212405 | TAGAP | T | 1·15 | 0·99, 1·35 | 0·07 | 1·17 | 0·93, 1·48 | 0·18 | 1·16 | 1·02, 1·32 | 0·03 | 1·15 | 1·11, 1·19 | <0·01 |
| 10 | rs2104286 | IL2RA | T | 1·22 | 1·03, 1·45 | 0·02 | 1·34 | 1·03, 1·73 | 0·03 | 1·26 | 1·09, 1·41 | <0·01 | 1·21 | 1·16, 1·26 | <0·01 |
| 10 | rs1782645 | ZMIZ1 | T | 1·19 | 1·02, 1·39 | 0·03 | 1·37 | 1·11, 1·70 | <0·01 | 1·25 | 1·09, 1·43 | <0·01 | 1·09 | 1·05, 1·13 | <0·01 |
| 10 | rs793108 | ZNF438 | A | 1·14 | 0·99, 1·32 | 0·07 | 1·24 | 1·00, 1·53 | 0·05 | 1·17 | 1·04, 1·31 | 0·01 | 1·09 | 1·06, 1·13 | <0·01 |
| 10 | rs2688608 | – | A | 1·23 | 1·06, 1·42 | 0·01 | 1·16 | 0·89, 1·50 | 0·28 | 1·20 | 1·05, 1·38 | 0·01 | 1·07 | 1·03, 1·10 | <0·01 |
| 11 | rs533646 | – | G | 1·13 | 0·96, 1·32 | 0·14 | 1·20 | 0·96, 1·51 | 0·12 | 1·15 | 1·01, 1·31 | 0·04 | 1·10 | 1·06, 1·14 | <0·01 |
| 12 | rs7132277 | PITPNM2 | A | 1·26 | 1·06, 1·49 | 0·01 | 1·05 | 0·81, 1·37 | 0·70 | 1·18 | 1·00, 1·40 | 0·05 | 1·10 | 1·06, 1·15 | <0·01 |
| 14 | rs74796499 | GALC | C | 1·59 | 1·04, 2·44 | 0·03 | 2·10 | 0·99, 4·46 | 0·05 | 1·71 | 1·18, 2·49 | 0·01 | 1·31 | 1·21, 1·42 | <0·01 |
| 16 | rs4780346 | CLEC16A | A | 1·15 | 0·98, 1·35 | 0·09 | 1·11 | 0·89, 1·40 | 0·36 | 1·14 | 1·00, 1·29 | 0·05 | 1·09 | 1·05, 1·13 | <0·01 |
| 16 | rs12927355 | CLEC16A | C | 1·33 | 1·14, 1·56 | <0·01 | 1·43 | 1·11, 1·84 | <0·01 | 1·36 | 1·19, 1·56 | <0·01 | 1·21 | 1·17, 1·26 | <0·01 |
CHR=chromosome; SNP=single nucleotide polymorphism; A= allele; OR=odds ratio; CI=confidence interval
Analyses adjusted for genetic ancestry
Figure 1.
Individual 104 risk variant comparisons between POMS findings and published IMSGC adult MS odds ratios.
A significantly higher wGRS was observed in POMS cases compared to controls in both populations even after controlling for HLA–DRB1*15 carrier status (ORmeta=2.77, 95% CI 2.33, 3.32, p<2.0 × 10−16), indicating strong, independent effects of both wGRS and HLA–DRB1 on disease susceptibility. No association was observed between wGRS and age of onset in POMS cases (data not shown). Further, we compared pediatric–onset with adult MS cases for differences in wGRS. Results showed the mean wGRS was higher in pediatric–onset compared to adult MS cases; this difference was significant in the Swedish dataset (p=0.01); similar results were observed in the U.S. dataset, but were not significant (p=0.31).
Major Histocompatibility Complex
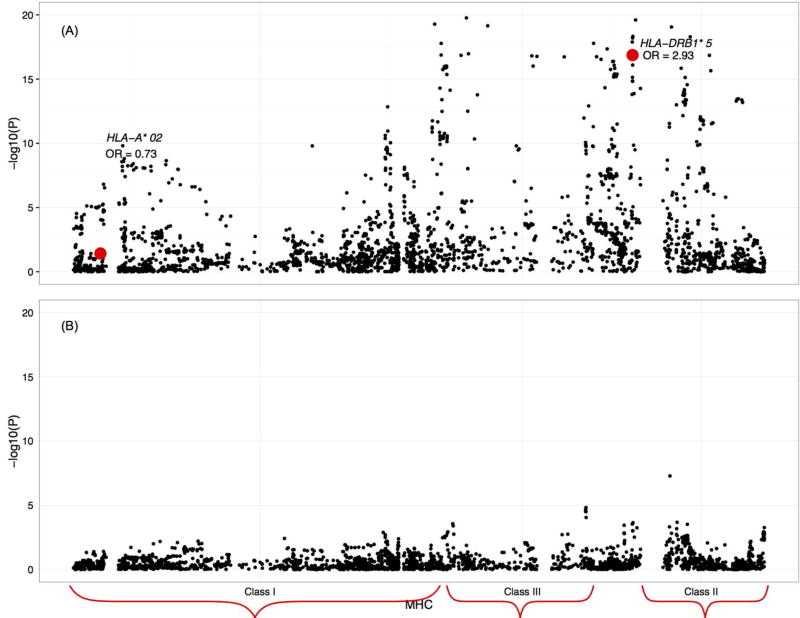
HLA–DRB1*15:01 was strongly associated with POMS cases compared to controls (ORmeta=2.95, 95% CI 2.33, 3.73, p<2.0 × 10−16; Figure 2A). Evidence for a protective effect of HLA–A2 was also present (ORmeta=0.73, 95% CI 0.54, 0.98, p=0.04). Adjustment for both HLA– DRB1*15:01 and HLA–A2 associations revealed more than 300 MHC variants associated with POMS (p<0.05) (Figure 2B; Supplementary Table 3); the top 20 SNPs are shown in Table 3 (p < 10−3) and suggest both class II and III effects, independent of HLA–A2 and HLA–DRB1*15:01, are present. No association was observed between HLA–DRB1*15:01 and age of onset in POMS cases (data not shown). When we compared pediatric–onset with adult MS cases, HLA– DRB1*15:01 allele distribution differences were present in the Swedish dataset (p=0.02); 68% of pediatric–onset Swedish cases carried one or two risk alleles, compared to 58% of adult–onset cases. No significant difference in HLA–DRB1*15:01 allele distribution between pediatric and adult–onset MS cases were found in the U.S. dataset (data not shown).
Figure 2.
Meta–analyses of the major histocompatibility complex (MHC) in POMS cases and controls: (A) unadjusted, and (B) adjusted for HLA–DRB1*15:01 and HLA–A*02. Individual variants were analyzed spanning 6p22.1 to 6p21.3. Evidence for MHC involvement, particularly in the Class III HLA region is shown. U.S. analyses additionally adjusted for genetic ancestry.
Table 3.
Top twenty meta–analysis results of MHC region in POMS cases and controls adjusted for HLA–DRB1*15:01 and HLA–A*02
| CHR | BP | SNP | GENE | SUBREGION | OR | p–value |
|---|---|---|---|---|---|---|
| 6 | 32595063 | rs28720766 | – | Class II | 2·16 | 5·19×10−8a |
| 6 | 32186245 | rs438475 | NOTCH4 | Class III | 0·59 | 1·51×10−5a |
| 6 | 32184345 | rs404860 | NOTCH4 | Class III | 0·63 | 2·18×10−5a |
| 6 | 32184574 | rs384247 | NOTCH4 | Class III | 0·64 | 3·11×10−5a |
| 6 | 32186193 | rs431541 | NOTCH4 | Class III | 0·64 | 3·14×10−5a |
| 6 | 32186872 | rs2256594 | NOTCH4 | Class III | 0·65 | 8·95×10−5 |
| 6 | 32629868 | rs1049088 | HLA–DQB1 | Class II | 0·67 | 2·11×10−4 |
| 6 | 32415109 | rs3135387 | HLA–DRA | Class II | 1·55 | 2·27×10−4 |
| 6 | 31537606 | rs2857706 | LTA | Class III | 1·36 | 2·74×10−4 |
| 6 | 32411726 | rs3129888 | HLA–DRA | Class II | 1·54 | 2·88×10−4 |
| 6 | 32679992 | rs12527228 | – | Class II | 1·78 | 3·03×10−4 |
| 6 | 32372032 | rs73400865 | BTNL2 | Class III | 1·55 | 3·60×10−4 |
| 6 | 31539767 | rs2844482 | LTA | Class III | 1·36 | 4·02×10−4 |
| 6 | 32592737 | rs28421666 | – | Class II | 1·86 | 4·80×10−4 |
| 6 | 33054457 | rs1042544 | HLA–DPB1 | Class II | 1·26 | 5·43×10−4 |
| 6 | 32434314 | rs12529318 | – | – | 1·75 | 5·69×10−4 |
| 6 | 32627700 | rs6689 | HLA–DQB1 | Class II | 0·73 | 6·66×10−4 |
| 6 | 32414420 | rs7382085 | HLA–DRA | Class II | 1·44 | 9·57×10−4 |
| 6 | 32413317 | rs2395182 | HLA–DRA | Class II | 1·43 | 9·83×10−4 |
| 6 | 33051683 | rs9277411 | HLA–DPB1 | Class II | 1·25 | 1·15×10−3 |
CHR=chromosome; BP=base pair; SNP=single nucleotide polymorphism; OR=odds ratio
Significant after multiple comparisons correction
DISCUSSION
This study comprehensively investigated the genetic component in MS cases for whom first clinical disease symptoms appeared before age 18. HLA–DRB1*15 was associated with an effect estimate (ORmeta= 2.95) similar to previous smaller studies of this age group (OR=2.28– 3.30).4,5,16 Additional MHC variants have been implicated in previous studies of adult MS patients, including a protective effect for HLA–A*02.17–20 Through fine mapping of the MHC region in the current study, we observed only modest evidence for HLA–A*02 effect in POMS.
Strong evidence for independent class II and III effects were detected, similar to findings from a previous study of adult MS cases and controls for which 11 independent associations within the MHC were reported.18 In the current study, several SNPs within/near NOTCH4 in the class III region provided the strongest evidence for association after accounting for HLA–DRB1*15:01 and correction for multiple comparisons, with one SNP (rs438475) demonstrating high evidence of functionality as an expression quantitative trait locus and transcription factor binding site. Nominal evidence for class II effects was also present, including two SNPs (rs3129888 and rs2395182) identified as expression quantitative trait loci and transcription factor binding sites. The human MHC class III region is the most gene–dense region of the human genome.21 Variation within NOTCH4 has been shown recently to decrease the risk for developing MS in Japanese adults,22,23 similar to our finding, but in contrast to another.24 Taken together, the involvement in POMS susceptibility of HLA–DRB1*15:01 and one or more gene variants within the class III MHC region is strongly supported. MHC region and classical HLA loci genotyping is needed in a larger data set to fully characterize the MHC contribution to POMS, which, similar to adult studies, appears to be complex.18,25
For the first time, we studied individual and collective associations between 104 of the 110 established non–MHC adult MS risk variants6 and susceptibility to POMS. Twenty–eight variants were significantly associated at p<0.05; several variants had larger effect estimates compared to published results from adult MS studies. Top SNPs included rs74796499 within GALC and rs9828629 in FOXP1; both are involved in nervous system development. We found a significant inverse association between a SNP in BACH2 and POMS, which has been shown to increase the risk of MS in adults; further work and replication is needed to confirm the difference in directionality between the two populations. A significantly higher wGRS was also observed in POMS cases compared to controls in both U.S. and Swedish datasets after adjusting for genetic ancestry and HLA–DRB1*15:01, in agreement with a small study that used a subset of the 110 MS risk variants to construct a 57 SNP wGRS.26 We did not find an association between wGRS and age of pediatric disease onset, which has been previously reported for adult–onset MS.27 One reason why an association with age of onset in children may not be found is that the range is limited (age 4–17 years included). Nevertheless, these findings warrant further investigation.
Whether HLA–DRB1*15:01 is associated with earlier age of symptom onset in adult MS has been the subject of several investigations. Strong evidence supports the hypothesis that HLA– DRB1*15:01 is indeed associated with an earlier onset in adults;28–30 though conflicting findings have been reported, including one large6 and two small studies with < 100 patients.4,31 In the current study, we did not observe a relationship between HLA–DRB1*15:01 and age of onset in POMS cases. When we compared the cases with symptom onset <18 years of age to cases with symptom onset >18 years of age, HLA–DRB1*15:01 allele distribution differences were present in the Swedish but not U.S. dataset.
Our study had several important strengths. Rigorously diagnosed and clinically well– characterized POMS cases with symptom onset ~ 20 years earlier than most adult MS cases were studied. In addition, a well–powered analysis of HLA–DRB1*15:01 and 104 established MS risk variants was considered for the first time in POMS, and a comparison with adult cases was included. We studied the MHC region with high density SNP mapping in POMS which has not be previously attempted. Genetic diversity among POMS cases is well established, and the potential for population stratification in a case–control study leading to false positive findings is high. Our initial patient group included representatives from non–white populations (n=320; 43% of cases). Comprehensive methods, including multi–dimensional scaling, were used to identify these subgroups, and we restricted the current study to white/European, non–Hispanic individuals for increased genetic homogeneity (Appendix 2). We also adjusted for genetic ancestry within Europeans in our models when necessary.
Some limitations of this study must also be acknowledged. POMS cases were identified through tertiary clinics and may not represent all MS cases with onset in childhood. It is possible that individuals with a more benign disease or without access to healthcare may have been missed. While many of our POMS cases were diagnosed as children in a clinical setting (n=130), some reported the onset of their first disease symptoms in childhood and/or data were extracted from medical records regarding timing of first symptoms but were ascertained as adults. We stratified these groups for comparison to detect any clinical or genetic differences that may have biased our findings (Supplementary Table 4). Notably, MS patients diagnosed as children in the clinic (U.S.) had a slightly lower mean age of symptom onset compared to patients with pediatric symptom onset who were recruited as adults (U.S. and Sweden); this patient group also had a lower frequency of HLA–DRB1*15:01 and average wGRS compared to other groups; though as reported, the patient group as a whole (<18 years of age at onset) had a significantly higher wGRS and HLA–DRB1*15:01 frequency when compared to controls. The possibility of two genetically distinct MS patient groups – one with symptom onset in early childhood – and one with symptom onset in late teen/early adulthood is one to strongly consider, given the findings presented in the current study. We were unable to complete well–powered stratified analyses based on adolescence due to the limited sample size of cases. Other factors may have a stronger influence on disease onset in younger MS patients, such as yet unidentified genetic loci or environmental factors, as previously suggested.3 Due to the unavailability of environmental exposures for a substantial portion of our control group, we were unable to additionally adjust for this information. These factors may be more influential in triggering symptoms of MS in early childhood, given that prenatal, perinatal and early childhood windows represents a period of vulnerability to exposures.
MS onset in childhood is a rare occurrence, and thus our case sample was small compared to genetic studies in adult disease. It is possible that very modest effects for less common alleles may have been missed due to our sample size. While this study had >85% power to detect associations with ORs >1.25 and <0.80 for the 110 candidate SNP analysis in the combined meta–analysis (minor allele frequency of 0.20, p=0.05), certain candidates may have been missed due to lack of power. The results presented here will need to be replicated in a larger data set. Finally, our study was restricted to white/European, non–Hispanics. Examination of rare variants and established European MS genetic risk variants in non–white patient groups is needed to further the understanding of MS pathogenesis, as well as focused studies to identify population– specific risk variants.
In conclusion, we confirmed a role for several previously identified adult–onset MS genetic risk factors in the development of pediatric–onset disease and identified stronger effects of some variants compared to adult–onset MS. Comprehensive assessment of genome–wide associations in POMS, as well as interaction between genetic and environmental exposures, will be important to further characterize POMS susceptibility and are currently underway.
Supplementary Material
Acknowledgments
This work was supported in part by the NIH NINDS: 1R01NS071463 (PI Waubant), R01NS049510 (PI Barcellos), NIH NIEHS: R01ES017080 (PI Barcellos), NIH NIAID: R01AI076544 (PI Barcellos), NIH RC2AG036607 (PI Schaefer), the National MS Society HC 0165 (PI Casper), the Robert Wood Johnson Foundation, the Wayne and Gladys Valley Foundation, and the Ellison Medical Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF CONFLICTING INTERESTS
Drs. Gianfrancesco, Stridh, Graves, Chitnis, Waldman, Lotze, Schreiner, Belman, Weinstock– Guttman, Aeen, Tillema, Ness, Rubin, Candee, Krupp, Gorman, Benson, Rodriguez, Mar, Kahn, Casper, Shen, Hillert, Hedstrom, Olsson, Kockum, and Schaefer report no disclosures.
Ms. Rhead, Shao, Hart, Caillier, Harris, Roalstad, D. Quach and H. Quach report no disclosures.
Dr. Waldman has research support from the NIH, NINDS K23NS069806 (PI Waldman).
Dr. Greenberg has research support from the NIH, Guthy Jackson Charitable Foundation, Medimmune, Chugai, Acorda, Genentech and PCORI, and consulting from Novartis and Alexion.
Dr. Rose has research support from VA, NMSS, Guthy Jackson Charitable Foundation, Arrien Pharmaceuticals, Teva Neuroscience and Biogen. He is a member of the Medical Advisory Board for the DECIDE trial, which is funded by Biogen and AbbVie.
Dr. Alfredsson received research support from the Swedish Medical Research Council (K2013– 69X–14973–10–4), the Swedish Council for Health, Working Life and Welfare (Dnr 2012–0325) and the Swedish Brain Foundation; has received speaker honoraria from Biogen Idec and TEVA.
Dr. Barcellos is funded by the NIH, the National MS Society, and the Race to Erase MS.
Dr. Waubant is funded by the NIH, the NMSS, and the Race to Erase MS. She volunteers on an advisory board for a Novartis trial. She has received honorarium or travel support from ACTRIMS, ECTRIMS, and AAN.
References
- 1.Chitnis T, Krupp L, Yeh A, et al. Pediatric multiple sclerosis. Neurol Clin. 2011;29:481–505. doi: 10.1016/j.ncl.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Yeh EA, Chitnis T, Krupp L, et al. Pediatric multiple sclerosis. Nat Rev Neurol. 2009;5:621–31. doi: 10.1038/nrneurol.2009.158. [DOI] [PubMed] [Google Scholar]
- 3.Waubant E, Ponsonby A, Pugliatti M, et al. Environmental and genetic factors in pediatric inflammatory demyeliating diseases. Neurol. 2016;87(Suppl 2):S20–7. doi: 10.1212/WNL.0000000000003029. [DOI] [PubMed] [Google Scholar]
- 4.Boiko AN, Gusev EI, Sudomoina MA, et al. Association and linkage of juvenile MS with HLA-DR2(15) in Russians. Neurol. 2002;58(4):658–60. doi: 10.1212/wnl.58.4.658. [DOI] [PubMed] [Google Scholar]
- 5.Banwell D, Bar-Or A, Arnold DL, et al. Clinical, environmental, and genetic determinants of multiple sclerosis in children with acute demyelination: a prospective national cohort study. Lancet Neurol. 2011;10(5):426–45. doi: 10.1016/S1474-4422(11)70045-X. [DOI] [PubMed] [Google Scholar]
- 6.Beecham AH, Patsopoulos NA, Xifara DK, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waubant E, Mowry EM, Krupp L, et al. Common viruses associated with lower pediatric multiple sclerosis risk. Neurol. 2011;76:1989–95. doi: 10.1212/WNL.0b013e31821e552a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gianfrancesco MA, Glymour MM, Walter S, et al. Causal effect of genetic variants associated with body mass index on multiple sclerosis susceptibility. Am J Epidemiol. 2017;185(3):162–171. doi: 10.1093/aje/kww120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krupp LB, Tardieu M, Amato MP, et al. International Pediatric Multiple Sclerosis Study Group criteria for pediatric multiple sclerosis and immune-mediated central nervous system demyelinating disorders: revisions to the 2007 definitions. Mult Scler. 2013;19(10):1261–7. doi: 10.1177/1352458513484547. [DOI] [PubMed] [Google Scholar]
- 10.McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50:121–7. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 11.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nourbakhsh B, Graves J, Casper TC, et al. Dietary salt intake and time to relapse in paediatric multiple sclerosis. J Neurol Neurosurg Psychiatry. 2016;87(12):1350–3. doi: 10.1136/jnnp-2016-313410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedstrom AK, Hillert J, Olsson T, Alfredsson L. Smoking and multiple sclerosis susceptibility. Eur J Epidemiol. 2013;28:867–74. doi: 10.1007/s10654-013-9853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zivkovic M, Stankovic A, Dincic E, et al. The tag SNP for HLA-DRB1*1501, rs3135388, is significantly associated with multiple sclerosis susceptibility: cost-effective high- throughput detection by real-time PCR. Clin Chim Acta. 2009;406:27–30. doi: 10.1016/j.cca.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 15.De Jager PL, Chibnik LB, Cui J, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1111–9. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Disanto G, Magalhaes S, Handel AE, et al. HLA-DRB1 confers increased risk of pediatric-onset MS in children with acquired demyelination. Neurol. 2011;76(9):781–6. doi: 10.1212/WNL.0b013e31820ee1cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barcellos LF, Sawcer S, Ramsay PP, et al. Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet. 2006;15:2813–24. doi: 10.1093/hmg/ddl223. [DOI] [PubMed] [Google Scholar]
- 18.Patsopoulos NA, Barcellos LF, Hintzen RQ, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013;9(11):e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo TW, De Jager PL, Gregory SG, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol. 2007;61:228–36. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link J, Kockum I, Lorentzen AR, et al. Importance of human leukocyte antigen (HLA) class I and II alleles on the risk of multiple sclerosis. PLoS One. 2012;7(5):e36779. doi: 10.1371/journal.pone.0036779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie T, Rowen L, Begona A, et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13(12):2621–36. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Yoshimura S, Isobe N, et al. A NOTCH4 missense mutation confers resistance to multiple sclerosis in Japanese. Mult Scler. 2013;19(13):1696–1703. doi: 10.1177/1352458513482512. [DOI] [PubMed] [Google Scholar]
- 23.McElroy JP, Isobe N, Gourraud PA, et al. SNP-based analysis of the HLA locus in Japanese multiple sclerosis patients. Genes Immun. 2011;12(7):523–30. doi: 10.1038/gene.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duvefelt K, Anderson M, Fogdell-Hahn A, Hillert J. A NOTCH4 association with multiple sclerosis is secondary to HLA-DR*1501. Tissue Antigens. 2004;63(1):13–20. doi: 10.1111/j.1399-0039.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 25.Ramagopalan SV, Knight JC, Ebers GC. Multiple sclerosis and the major histocompatibility complex. Curr Opin Neurol. 2009;22(3):219–25. doi: 10.1097/WCO.0b013e32832b5417. [DOI] [PubMed] [Google Scholar]
- 26.van Pelt ED, Mescheriakova JY, Makhani N, et al. Risk genes associated with pediatric- onset MS but not with monophasic acquired CNS myelination. Neurol. 2013;81(23):1996–2001. doi: 10.1212/01.wnl.0000436934.40034eb. [DOI] [PubMed] [Google Scholar]
- 27.Sorosina M, Esposito F, Guaschino C, et al. Inverse correlation of genetic risk score with age at onset in bout-onset and progressive-onset multiple sclerosis. Mult Scler. 2015;21(11):1463–7. doi: 10.1177/1352458514561910. [DOI] [PubMed] [Google Scholar]
- 28.Bove R, Chua AS, Xia Z, et al. Complex relation of HLA-DRB1*1501, age at menarche, and age at multiple sclerosis onset. Neurol Genet. 2016;2(4):e88. doi: 10.1212/NXG.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramagopalan SV, Byrnes JK, Dyment DA, et al. Parent-of-origin of HLA-DRB1*1501 and age of onset of multiple sclerosis. J Hum Genet. 2009;54:547–9. doi: 10.1038/jhg.2009.69. [DOI] [PubMed] [Google Scholar]
- 30.Masterman T, Ligers A, Olsson T, et al. HLA-DR15 is associated with lower age at onset in multiple sclerosis. Ann Neurol. 2000;48:211–9. [PubMed] [Google Scholar]
- 31.Anagnostouli MC, Manouseli A, Artemiadis AK, et al. HLA–DRB1* allele frequencies in pediatric, adolescent, and adult–onset multiple sclerosis patients, in a hellenic sample. Evidence for new and established associations. J Mult Scler. 2014;1:104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.