Abstract
The secondary structure of a 55-residue fragment of the mouse prion protein, MoPrP(89–143), was studied in randomly aggregated (dried from water) and fibrillar (precipitated from water/acetonitrile) forms by 13C solid-state NMR. Recent studies have shown that the fibrillar form of the P101L mutant of MoPrP(89–143) is capable of inducing prion disease in transgenic mice, whereas unaggregated or randomly aggregated samples do not provoke disease. Through analysis of 13C chemical shifts, we have determined that both wild-type and mutant sequence MoPrP(89–143) form a mixture of β-sheet and α-helical conformations in the randomly aggregated state although the β-sheet content in MoPrP(89–143, P101L) is significantly higher than in the wild-type peptide. In a fibrillar state, MoPrP(89–143, P101L) is completely converted into β-sheet, suggesting that the formation of a specific β-sheet structure may be required for the peptide to induce disease. Studies of an analogous peptide from Syrian hamster PrP verify that sequence alterations in residues 101–117 affect the conformation of aggregated forms of the peptides.
It is now widely accepted that the prion diseases, such as Creutzfeldt-Jakob disease and Gerstmann-Straussler-Scheinker disease in humans and bovine spongiform encephalopathy, are caused by a conformational change of the prion protein (PrP) from a normally folded cellular form, PrPC, to an alternate, aggregation-prone, pathogenic scrapie form, PrPSc (1, 2). In sporadic and inherited forms of these diseases, initiation is thought to occur through spontaneous conversion of endogenous PrPC to PrPSc, which then autocatalytically propagates. Remarkably, PrPSc can be introduced into a naive host where it can initiate this autocatalytic propagation; this paradigm describes the infectious forms of prion diseases. Although PrPC and PrPSc have identical amino acid sequences, the two isoforms have profoundly different secondary, tertiary, and quaternary structures as observed by CD, Fourier-transform IR, and limited proteolysis (3–7). PrPC is primarily α-helical and monomeric whereas PrPSc is much richer in β-sheet and is oligomeric. It is known that a variety of sequence variants lead to a much higher probability of disease, presumably by facilitating the initial conversion of PrPC to PrPSc. The sites of variation are distributed rather widely through the protein, including some in the well-folded regions of the solution structure of PrPC whereas others are in poorly ordered regions. Knowing the structures of both isoforms and understanding the relationship between sequence and the ability to convert between structures are vital to understanding the process of structural transformation from PrPC to PrPSc.
The structure of an unglycosylated form of PrPC has been determined by solution NMR (3, 5, 6) (Fig. 1); however, less is currently known about the structure of PrPSc, primarily because purified PrPSc is isolated as insoluble aggregates. When PrPSc is subjected to limited proteolysis, it becomes N-terminally truncated to form PrP 27–30. Preparations of PrP 27–30 are infectious and contain para-crystalline fibrils exhibiting the properties of amyloid, which cannot be studied by conventional techniques for structure determination such as x-ray crystallography or solution NMR. Attempts to carry out in vitro conversion of PrPC to PrPSc on a large scale have been unsuccessful. For this reason there have been attempts to recapitulate the behavior of the whole protein with fragments (peptides). Recently, Kaneko et al. (8) have worked with mice expressing low levels of PrP (MoPrP) carrying the P101L mutation, which causes Gerstmann-Straussler-Scheinker disease in humans, and causes disease in mice when expressed at high levels. They showed that a synthetic 55-aa fragment of MoPrP, residues 89–143 with the P101L mutation, can form β-rich, fibrillar aggregates that stimulate disease when inoculated into these animals, whereas the peptide in nonfibrillar form does not (8). Thus, MoPrP (89–143, P101L) presents an opportunity to gain insight into the structural features associated with the ability to cause disease and to learn about the mechanism of conversion from PrPC to PrPSc.
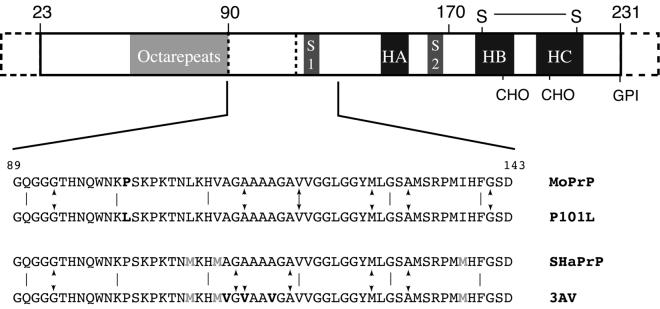
Figure 1.
A schematic diagram of the PrP showing the sequence motifs and secondary structure of PrPC. The segment marked octarepeats contains six copies of an eight-residue repeat sequence, S1 and S2 are the two small β-strands, and HA, HB, and HC designate the three helices. Residues 1–23 are processed off during transport. The points marked CHO indicate the sites at which oligosaccharides are attached in vivo, and GPI indicates the attachment point of the membrane anchor. The region 23–124 is unstructured in PrPC in solution. Below the sequences of peptides studied are given, and the sites of sequences variations are highlighted. Sites of incorporation of isotope labels are indicated by arrows.
Numerous studies support the notion that the key region of PrP in the conversion process includes residues 89–143. The N terminus of protease-resistant PrP 27–30 is at residue 90, defining the approximate start of the infectious domain (9). A Japanese male with an amber mutation at residue 145 died of prion disease, thereby suggesting that the last 87 residues of PrP are not crucial for the initial formation or propagation of PrPSc (10), although multiple attempts to transmit prions form the brain of this patient to experimental animals have been unsuccessful. A mini-prion has been created containing residues 89–140 linked to 176–231 (11) and demonstrated to cause a transmissible neuropathologically accurate prion disease in transgenic mice. Moreover, deletion of residues 90–145 has been shown to prevent PrPSc formation (12). Various fragments in this region have been shown to fold into α-helix- or β-sheet-rich conformations depending on the conditions under which they were prepared (13–20). Thus, there is considerable data to suggest that the region from residue 90 to 143 has substantial conformational lability and plays a significant role in the conversion from PrPC to PrPSc.
In this work, we describe experiments using solid-state NMR to study the secondary structure of PrP (89) before and after conversion to the active, fibril-rich, aggregated state. Unlike solution NMR or x-ray crystallography, solid-state NMR can provide structural information for samples in ordered states, such as aggregated peptides and proteins, through the use of chemical-shift information and the determination of internuclear distances. It is well established that 13C and 15N chemical shifts in peptides and proteins are quite sensitive to local secondary structure (21–24). For example, the isotropic chemical shifts of a Cα in an α-helix versus a β-sheet can differ by more than 8 ppm. Thus, 13C chemical shifts can provide insight into the local secondary structure and can be used in characterizing the secondary structure of the PrP fragments. By placing 13C labels in six residues spread throughout the 55-residue peptide, the secondary structure was monitored in both the wild-type (wt) and P101L mutant forms of MoPrP(89–143). Solid-state 13C NMR spectra of both the wt and P101L peptides exhibit a mixture of helical and extended conformation resonances, indicating substantial conformational heterogeneity and lability in the nonfibrillar state. After aggregation in an acetate-buffered acetonitrile/water solution, the solid-state 13C NMR spectrum of the P101L peptide loses all of the helical peaks, suggesting that the mutation affects not only the local secondary structure, but also the overall structure of the fragment. Similar structural plasticity and long-range conformational effects caused by mutations also were observed in the equivalent hamster sequence fragment with three alanine-to-valine (3AV) mutations at positions 112, 114, and 117, a sequence region previously shown to have structural plasticity (19). Mice carrying the 3AV transgene develop a spontaneous and rapidly lethal condition with neuropathology consistent with a prion disease.
Experimental Procedures
The 13C isotopic labels incorporated into PrP peptides were selected on the basis of their isotropic chemical-shift ranges (22) so that the 13C sites would be resolvable in a one-dimensional NMR spectrum. All peptides were synthesized by using optimized fluorenylmethoxycarbonyl (Fmoc) chemistry on an Applied Biosystems 433 peptide synthesizer. Fmoc amino acid derivatives were preactivated by reaction with 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluoro-phosphate and diisopropylethylamine. After the coupling of each amino acid, a capping step was performed by using N-(2-chlorobenzyloxycarbonyloxy) succinimide (Nova Biochem). Labeled residues were coupled manually by using a 1.5-fold excess of amino acid, and coupling efficiency was monitored by using the quantitative ninhydrin test. The peptide was deprotected and cleaved from the resin in 1:2:2:35 ethanedithiol/thiophenol/thioanisole/95% trifluoroacetic acid after removal of the N-terminal Fmoc group. The crude peptide was precipitated with diethyl ether and then redissolved in 8 M guanidine hydrochloride before purification by semipreparative C4 reversed-phase HPLC at 50°C on a Rainin Instruments liquid chromatography system. The identity of the purified peptides was confirmed by electrospray MS using a Perkin–Elmer-Sciex API-300 instrument.
Randomly aggregated peptide was generated by drying a water solution of the peptide by blowing a stream of dry nitrogen over it until it was macroscopically dry. To convert the P101L and 3AV peptides to a fibrillar state, the samples were dissolved in a 50:50 (vol/vol) mixture of acetonitrile and acetate-buffered saline (100 mM NaCl, 20 mM NaOAc, pH 5.0). The solution then was allowed to sit undisturbed for ≈3 weeks. The solution gradually grew cloudy as an insoluble aggregate collected. Aggregation was judged complete at ≈3 weeks, after which the suspension was sedimented in an ultracentrifuge to collect the solid material. The supernatant was removed and the gelatinous pellet was dried overnight by blowing gaseous nitrogen over the sample. Electron microscopy was used as described (20) to verify the presence of fibrils in the aggregated forms.
All 13C NMR spectra were obtained either at 7.07 Tesla (corresponding to a 13C Larmour frequency of 75.74 MHz) on a custom spectrometer based on a Tecmag (Houston) pulse programmer with a Chemagnetics–Otysuka Electronics (Fort Collins, CO) 4-mm double-resonance magic-angle spinning (MAS) probe, or at 11.72 Tesla (corresponding to a 13C Larmor frequency of 125.75 MHz) on a triple-resonance Varian/Chemagnetics Infinity spectrometer with a 4-mm T3 triple-resonance MAS Chemagnetics probe. For spectra obtained at 7.07 Tesla, the cross-polarization (CP) contact time was 2.0 ms, the 1H decoupling field strength was 91 kHz, and the recycle delay was 1.0 sec. For spectra obtained at 11.72 Tesla, the CP contact time was 2.0 ms, the 1H decoupling field strength was 114 kHz, and the recycle delay was 1.0 sec. A spectrum of the fibrillar MoPrP 89–143 P101L also was obtained with two-pulse phase-modulation decoupling (25). Isotropic shift values were measured relative to the carbonyl carbon of glycine at 176.04 ppm in an external reference sample of glycine.
Results and Discussion
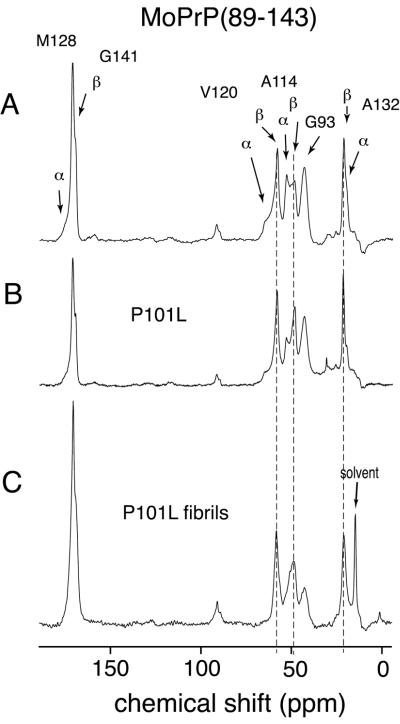
The 13C labels that were incorporated into MoPrP(89–143) are listed in Table 1 along with the expected chemical-shift ranges for each label. Different amino acid labels were used to minimize spectral overlap and to allow assignment of the different resonances. The locations of the isotopic labels were chosen so that the secondary structure of the peptide could be monitored at points distributed although much of the peptide. Fig. 2A shows a 13C CPMAS spectrum of the wt MoPrP(89–143) peptide. Resonance assignments were based on published 13C solid-state chemical-shift ranges (22). The distribution of shifts in the spectrum, including multiple lines for some sites, of the wt MoPrP(89–143) peptide indicates a mixture of helical and extended (β-sheet-like) conformers (identified in Fig. 2A). For V120 it is quite clear that there is a continuous distribution of shifts, indicating substantial conformational heterogeneity although extended conformers are predominant, which also seems to be the case for A132. For A114, both helical and extended conformation resonances are present, but with nearly equal intensity. Together these results indicate that the sample was not merely a mixture of helical and extended peptides, but peptides that adopt a variety of mixed secondary structures, i.e., microscopically disordered. This conformational heterogeneity in the solid state is not unexpected, given that solution NMR results suggest that PrP(89–143) is generally unstructured in solution (data not shown), as is the equivalent Syrian hamster (SHa) peptide (17) and smaller peptides from this part of the protein also had displayed conformational variability (19).
Table 1.
13C labels used to study the secondary structure of PrP peptides
| Label | Shift range, ppm |
|---|---|
| G93 Cα | 43 —44 |
| G113, 141 C0 | 168 —172 |
| V114, 120 Cα | 56 —66 |
| A114, 119 Cα | 46 —54 |
| A132 Cβ | 14 —22 |
| M128 C0 | 170 —176 |
Figure 2.
One hundred twenty-six-megahertz solid-state 13C CPMAS spectra of wt mouse PrP(89–143) obtained at a spinning speed of 10 kHz. (A) wt MoPrP(89–143) spectrum, with the sample dried from water. Arrows indicate the resonances, which occur at characteristic shifts for helical and extended (sheet) conformations. The resonance from the carbonyls of G141 in a helical conformation and M128 in the extended conformation overlap, and so cannot be distinguished. (B) Spectrum of MoPrP(89–143, P101L) dried from water. (C) Spectrum of MoPrP(89–143, P101L) peptide after conversion to fibrillar form by precipitation from acetonitrile/water. The lines superimposed over the resonances for V120, A114, and A132 mark the β-sheet chemical shifts. The higher ratio of β-sheet to helical resonances in the unaggregated mutant MoPrP(89–143, P101L) indicates the preference for this mutant to adopt β-sheet structures. This preference for β-sheet structures was enhanced by exposure of the mutant peptide to acetonitrile/water (C) where the helical resonances have almost disappeared.
In Fig. 2, the solid-state 13C NMR spectrum of wt MoPrP(89–143) is compared with the equivalent spectra of the mutant MoPrP(89–143, P101L) peptide both before (Fig. 2B) and after (Fig. 2C) conversion to the aggregated, fibrillar state. The chemical shifts of peaks in these spectra are listed in Table 2. The effects of the point mutation on the secondary structure of the peptide are quite significant; the intensities of the helical resonances for all of the 13C labels decrease, regardless of their location in the peptide. This effect is most striking for the 13Cα label in A114, for which the ratio of helix to β-sheet changes from ≈1:1 in the spectrum of the wt peptide to ≈1:2 in the spectrum of the P101L mutant. These results confirm that the PrP(89–143) fragment is structurally pliable and highly sensitive to mutations because a single amino acid substitution gives rise to conformational changes throughout the peptide, including amino acids over 30 residues away from the P101L substitution, as evidenced by the changes at A132.
Table 2.
Comparison of measured chemical shifts in wt and mutant MoPrP(89–143) peptides
| Label | Expected chemical
shifts
|
Experimental chemical shifts
|
||
|---|---|---|---|---|
| α-Helix | β-Sheet | wt* | P101L, converted | |
| G93 Cα | Unknown | 43.2 | 42.8 | 42.3 |
| A114 Cα | 52.5 | 48.7 | 52.6; 48.3 | 48.4 |
| V120 Cα | 65.5 | 58.3 | 64.1; 57.8 | 58.1 |
| A132 Cβ | 15.1 | 20.1 | 15.05; 20.88 | 20.6 |
| M128 C0 | 175.1 | 170.6 | 174.9; 170.6† | 170.4 |
| G141 C0 | 171.8 | 168.5 | 170.6†; 169.3 | 168.9 |
Typical α-helical and β-sheet chemical shifts of peptides in solids are listed. All chemical shifts are in ppm, externally referenced to tetramethylsilane via the 13C0 resonance in glycine (176.04 ppm).
Two conformations were present in the wt peptide. Shifts for both conformations are listed, with the α-helical chemical shifts listed first.
The M128 C0 sheet chemical shift overlaps with the G141 C0 helix chemical shift, and so the two cannot be distinguished in the spectra.
Upon aggregation, the P101L mutant peptide appears to convert completely to an extended, β-sheet-like form, as seen in Fig. 2C. However, the resonances in the spectrum of the fibrillar P101L peptide (Fig. 2C) appear broader than the randomly aggregated P101L peptide (Fig. 2B). The most likely explanation is that there is a distribution of extended conformations present, which would account for what appear to be shoulders on the A114 and G93 13Cα resonances. In an attempt to better resolve these resonances, another spectrum of the aggregated P101L peptide also was obtained by using two-pulse phase-modulation (25) to improve 1H decoupling and narrow the 13C resonances. The A114 13Cα resonance remained broadened, supporting the idea that there are multiple extended conformations. The downfield shoulder of the A114 13Cα resonance has a chemical shift that lies between those expected for a random coil and a β-sheet conformation (Table 2), which may reflect the presence of peptides in the aggregate that is irregular in structure. This finding is consistent with the observation of Kaneko et al. (8) that not all of the aggregated peptide is present in fibrillar form, and that a distribution of fibril lengths was present in the sample of the aggregated P101L peptide used for this study. Despite the presence of nonfibrillar peptide, the spectra show that β-sheet conformers are most highly populated and must reflect the dominant fibrillar form.
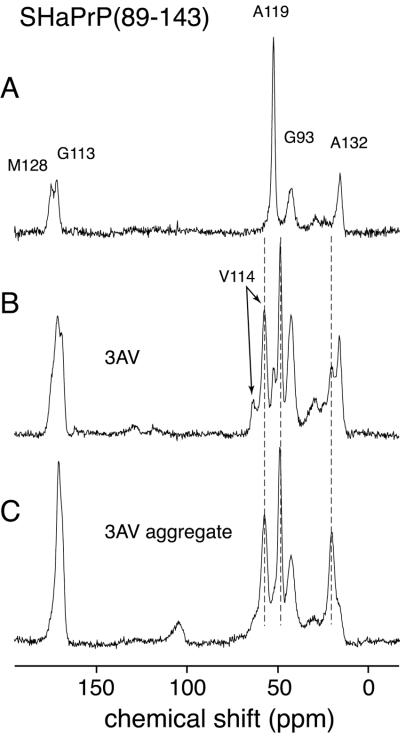
In addition to the P101L mutant of MoPrP(89–143), we examined the equivalent peptide from the SHaPrP sequence corresponding to residues 90–144. In this article we have chosen to use the equivalent mouse residue numbers to facilitate comparison with the mouse peptides. The SHa(89–143) peptide is relatively homogeneous in conformation when dried from water, with probes at positions A119 and A132 indicating helical conformations (Fig. 3A). This peptide did not aggregate or form fibrils upon treatment with acetonitrile/water and hence could not be studied in that form. A variant of the peptide carrying three alanine to valine mutations, at positions 112, 114, and 117, also was made, dramatically reducing the intrinsic helical propensity of the peptide. When dried from water, the SHa PrP(89–143, 3AV) peptide showed doubling of resonances, indicating two conformers, an extended form in addition to the helical form (Fig. 3B). The Cα labels at V114 and A119 indicate a predominant β-sheet conformation, with a smaller population of α-helix whereas A132 has a higher population of helical conformers than β-sheet, although β-sheet is substantially increased relative to the wt peptide. The mutant SHaPrP(89–143, 3AV) peptide has been shown to aggregate when left in cold acetonitrile/acetate-buffered saline solution, as does MoPrP(89–143,P101L), although long fibrils were not seen with electron microscopy. The spectra of the mutant SHaPrP(89–143, 3AV) peptide in the aggregated form show a further increase in the population of the extended conformers, very analogous to what was found for the MoPrP(89–143, P101L) peptide. Because of the sequence differences, some of the 13C isotopic labels were different from those used for the P101L peptide. The labels used are listed in Table 1. The chemical shifts of all of the 13C resonances in Fig. 3 are listed in Table 3. These results confirm the lability of the region in PrP containing residues 89–143 and show that the conformation and ability to form β-rich fibrils and aggregates are highly sensitive to point mutations. The mutations affect the global peptide conformation because sites far from the mutations show altered conformations. Although the conformational preference could be propagated through the peptide, it seems more likely that packing of peptides into aggregates is responsible for the long-range effects, and that the sequence effects reflect the ability of the peptides to form ordered quaternary structures.
Figure 3.
Seventy-six-megahertz solid-state 13C CPMAS spectra of the SHaPrP(89–143) peptide, acquired at a spinning speed of 10 kHz. (A) Spectrum of the wt SHaPrP(89–143) peptide dried from water. (B) Spectrum of the mutant SHaPrP(89–143, 3AV) peptide dried from water. (C) Spectrum of mutant SHaPrP(89–143, 3AV) peptide after conversion to an aggregated form by precipitation from acetonitrile/water. The lines superimposed over the resonances for V114, A119, and A132 mark the β-sheet chemical shifts.
Table 3.
Comparison of measured chemical shifts in wt and mutant SHaPrP(89–143)
| Label | Expected chemical
shifts
|
Experimental chemical shifts
|
||
|---|---|---|---|---|
| α-Helix | β-Sheet | wt | 3AV, converted | |
| G93 Cα | Unknown | 43.2 | 42.8 | 42.8 |
| G113 C0 | 171.8 | 168.5 | 171.7 | 169.5 |
| V114 Cα | 65.5 | 58.3 | — | 57.6 |
| A119 Cα | 52.5 | 48.7 | 52.0 | 49.2 |
| A132 Cα | 15.1 | 20.1 | 15.6 | 20.6 |
| M128 C0 | 175.1 | 170.6 | 174.7 | 170.8 |
Typical α-helical and β-sheet chemical shifts in solid-state peptides are listed. All chemical shifts are in ppm, externally referenced to tetramethylsilane via the 13C0 resonance in glycine (176.04 ppm).
Conclusions
Using solid-state 13C NMR, the secondary structure in PrP(89–143) peptides was probed in different aggregated states. The wt MoPrP(89–143) peptide exists in a variety of helical and extended conformations. Although the mutant MoPrP(89–143, P101L) also exhibits some degree of conformational heterogeneity, it is clear that the mutation drives the overall conformation of the peptide toward extended, β-sheet-like conformations. Furthermore, the fact that this effect occurs throughout the peptide indicates that residues 89–143 of the PrP are structurally pliable and that mutations within this region have long-range effects on the conformation in ordered states, which is very likely through cooperative formation of the fibrillar aggregates. The increased linewidth in the spectra of the fibrillar form suggests that some conformational heterogeneity may still exist, at least for some sections of the peptide. Analogous results were found for the wt and mutant SHaPrP peptides. When dried from water, the sites probed in wt SHaPrP(89–143) were found to be substantially helical; however, introduction of the valine to alanine mutations profoundly changed the structure of the peptide. Probing sites throughout the mutant SHaPrP(89–143, 3AV) peptide indicated an increased population of extended conformers with these β-rich forms dominating strongly in the fibrillar state.
The in vitro-prepared, fibrillar form of the MoPrP(89–143, P101L) peptide stimulates disease in transgenic mice whereas other forms do not, indicating that the formation of a specific β-sheet conformation may be necessary for the peptide to become “infectious.” This behavior of the mutant peptide correlates with observations on PrP in transgenic mice and humans with Gerstmann-Straussler-Scheinker disease. The mutant SHaPrP(89–143, 3AV) peptide formed aggregates rich in β-sheet like the MoPrP(89–143, P101L) peptide, whereas the wt SHaPrP(89–143) did not. The mutant SHaPrP(89–143, 3AV) peptide has not been found to cause disease in animals; however, introduction of mutations may cause an artificial “species barrier” and animals expressing a transgene with these mutations develop neurologic disease spontaneously (26). The present work makes clear that mutations strongly affect the conformational preferences of the peptides in ordered forms, and it seems likely that analogous effects occur through other disease-causing mutations. Even if the stimulation of neurodegeneration by the mutant MoPrP(89–143, P101L) peptide does not perfectly recapitulate all aspects of prion diseases, it seems to be a valuable model system for understanding features of the conformational change that PrPc undergoes as it is converted into PrPSc.
Acknowledgments
D.D.L. was supported by a Howard Hughes Medical Institute predoctoral fellowship. We also acknowledge funding through National Institutes of Health Grant AG-10770, and support through the Director, Office of Energy Research, Office of Basic Energy Sciences, Materials Sciences Division of the U.S. Department of Energy under Contract No. DE-AC03–76SF00098.
Abbreviations
- PrP
prion protein
- MoPrP
mouse PrP
- SHaPrP
Syrian hamster PrP
- wt
wild type
- PrPC
Prp cellular form
- PrPSc
PrP scrapie form
- MAS
magic-angle spinning
- CP
cross-polarization
References
- 1.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 4.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 6.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko K, Ball H L, Wille H, Zhang H, Groth D, Torchia M, Trembley P, Safar J, Prusiner S B, De Armond S J, et al. J Mol Biol. 2000;295:997–1007. doi: 10.1006/jmbi.1999.3386. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner S B, Groth D, Bolton D C, Kent S B, Hood L E. Cell. 1984;38:127–134. doi: 10.1016/0092-8674(84)90533-6. [DOI] [PubMed] [Google Scholar]
- 10.Kitamoto T, Iizuka R, Tateishi J. Biochem Biophys Res Commun. 1993;192:525–531. doi: 10.1006/bbrc.1993.1447. [DOI] [PubMed] [Google Scholar]
- 11.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen H O, Heinrich C, Torchia M, Safar J, et al. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 12.Muramoto T, Scott M, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasset M, Baldwin M, Lloyd D H, Gabriel J-M, Holtzman D M, Cohen F E, Fletterick R, Prusiner S B. Proc Natl Acad Sci USA. 1992;89:10940–10944. doi: 10.1073/pnas.89.22.10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Come J H, Fraser P E, Lansbury P T. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forloni G, Angeretti N, Chiesa R, Monzani E, Salmoni M, Bugiani O, Tagliavini F. Nature (London) 1993;362:543–546. doi: 10.1038/362543a0. [DOI] [PubMed] [Google Scholar]
- 16.Tagliavini F, Prelli F, Verga L, Giaccone G, Sarma R, Gorevic P, Ghetti B, Passerinin F, Ghibaudi E, Forloni G, et al. Proc Natl Acad Sci USA. 1993;90:9678–9682. doi: 10.1073/pnas.90.20.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Kaneko K, Nguyen J, Livshits T L, Baldwin M, Cohen F E, James T L, Prusiner S B. J Mol Biol. 1995;250:514–526. doi: 10.1006/jmbi.1995.0395. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen J, Baldwin M, Cohen F E, Prusiner S B. Biochemistry. 1995;34:4186–4192. doi: 10.1021/bi00013a006. [DOI] [PubMed] [Google Scholar]
- 19.Heller J, Kolbert A C, Larsen R, Ernst M, Bekker T, Baldwin M, Prusiner S B, Pines A, Wemmer D E. Protein Sci. 1996;5:1655–1661. doi: 10.1002/pro.5560050819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko K, Wille H, Mehlhorn I, Zhang H, Ball H L, Cohen F E, Baldwin M, Prusiner S B. J Mol Biol. 1997;270:574–586. doi: 10.1006/jmbi.1997.1135. [DOI] [PubMed] [Google Scholar]
- 21.Saito H, Tabeta R, Shojoi A, Ozaki T. Macromolecules. 1983;16:1050–1057. [Google Scholar]
- 22.Saito H. Magn Reson Chem. 1986;24:835–852. [Google Scholar]
- 23.Spera S, Bax A. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 24.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 25.Bennett A E, Rienstra C M, Auger M, Lakshmi K V, Griffin R G. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 26.Hegde R S, Tremblay P, Groth D, DeArmond S J, Prusiner S B, Lingappa V R. Nature (London) 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]