Abstract
The ALK gene encodes a transmembrane tyrosine kinase receptor. ALK is physiologically expressed in the nervous system during embryogenesis, but its expression decreases postnatally. ALK first emerged in the field of oncology in 1994 when it was identified to fuse to NPM1 in anaplastic large‐cell lymphoma. Since then, ALK has been associated with other types of cancers, including non‐small‐cell lung cancer (NSCLC). More than 19 different ALK fusion partners have been discovered in NSCLC, including EML4, KIF5B, KLC1, and TPR. Most of these ALK fusions in NSCLC patients respond well to the ALK inhibitor, crizotinib. In this paper, we reviewed fusion partner genes with ALK, detection methods for ALK‐rearrangement (ALK‐R), and the ALK‐tyrosine kinase inhibitor, crizotinib, used in NSCLC patients.
Keywords: ALK‐rearrangement (ALK‐R), ALK‐tyrosine kinase inhibitor (TKI), anaplastic lymphoma kinase (ALK), detection platforms, non‐small‐cell lung cancer (NSCLC)
The ALK gene
The ALK gene is located on the short arm of chromosome 2 (2p23), belongs to the insulin receptor superfamily, and encodes for the ALK protein (Fig 1a). ALK is a transmembrane tyrosine kinase receptor, and like other receptor tyrosine kinases, it has an extracellular domain, a transmembrane segment, and a cytoplasmic receptor kinase segment (Fig 1a–c).1, 2 ALK expression occurs in the nervous system during embryo genesis and decreases in postnatal life. Therefore, in human adults, low levels of ALK protein are produced only in rare, scattered neural and endothelial cells and in pericytes in the brain.3, 4
Figure 1.
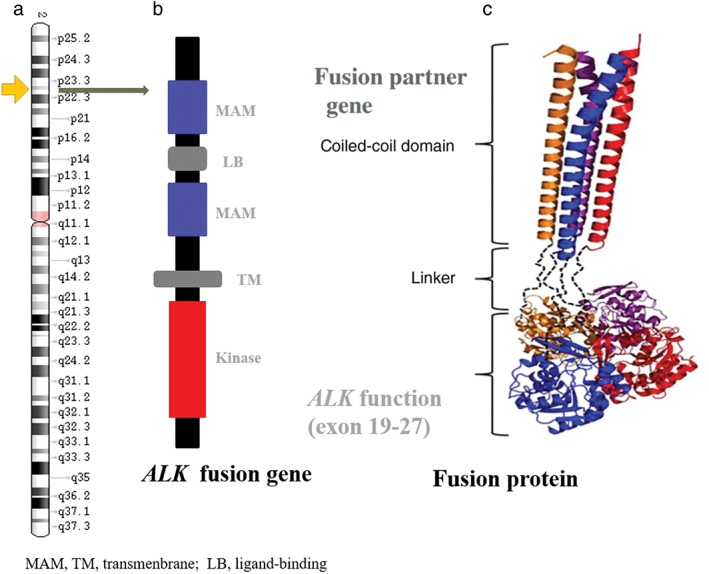
(a) The ALK gene location in the genome; (b) structural organization of ALK protein; and (c) the domain of the fusion protein.
Types of oncogenesis in ALK
There are three types of ALK gene mutations: rearrangement (ALK‐R), amplification (ALK‐A), and point mutation.
Most mutations of the ALK gene are in the form of a translocation with another partner gene leading to a fusion oncogene. This fusion gene then becomes overly expressed in cancers. In 1994, ALK was originally identified in anaplastic large‐cell lymphoma as a fusion partner of nucleophosmin (NPM‐ALK) resulting from a chromosomal translocation.5 Subsequently, ALK‐rearrangement (ALK‐R) was identified in many different cancers, including inflammatory myofibroblastic tumors, diffuse large B‐cell lymphoma, non‐small‐cell lung cancer (NSCLC), and esophageal squamous cell, colorectal, and breast carcinomas.6, 7 ALK rearrangements create an oncogenic ALK tyrosine kinase that activates many downstream signaling pathways resulting in increased cell proliferation and survival.8 Additional gene partners have been discovered in fusion oncogenes with the ALK gene, including TPM3, TFG, CLTCL1, and ATIC (Table 1).9
Table 1.
ALK gene mutations and the disease they represent
| ALK‐R | ALK‐A (disease) | Main point mutation | |
|---|---|---|---|
| Disease | Partner Gene | ||
| Anaplastic large cell lymphoma | NPM1 | Inflammatory breast cancer | L1196M |
| Inflammatory myofibroblastic tumors | TPM3/4 | Small cell lung cancer | C1156Y |
| Diffuse large B‐cell lymphoma | TFG | Anaplastic large cell lymphoma | G1269A |
| Non‐small cell lung cancer | EML4 | Pulmonary sarcomatoid carcinoma | F1174L |
| Esophageal squamous cell carcinoma | CLTCL1 | Rhabdomyosarcoma | L1152R |
| Colorectal carcinoma | ATIC | Carcinoma of the esophagus | F1245C |
| Renal medullary carcinoma | VCL | Adult renal cell carcinoma | G1201E |
Another type of ALK gene mutation is ALK‐A. The oncogenic mechanism of ALK‐A was first described in NB cell lines in 2002. The study showed that ALK‐A leads to constitutive activation, resulting in the selective activation of SHcC, a docking protein close to the substrate of the ALK receptor.10 Several studies have reported extra copies of the ALK gene in inflammatory breast cancer, NSCLC, anaplastic large‐cell lymphoma, and pulmonary sarcomatoid carcinoma.
The last type of ALK gene mutation is point mutation. Secondary resistance is an acquired mechanism after the tumor has been exposed to an ALK inhibitor2 and most types of resistance are caused by mutations in the target ALK gene, resulting in an inability to inhibit the encoded tyrosine kinase.11 The first drug resistance point mutations identified were C1156Y and L1196M.12 Subsequently, several other point mutations conferring drug resistance have been identified, including: G1269A, F1174L, 1151Tins, L1152R, S1206Y, I1171T, G1202, D1203N, and V1180L.11, 12, 13, 14
ALK rearrangement in non‐small cell lung cancer (NSCLC)
Non‐small‐cell lung cancer accounts for approximately 80–85% of lung cancers and is a leading cause of cancer‐related mortality in both men and women worldwide.15, 16, 17, 18 ALK gene rearrangement is a driving mutation underlying the development of NSCLC, and has been identified in 5–6% of NSCLC cases.19 Notwithstanding the substantial evidence linking activated ALK to tumor genesis in these rare tumors, it is fair to say that the considerable current enthusiasm for ALK as a target for cancer therapy is largely driven by the relatively recent finding of a recurring ALK gene translocation in a significant subset of NSCLC.20, 21 ALK rearrangement appears to be more common in younger patients and never or light smokers diagnosed with adenocarcinoma. Data from several patient series has shown that the median age of ALK positive NSCLC patients is 55 years and approximately 70% of these patients are never smokers. The incidence of ALK positive NSCLC among men and women is similar across the world.22, 23
ALK mutations were first described in NSCLC in 2007 when a subset (7%) of Japanese patients were found to have EML4 rearrangement with ALK leading to the fusion oncogene EML4‐ALK.24 This rearrangement was an inversion rearrangement from inv.(2) (p21;p23) that results in EML4 replacing the extracellular and intramembranous parts of ALK and fusing with the juxtamembrane domain. The EML4‐ALK fusion gene represents a new molecular target. It has been reported that the incidence of ALK rearrangement ranges from approximately 3% to 13% in unselected or selected patients with NSCLC.23, 25, 26, 27 Because of the different breakpoints on EML4, several variants of the EML4‐ALK mutation have been described (Table 2).27, 28, 29 EML4‐ALK variants with differing frequencies are V1 (54.5%), V2 (10%), V3a/V3b (34%), and V5a (1.5%).28, 29
Table 2.
EML4‐ALK variant fusions
| Variants | EML4‐ALK Fusion Types | Number of types | Frequency (%) |
|---|---|---|---|
| E13;A20 | E13;A20(variant 1), E13; ins69A20, E13;ins69A20, E13;exoc6bA20 | 4 | 33 |
| E6;A20 | E6;A20(variant 3a), E6ins33;A20(variant 3b), E6;ins18A20 | 3 | 29 |
| E20;A20 | E20;A20(variant 2), E20;ins18A20 | 2 | 9 |
| E18;A20 | E18;A20(variant 5′) | 1 | 2 |
| E14;A20 | E14;ins11del49A20 (variant 4), E14del12A20 (variant 7), E14;del14A20, E14;del36A20,E14;del38A20, E14ins21;del113A20 | 5 | 3 |
| E15;A20 | E15del19;del20A20 (variant 4′), E15del60;del71A20 | 2 | 2 |
| E2;A20 | E2;A20(variant 5a), E2;ins117A20(variant 5a/b) | 2 | 2 |
| E17;A20 | E17;ins68A20, E17ins65;A20, E17;ins30A20 (variant 8a), E17del58;ins39A20, E17ins61;ins34A20 (variant 8b) | 5 | 1 |
| E3;A20 | E3;ins69A20(variant 6), E3;ins53A20 | 2 | 1 |
| E6;A19 | E6;A19 | 1 | < 1 |
| E21;A20 | E21;A20 | 1 | < 1 |
| E10del54E13;A20 | E10del54E13;A20 | 1 | < 1 |
| E6;A17 | E6;A17 | 1 | < 1 |
| Total | 30 |
EML4‐ALK translocation can result in constitutive ALK kinase activity and represents an oncogenic addiction pathway in lung cancer. The EML4‐ALK gene induced tumor formation in nude mice.24, 30 EML4‐ALK possesses potent oncogenic activity both in vitro and in vivo, and the tumor can quickly be reduced after the administration of ALK‐tyrosine kinase inhibitors (TKIs).24, 31
EML4‐ALK fusion protein serves as a therapeutic target for an ALK‐TKI, and has shown promising results when used to treat NSCLC patients carrying ALK rearrangement.32, 33, 34, 35 Over the last few years, ALK inhibitors have shown significant benefits in the management of ALK‐positive NSCLC compared to conventional chemotherapy.21, 34, 36
Rearrangements of the ALK gene with partner genes other than EML4 have been described, namely, KIF5B, KLC1, TFG, TPR, HIP1, STRN, DCTN1, SQSTM1, NPM1, BCL11A, and BIRC6 (Table 3).37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50 Targeted therapeutic agents, including the TKI crizotinib, have shown clinical efficacy in treating NSCLC patients harboring EML4‐ALK gene fusion.34 Furthermore, a previous study demonstrated that crizotinib is also effective at treating tumors harboring ALK fused with other partner genes, including NPM1 and BCL11A.34 In addition, other not‐yet‐characterized fusions may also exist in solid tumors, including lung cancer.51
Table 3.
Fusion details of ALK partner genes
| Fusion partner gene | Reported year | Oncogenetic driver | TKI PFS | Variants | FISH result | IHC result | First report (ref) |
|---|---|---|---|---|---|---|---|
| EML4 | 2007 | Yes | — | > 30 types | — | — | 24 |
| TFG | 2007 | — | — | T6;A20 | — | — | 39 |
| KIF5B | 2009 | Yes | — | K24;A20/K15:A20 | Positive | Positive | 40 |
| KLC1 | 2012 | Yes | — | K9:A20 | Positive | Positive | 41 |
| PTPN3 | 2012 | — | Unknown | P2;A10–11;P3 | — | — | 45 |
| HIP1 | 2014 | Yes | 5M | H21;A20/H28;A20/H30:A20 | — | — | 50 |
| TPR | 2014 | — | Unknown | T15;A20 | Positive | Positive | 49 |
| BIRC6 | 2015 | Yes | > 9M | B10;A20 | Negative | Positive | 48 |
| DCTN1 | 2015 | — | Unknown | D26;A20 | Negative | — | 37 |
| SQSTM1 | 2015 | — | Unknown | S5;A20 | Negative | — | 37 |
| PRKAR1A | 2016 | Yes | 7M | P5;A20 | Positive | Positive | 44 |
| PPM1B | 2016 | Yes | Sensitivity | P1;A20 | Negative | — | 44 |
| EIF2AK3 | 2016 | Yes | 28M | E2;A20 | Negative | Negative (D5F3 and 5A4) | 44 |
| BCL11A | 2017 | Yes | > 6M | B4;A20 | — | — | 43 |
| CEBPZ | 2017 | Yes | Unknown | C3;A20 | Negative/fused signals | Positive | 42 |
| PICALM | 2017 | Yes | Unknown | P19;A20 | Negative/fused signals | Positive | 42 |
| GCC2 | 2017 | — | — | G12;A20 | Positive | Positive | 47 |
| LMO7 | 2017 | — | — | L15;A20 | Positive | Positive | 47 |
| PHACTR1 | 2017 | — | — | PH7;A20 | Positive | Positive | 47 |
| CMTR1 | 2017 | No | Drug resistant | C2;A20 | Negative | Positive | Under review |
FISH, fluorescence in situ hybridization IHC, immunohistochemical.
ALK rearrangement detection methods in NSCLC patients
ALK rearrangements may involve distinct break points and multiple fusion partners. Therefore, routine ALK testing presents a significant technical challenge. There are four primary methods of detecting ALK rearrangement: fluorescence in situ hybridization (FISH), immunohistochemical (IHC), reverse transcriptase‐PCR (RT‐PCR) and next generation sequencing (NGS). Each of these methods has both advantages and limitations.
Fluorescence in situ hybridization break‐apart assay is considered the gold standard for the evaluation of ALK status and is the first approved diagnostic test for ALK rearrangement to detect break‐apart signals, although IHC and RT‐PCR have also been evaluated for this purpose, with the former approved by the United States Food and Drug Administration (US FDA) in June 2015 (Table 4).19, 52 FISH relies on a spatial separation of the 5′‐ and 3′‐ portions of the ALK gene upon rearrangement, and produces characteristic spilt ALK‐specific signals in case of the translocation. The FISH break‐apart assay is currently the most reliable approach to ALK testing, but has a number of critical disadvantages. In particular, FISH requires significant time input of extensively trained personnel and cannot be subjected to reasonable automation; furthermore, it demonstrates relatively high failure rates in some sample series and may provide poorly interpretable results in a noticeable fraction of NSCLC cases.53, 54, 55 Despite these challenges, FISH is still regarded as the gold standard assay for the detection of ALK rearrangements and a comparator for the other ALK detection methods.
Table 4.
Comparison of the four methods used to detect ALK fusion
| FISH | IHC | RT‐PCR | NGS | |
|---|---|---|---|---|
| Fusion types detectable | No fusion specification | No fusion specification | Only EML4‐ALK fusion | All kinds of fusion |
| Sensitivity | 10–15% | 5–10% | 1–5% | 1–5% |
| Time used for analysis | 2–3 days | 0.5 days | 1 days | 5–7 days |
| Cost | Medium (~$349) | Low (~$31.5) | Medium (~$879) | High (~$945) |
| Is FFPE material applicable? | Yes | Yes | Yes | Yes |
| Is fresh tissue material applicable? | No | No | Yes | Yes |
| Amount of material required | One tissue section (3 μm thick) | One tissue section (3 μm thick) | 0.1–0.5 μg of RNA | 2–3 μg of DNA |
| Possibility to see large range of other gene mutations in one analysis | No | No | No | Yes |
| Requirement for technical skill | Medium | Low | Medium | High |
| Requirement for diagnostician | High | Medium | Medium | High |
| Applicability to average pathology laboratory | Most laboratories | All laboratories | Some laboratories | Some laboratories |
FFPE, formalin fixed paraffin‐embedded; FISH, fluorescence in situ hybridization IHC, immunohistochemical; NGS, next generation sequencing; RT, reverse transcriptase.
The development of highly sensitive ALK diagnostic antibodies has offered an opportunity to detect ALK‐driven tumors by a standard IHC method. One of the main advantages of IHC in comparison to FISH and RT‐PCR is the detection of the ALK protein, which is the target of ALK inhibitors. Other advantages of IHC are its low cost, short turnaround time, and ease of operation for users. The principle of IHC is based on the fact that activating ALK rearrangements are accompanied by significant overexpression of the catalytic portion of this tyrosine kinase. IHC is generally capable of producing highly reliable results when performed in reference laboratories; however, it requires the standardization of reagents and protocols across pathology laboratories.55, 56, 57, 58 The Ventana ALK assay used D5F3 antibody is a resultful method of detecting ALK rearrangement. The Ventana ALK (D5F3) CDx Assay (Ventana Medical Systems, Tucson, AZ, USA) was approved by the US FDA in 2015 as a companion detection test for the use of crizotinib.59 Several studies have found that there is high concordance between Ventana IHC and FISH.60, 61 A research analysis of 46 ALK‐positive patients reported sensitivity and specificity of the Ventana IHC of 100% and 98.2%, respectively, and the concordance rate between FISH and Ventana IHC was 98.4%.62 Although the sensitivity of IHC is high for detecting ALK fusion, FISH‐positive/IHC‐negative cases responding to ALK inhibitors have been reported in the literature.63
Reverse transcriptase‐PCR based assays have not been as widely used as FISH and IHC for ALK testing in NSCLC. However, conventional RT‐PCR has significant advantages compared to FISH and IHC. First of all, while FISH and IHC detect relatively indirect signs of the presence of ALK translocation, RT‐PCR usually reveals the exact variant of the rearrangement and therefore provides definitive evidence of ALK fusion. Furthermore, RT‐PCR has high sensitivity and specificity, with a rapid turnaround time and ease of analysis, and can detect a small number (1%) of ALK‐driven NSCLC cells in the presence of normal tissues.64, 65, 66 In addition, RT‐PCR analysis utilizes the same technical platform as other kinds of molecular NSCLC diagnosis, for example EGFR testing. Finally, a number of commercial RT‐PCR kits for detection of ALK rearrangements have been developed recently, including: the ALK RGQ RT‐PCR Kit (Qiagen, Valencia, CA, USA); the EML4‐ALK Fusion Gene Detection Kit (Amoy Diagnostics, Xiamen, China); and EML4 ALK Gene Fusion, PCR (Quest Diagnostics, Secaucus, NJ, USA).37, 67 However, there are some disadvantages of this platform. First, this method of analysis of RNA samples yields a poor quality of RNA obtained from formalin fixed paraffin‐embedded specimens and can only detect known fusion variants. Second, the high sensitivity may lead to a false‐positive result. In a previous study, the sensitivity and specificity of RT‐PCR were 95.5% and 87.0%, respectively, and the concordance rate between FISH and RT‐PCR was 89.0%.62
Next generation sequencing is a promising method for detecting ALK gene rearrangements. The great advantage of the NGS platform is the detection of known ALK gene fusions. NGS is also superior to other methods because it allows for simultaneous screening of novel ALK fusion partners as well as other lung cancer related gene mutations, fusions, and amplifications. However, there are still many challenges to overcome before this method can be applied to normal laboratory diagnosis of pathology. For example, expertise is needed to analyze and interpret the results, and the cost and turnaround time are high.68 As NGS has not yet been approved by the US FDA, it can only be used in conjunction with other methods.
All four methodologies show good sensitivity, specificity, and concordance when artifacts were characterized and excluded. However, the choice of diagnostic methodology for ALK rearrangement detection in clinical practice remains a matter of debate. In ambiguous cases at least two of the four methods should be used to confirm ALK rearrangement.
ALK inhibitors
Crizotinib
ALK rearrangements in NSCLC have introduced new treatment options for advanced NSCLC with the use of ALK‐TKIs.24 ALK fusion proteins can activate many different interconnected and overlapping pathways, such as Ras/Raf/MEK/ERK1/2, JAK/STAT, PI3K/Akt, and PLC‐γ pathways, all of which are involved in cell migration, proliferation, and survival.8 In addition, several ALK fusion partners have been identified. However, regardless of the involved partners, all chimeras retain the ALK gene kinase domain responsible for the constitutive activation of ALK signaling pathways.8
Crizotinib is the first ALK inhibitor to enter clinical trials. Crizotinib is a multi‐targeted TKI69 with activity against MET, ALK, and ROS1, and was approved by the US FDA in 2011 for metastatic NSCLC positive for ALK rearrangements.34, 70, 71 Crizotinib has a reported response rate of over 60% and a disease control rate of up to 90%.34 Furthermore, median progression‐free survival (PFS) exceeds nine months, and median overall survival is almost 75% after one year in ALK‐rearranged NSCLC.72 Comparison of crizotinib treatment with a historical control is instructive.38 Therefore, from identification to inhibitor approval, the story of ALK in NSCLC stands as a testament of the promises of targeted molecular medicine.22
Next generation ALK inhibitors
Unfortunately, almost all patients treated with crizotinib develop tumor progression. As such, potent inhibitors of ALK that can overcome resistance to crizotinib are needed. Several agents have been evaluated in patients with crizotinib refractory NSCLC. Ceritinib and alectinib are currently approved in the US, and brigatinib has received breakthrough designation by the US FDA. The response rates with these agents in patients with crizotinib drug resistance are 50–55%, and the median PFS rates are 6.9 for ceritinib, 8.9 for alectinib, and 15.6 months for brigatinib.73, 74, 75 Ensartinib is another next generation ALK inhibitor. Ensartinib activity was not only observed in crizotinib‐resistant patients but also yielded results in patients who had previously been administered more than two alternate ALK inhibitors.76
The ASCEND‐4 trial evaluated the effects of ceritinib or chemotherapy in randomized ALK‐positive treatment naïve patients.77 The median PFS with ceritinib was 16.6 versus 8.1 months in patients treated with chemotherapy. In the ASCEND‐5 trial, patients who initially received chemotherapy and crizotinib were randomized for further treatment of ceritinib or chemotherapy. Results demonstrated a significant improvement in PFS, with a median of 5.4 months after the administration of ceritinib compared to 1.6 months with chemotherapy.78 These data reveal that ceritinib is the preferred treatment for ALK‐positive NSCLC patients.
Disclosure
No authors report any conflict of interest.
Acknowledgment
This research was supported by the National Natural Science Foundation of China (grant number 81372834).
Contributor Information
Yan‐Hong Tai, Email: taiyanhong29@163.com.
Hong‐Jun Gao, Email: gaohj_307@163.com.
References
- 1. Duyster J, Bai RY, Morris SW. Translocations involving anaplastic lymphoma kinase (ALK). Oncogene 2001; 20: 5623–37. [DOI] [PubMed] [Google Scholar]
- 2. Wu JJ, Savooji J, Liu DL. Second‐ and third‐generation ALK inhibitors for non‐small cell lung cancer. J Hematol Oncol 2016; 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Motegi A, Fujimoto J, Kotani M, Sakuraba H, Yamamoto T. ALK receptor tyrosine kinase promotes cell growth and neurite outgrowth. J Cell Sci 2004; 117: 3319–29. [DOI] [PubMed] [Google Scholar]
- 4. Iwahara T, Fujimoto J, Wen D et al Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 1997; 14: 439–49. [DOI] [PubMed] [Google Scholar]
- 5. Morris SW, Kirstein MN, Valentine MB et al Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non‐Hodgkins‐lymphoma. Science 1994; 263: 1281–4 (Published erratum appears in Science 1995;267:316–7).8122112 [Google Scholar]
- 6. Barreca A, Lasorsa E, Riera L et al Anaplastic lymphoma kinase in human cancer. J Mol Endocrinol 2011; 47: R11–23. [DOI] [PubMed] [Google Scholar]
- 7. Franco R, Rocco G, Marino FZ et al Anaplastic lymphoma kinase: A glimmer of hope in lung cancer treatment? Expert Rev Anticancer Ther 2013; 13: 407–20. [DOI] [PubMed] [Google Scholar]
- 8. Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer 2008; 8: 11–23. [DOI] [PubMed] [Google Scholar]
- 9. Boi M, Zucca E, Inghirami G, Bertoni F. Advances in understanding the pathogenesis of systemic anaplastic large cell lymphomas. Br J Haematol 2015; 168: 771–83. [DOI] [PubMed] [Google Scholar]
- 10. Miyake I, Hakomori Y, Shinohara A et al Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene 2002; 21: 5823–34. [DOI] [PubMed] [Google Scholar]
- 11. Toyokawa G, Seto T. Updated evidence on the mechanisms of resistance to ALK inhibitors and strategies to overcome such resistance: Clinical and preclinical data. Oncol Res Treat 2015; 38: 291–8. [DOI] [PubMed] [Google Scholar]
- 12. Choi YL, Soda M, Yamashita Y et al EML4‐ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010; 363: 1734–9. [DOI] [PubMed] [Google Scholar]
- 13. Heuckmann JM, Hölzel M, Sos ML et al ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res 2011; 17: 7394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasaki T, Okuda K, Zheng W et al The neuroblastoma‐associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK‐translocated cancers. Cancer Res 2010; 70: 10038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 16. Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev 2010; 19: 1893–907. [DOI] [PubMed] [Google Scholar]
- 17. Wen M, Wang X, Sun Y et al Detection of EML4‐ALK fusion gene and features associated with EGFR mutations in Chinese patients with non‐small‐cell lung cancer. Onco Targets Ther 2016; 9: 1989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 19. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015; 16: E342–51. [DOI] [PubMed] [Google Scholar]
- 20. Toyokawa G, Seto T. ALK inhibitors: What is the best way to treat patients with ALK+ non‐small‐cell lung cancer? Clin Lung Cancer 2014; 15: 313–9. [DOI] [PubMed] [Google Scholar]
- 21. Shaw AT, Kim DW, Nakagawa K et al Crizotinib versus chemotherapy in advanced ALK‐positive lung cancer. N Engl J Med 2013; 368: 2385–94. [DOI] [PubMed] [Google Scholar]
- 22. Shaw AT, Engelman JA. ALK in lung cancer: Past, present, and future. J Clin Oncol 2013; 31: 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shaw AT, Yeap BY, Mino‐Kenudson M et al Clinical features and outcome of patients with non‐small‐cell lung cancer who harbor EML4‐ALK. J Clin Oncol 2009; 27: 4247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soda M, Choi YL, Enomoto M et al Identification of the transforming EML4‐ALK fusion gene in non‐small‐cell lung cancer. Nature 2007; 448: 561–6. [DOI] [PubMed] [Google Scholar]
- 25. Sun YH, Ren Y, Fang Z et al Lung adenocarcinoma from East Asian never‐smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 2010; 28: 4616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Inamura K, Takeuchi K, Togashi Y et al EML4‐ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008; 3: 13–7. [DOI] [PubMed] [Google Scholar]
- 27. Horn L, Pao W. EML4‐ALK: Honing in on a new target in non‐small‐cell lung cancer. J Clin Oncol 2009; 27: 4232–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi YL, Takeuchi K, Soda M et al Identification of novel isoforms of the EML4‐ALK transforming gene in non‐small cell lung cancer. Cancer Res 2008; 68: 4971–6. [DOI] [PubMed] [Google Scholar]
- 29. Li TH, Maus MK, Desai SJ et al Large‐scale screening and molecular characterization of EML4‐ALK fusion variants in archival non‐small‐cell lung cancer tumor specimens using quantitative reverse transcription polymerase chain reaction assays. J Thorac Oncol 2014; 9: 18–25. [DOI] [PubMed] [Google Scholar]
- 30. Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013; 13: 685–700 (Published erratum appears in Nat Rev Cancer 2013; 13:820). [DOI] [PubMed] [Google Scholar]
- 31. Soda M, Takada S, Takeuchi K et al A mouse model for EML4‐ALK‐positive lung cancer. Proc Natl Acad Sci U S A 2008; 105: 19893–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koivunen JP, Mermel C, Zejnullahu K et al EML4‐ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008; 14: 4275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McDermott U, Iafrate AJ, Gray NS et al Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res 2008; 68: 3389–95. [DOI] [PubMed] [Google Scholar]
- 34. Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non‐small‐cell lung cancer. N Engl J Med 2010;363:1693–703. (Published erratum appears in N Engl J Med 2011; 364: 588). [DOI] [PMC free article] [PubMed]
- 35. Shaw AT, Yeap BY, Solomon BJ et al Effect of crizotinib on overall survival in patients with advanced non‐small‐cell lung cancer harbouring ALK gene rearrangement: A retrospective analysis. Lancet Oncol 2011; 12: 1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iragavarapu C, Mustafa M, Akinleye A et al Novel ALK inhibitors in clinical use and development. J Hematol Oncol 2015; 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iyevleva AG, Raskin GA, Tiurin VI et al Novel ALK fusion partners in lung cancer. Cancer Lett 2015; 362: 116–21. [DOI] [PubMed] [Google Scholar]
- 38. Mano H. ALKoma: A cancer subtype with a shared target. Cancer Discov 2012; 2: 495–502. [DOI] [PubMed] [Google Scholar]
- 39. Rikova K, Guo A, Zeng Q et al Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007; 131: 1190–203. [DOI] [PubMed] [Google Scholar]
- 40. Takeuchi K, Choi YL, Togashi Y et al KIF5B‐ALK, a novel fusion oncokinase identified by an immunohistochemistry‐based diagnostic system for ALK‐positive lung cancer. Clin Cancer Res 2009; 15 (9): 3143. [DOI] [PubMed] [Google Scholar]
- 41. Togashi Y, Soda M, Sakata S et al KLC1‐ALK: A novel fusion in lung cancer identified using a formalin‐fixed paraffin‐embedded tissue only. PLoS One 2012; 7: e31323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li WB, Zhang J, Guo L, Chuai S, Shan L, Ying J. Combinational analysis of FISH and immunohistochemistry reveals rare genomic events in ALK fusion patterns in NSCLC that responds to crizotinib treatment. J Thorac Oncol 2017; 12: 94–101. [DOI] [PubMed] [Google Scholar]
- 43. Tian Q, Deng WJ, Li ZW. Identification of a novel crizotinib‐sensitive BCL11A‐ALK gene fusion in a nonsmall cell lung cancer patient. Eur Respir J 2017; 49: pii:1602149. [DOI] [PubMed] [Google Scholar]
- 44. Ali SM, Hensing T, Schrock AB et al Comprehensive genomic profiling identifies a subset of crizotinib‐responsive ALK‐rearranged non‐small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist 2016; 21: 762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jung Y, Kim P, Jung Y et al Discovery of ALK‐PTPN3 gene fusion from human non‐small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer 2012; 51: 590–7. [DOI] [PubMed] [Google Scholar]
- 46. Drilon A, Wang L, Arcila ME et al Broad, hybrid capture‐based next‐generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res 2015; 21: 3631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noh KW, Lee MS, Lee SE et al Molecular breakdown: A comprehensive view of anaplastic lymphoma kinase (ALK)‐rearranged non‐small cell lung cancer. J Pathol 2017; 243: 307–19. [DOI] [PubMed] [Google Scholar]
- 48. Shan L, Jiang P, Xu F et al BIRC6‐ALK, a novel fusion gene in ALK break‐apart FISH‐negative lung adenocarcinoma, responds to crizotinib. J Thorac Oncol 2015; 10: E37–9. [DOI] [PubMed] [Google Scholar]
- 49. Choi YL, Lira ME, Hong M et al A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol 2014; 9: 563–6. [DOI] [PubMed] [Google Scholar]
- 50. Fang DD, Zhang B, Gu Q et al HIP1‐ALK, a novel ALK fusion variant that responds to crizotinib. J Thorac Oncol 2014; 9: 285–94. [DOI] [PubMed] [Google Scholar]
- 51. Mano H. Non‐solid oncogenes in solid tumors: EML4‐ALK fusion genes in lung cancer. Cancer Sci 2008; 99: 2349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol 2015; 12: 511–26. [DOI] [PubMed] [Google Scholar]
- 53. Iwama E, Okamoto I, Harada T, Takayama K, Nakanishi Y. Development of anaplastic lymphoma kinase (ALK) inhibitors and molecular diagnosis in ALK rearrangement‐positive lung cancer. Onco Targets Ther 2014; 7: 375–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thunnissen E, Bubendorf L, Dietel M et al EML4‐ALK testing in non‐small cell carcinomas of the lung: A review with recommendations. Virchows Arch 2012; 461: 245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tembuyser L, Tack V, Zwaenepoel K et al The relevance of external quality assessment for molecular testing for ALK positive non‐small cell lung cancer: Results from two pilot rounds show room for optimization. PLoS One 2014; 9: e112159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marchetti A, Ardizzoni A, Papotti M et al Recommendations for the analysis of ALK gene rearrangements in non–small‐cell lung cancer: A consensus of the Italian Association of Medical Oncology and the Italian Society of Pathology and Cytopathology. J Thorac Oncol 2013; 8: 352–8. [DOI] [PubMed] [Google Scholar]
- 57. Cooper W, Fox S, O'Toole S et al National Working Group Meeting on ALK diagnostics in lung cancer. Asia Pac J Clin Oncol 2014; 10 (Suppl 2): 11–7. [DOI] [PubMed] [Google Scholar]
- 58. Cabillic F, Gros A, Dugay F et al Parallel FISH and Immunohistochemical studies of ALK status in 3244 non‐small‐cell lung cancers reveal major discordances. J Thorac Oncol 2014; 9: 295–306. [DOI] [PubMed] [Google Scholar]
- 59. Conde E, Hernandez S, Prieto M, Martinez R, Lopez‐Rios F. Profile of Ventana ALK (D5F3) companion diagnostic assay for non‐small‐cell lung carcinomas. Expert Rev Mol Diagn 2016; 16: 707–13. [DOI] [PubMed] [Google Scholar]
- 60. Minca EC, Portier BP, Wang Z et al ALK status testing in non–small cell lung carcinoma. A correlation between ultrasensitive IHC and FISH. J Mol Diagn 2013; 15: 341–6. [DOI] [PubMed] [Google Scholar]
- 61. Ying J, Guo L, Qiu T et al Diagnostic value of a novel fully automated immunochemistry assay for detection of ALK rearrangement in primary lung adenocarcinoma. Ann Oncol 2013; 24: 2589–93. [DOI] [PubMed] [Google Scholar]
- 62. Wang J, Cai Y, Dong Y et al Clinical characteristics and outcomes of patients with primary lung adenocarcinoma harboring ALK rearrangements detected by FISH, IHC, and RT‐PCR. PLoS One 2014; 9: e101551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mattsson JSM, Brunnström H, Jabs V et al Inconsistent results in the analysis of ALK rearrangements in non‐small cell lung cancer. BMC Cancer 2016; 16: 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marchetti A, Pace MV, di Lorito A et al Validation of a new algorithm for a quick and easy RT‐PCR‐based ALK test in a large series of lung adenocarcinomas: Comparison with FISH, immunohistochemistry and next generation sequencing assays. Lung Cancer 2016; 99: 11–6. [DOI] [PubMed] [Google Scholar]
- 65. Weber B, Liu M, Sobkin P et al Successful treatment of hepatic oligometastases with stereotactic ablative radiotherapy and radiofrequency ablation in an anaplastic lymphoma kinase fusion‐positive lung cancer patient. J Med Radiat Sci 2016; 63: 67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hout D, Schweitzer B, Lawrence K et al Performance of a RT‐PCR assay in comparison to FISH and immunohistochemistry for the detection of ALK in non‐small cell lung cancer. Cancer 2017; 9: pii. E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mitiushkina NV, Iyevleva AG, Poltoratskiy AN et al Detection of EGFR mutations andEML4‐ALK rearrangements in lung adenocarcinomas using archived cytological slides. Cancer Cytopathol 2013; 121: 370–6. [DOI] [PubMed] [Google Scholar]
- 68. Tuononen K, Sarhadi VK, Wirtanen A et al Targeted resequencing reveals ALK fusions in non‐small cell lung carcinomas detected by FISH, immunohistochemistry, and real‐time RT‐PCR: A comparison of four methods. Biomed Res Int 2013; 2013: 757490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yasuda H, de Figueiredo‐Pontes LL, Kobayashi S, Costa DB. Preclinical rationale for use of the clinically available multitargeted tyrosine kinase inhibitor crizotinib in ROS1‐translocated lung cancer. J Thorac Oncol 2012; 7: 1086–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ou SH, Bartlett CH, Mino‐Kenudson M, Cui J, Iafrate AJ. Crizotinib for the treatment of ALK‐rearranged non‐small cell lung cancer: A success story to usher in the second decade of molecular targeted therapy in oncology. Oncologist 2012; 17: 1351–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Camidge DR, Doebele RC. Treating ALK‐positive lung cancer‐‐early successes and future challenges. Nat Rev Clin Oncol 2012; 9: 268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Camidge DR, Bang YJ, Kwak EL et al Activity and safety of crizotinib in patients with ALK‐positive non‐small‐cell lung cancer: Updated results from a phase 1 study. Lancet Oncol 2012; 13: 1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim DW, Mehra R, Tan DS et al Activity and safety of ceritinib in patients with ALK‐rearranged non‐small‐cell lung cancer (ASCEND‐1): Updated results from the multicentre, open‐label, phase 1 trial. Lancet Oncol 2016; 17: 452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ou SH, Ahn JS, De Petris L et al Alectinib in crizotinib‐refractory ALK‐rearranged non‐small‐cell lung cancer: A phase II global study. J Clin Oncol 2016; 34: 661–8. [DOI] [PubMed] [Google Scholar]
- 75. Kim DW, Tiseo M, Ahn MJ et al Brigatinib in patients with crizotinib‐refractory anaplastic lymphoma kinase‐positive non‐small‐cell lung cancer: A randomized, multicenter phase II trial. J Clin Oncol 2017; 35: 2490–8. [DOI] [PubMed] [Google Scholar]
- 76. Horn L, Wakelee H, Reckamp KL et al MINI01.02: Response and plasma genotyping from phase i/ii trial of ensartinib (X‐396) in patients (pts) with ALK+ NSCLC: Topic: Medical oncology . J Thorac Oncol 2016; 11 (11 Suppl): S256–7. [Google Scholar]
- 77. Soria JC, Tan DSW, Chiari R et al First‐line ceritinib versus platinum‐based chemotherapy in advanced ALK‐rearranged non‐small‐cell lung cancer (ASCEND‐4): A randomised, open‐label, phase 3 study. Lancet 2017; 389: 917–29. [DOI] [PubMed] [Google Scholar]
- 78. Shaw AT, Kim TM, Crinò L et al Ceritinib versus chemotherapy in patients with ALK‐rearranged non‐small‐cell lung cancer previously given chemotherapy and crizotinib (ASCEND‐5): A randomised, controlled, open‐label, phase 3 trial. Lancet Oncol 2017; 18: 874–86. [DOI] [PubMed] [Google Scholar]


