In the early 1990s, the team of Dr. Honjo identified a new gene that was upregulated during T-cell activation, supposedly as an apoptosis-associated molecule (1). This gene was named programmed cell death-1 (PD-1 or CD279 or PDCD1). The PD-1 protein turned out to be a receptor delivering a co-inhibitory signal, playing a decisive role in maintaining peripheral tolerance and impeding autoimmunity, but with no effect on cell apoptosis. In 2000, Drs. Freeman and Sharpe discovered the natural ligands for the PD-1 receptor, called programmed cell death-1 ligand-1 (PD-L1 or CD274 or PDCD1L1) and programmed cell death-1 ligand-2 (PD-L2 or CD273 or PDCD1L2) (2,3). Clues to their respective functions and their potential to be exploited in cancer immunotherapy were elucidated during the next decade, notably when they found out that these ligands inhibited T-cell responses and were expressed by some cancer cell lines. Today, the PD-1/PD-L1 axis has become one of the most important negative regulators of the immune response. This axis is targeted by the so-called “immune checkpoint inhibitors” in the treatment of solid tumors. Those inhibitors are currently evaluated in approximately 800 clinical trials and have been approved for seven different tumor types (4). For the first time in decades, this therapeutic approach has demonstrated unprecedented clinical efficacy in some patients in more than 15 cancer types, including difficult-to-treat cancers such as metastatic melanoma, non-small cell lung cancer, renal cell carcinoma, and bladder carcinoma (5).
However, as with most anti-cancer treatments, many patients are not responsive to PD-1/PD-L1 inhibitors, and more worrying, some patients who demonstrate initial response acquire resistance over time (6). Expression of PD-L1 on either tumor cells or on tumor-infiltrating immune cells does not accurately predict for the patient’s response to PD-1/PD-L1 inhibitors. Today, one major goal is to better understand the regulation of PD-L1 expression. We need a better understanding of its role in tumor biology, markers to better predict the patient’s response to immune checkpoint inhibition, and the development of combinatorial therapeutic strategies for improving the results (7). Several mechanisms regulate PD-L1 expression (Table 1). Most of them are related to immune evasion strategies and lead to the overexpression of PD-L1 at the surface of cancer cells (23), dendritic cells (DCs) (24), and macrophages (15). First, intrinsic modifications of cancer cells could alter PD-L1 expression: some tumor display genomic rearrangements and subsequent amplification of the PD-L1 gene on chromosomal region 9p24.1. The regulation of PD-L1 also depends on microRNAs (miR-20b, miR-21, miR-130b, miR-200, and miR-197) (10) and PD-L1 3’ untranslated region (UTR) itself, which represses own PD-L1 expression (8). Other molecular events, including constitutive oncogenic signaling such as loss of PTEN, CDK5 disruption, MYC and PI3K pathways activation, ALK rearrangements and EGFR mutations can lead to increased PD-L1 expression (9,20). However, the microenvironment remains the major source of signals regulating PD-L1 expression. The main regulator of PD-L1 is the interferon (IFN)-pathway: type I (IFNA, IFNB), type II (IFNG), type III (IFNL) interferons and interferon regulatory factor 1 (IRF1). PD-L1 expression is however primarily induced in response to INFG (12). The response to IFNG is part of a homeostatic feedback loop activated to control local cytotoxicity (13,14). This mechanism explains why, in some specific tumors, PD-L1 upregulation is associated with a good prognosis, and in these cases, is always correlated with IFNG production (25-27). JAK/STAT members (JAK1, JAK2, STAT1, STAT2, STAT3, IRF1) are downstream actors of the IFN-pathway and their activation leads to PD-L1 upregulation (16). Other signals, notably key upstream mediators linking inflammation to cancer, such as hypoxia [hypoxia-inducible factor 1 alpha (HIF1α)], interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), nuclear factor κB (NF-κB), inducible nitric oxide synthase, cyclooxygenase 2, and multiple growth factors are other known modulators of PD-L1 expression: the related receptor-mediated signaling molecules that also affect the cell cycle, proliferation, apoptosis, and survival (including NF-κB, MAPK, PI3K, and mTOR) are thus involved in the induction of PD-L1 (17,18). Finally, the epithelial-to-mesenchymal transition status of the tumor cells is also correlated to PD-L1 expression, with increased PD-L1 expression on ZEB1 + cells, i.e., on more mesenchymal tumor cells (19). In all these cases, PD-L1 expression is mostly regulated at the transcriptional level, and involves fixation of transcription factors or HIFα to the PD-L1 promoter. Post-translation modifications have also been described, notably glycosylation (stabilization) and ubiquitination (degradation) of PD-L1 (11,28).
Table 1. Mechanisms regulating PD-L1 expression.
| Regulatory signals | Mechanisms | Impact on PD-L1 expression | Ref. |
|---|---|---|---|
| Genomic alterations | |||
| Amplification of PD-L1 (chromosome 9p21) | Up | (8) | |
| G > C mutation in the 3’-untranslated region (UTR) of PD-L1 RNA leads to a change in the binding site for miR-57 | Up | (8) | |
| Truncation of the 3’-UTR = stabilisation of PD-L1 mRNA | Up | (8) | |
| Repression of PD-L1 expression by PD-L1 3'UTR | Down | (8) | |
| Constitutive oncogenic signaling (MYC, MAPK activation), loss of PTEN expression, PI3K-pathway activation, ALK fusion with EML4, EGFR mutations, BRAF inhibitory mutations | Up | (9) | |
| Post-transcriptional regulation | |||
| miR-15b, miR-16, miR-34, miR-193a-3p, miR-195, miR-197 and miR-200c | Down | (10) | |
| miR-20b, miR-21, miR-130b | Up | (10) | |
| Ubiquitination (degradation) | Down | (11) | |
| Glycosylation (stabilization) | Up | (11) | |
| Extrinsic factors | |||
| IFN-pathway (IFNG, IRF1, IFN-types I and III) | Up | (12-14) | |
| Cytokines leading to STAT1-STAT3 activation (IL-6, TNFA) | Up | (12-14,15) | |
| TLR4-pathway | Up | (16-18) | |
| Hypoxia (via HIF1α) | Up | (14) | |
| Others | |||
| Epithelial-mesenchymal transformation (up-regulation of ZEB1) | Up | (19) | |
| Cell cycle (CDK4-5), proliferation (MYC), apoptosis (TP53) regulation | Up | (20) | |
| CMTM4/6 stabilize PD-L1 expression (prevent ubiquitination) | Up | (21,22) | |
Two papers recently published in Nature on September 2017 (21,22) have identified novel PD-L1 protein regulators through a haploid genetic screen and whole-genome CRISPR-Cas9 deletion library screen. In addition to the known factors mentioned above (IFNGR1, IFNGR2, STAT1, JAK1, JAK2), both studies came out with the same molecule, CMTM6 [Chemokine-like factor-like (CKLF) MARVEL Transmembrane domain containing family Member 6]. CMTM6 enhanced both constitutive and induced PD-L1 expression at the cell membrane, without compromising antigen presentation by reducing cell surface MHC class I expression levels. Exogenous expression of CMTM6 in CMTM6-knockout cells restored PD-L1 expression in a dose-dependent manner. CMTM6 did not modify the constitutive or INFG-induced mRNA expression levels of PD-L1, suggesting post-transcriptional regulation. The effect of CMTM6 knock-down was variable on different cell lines, suggesting the possible existence of additional regulators. CMTM4, another member of the CMTM family with 55% sequence similarity to CMTM6, was indeed identified as a “back-up” positive regulator of PD-L1 expression, only when CMTM6 was absent and with less efficiency than CMTM6. The fact that this dual strategy was selected during evolution suggests a major role for CMTM6 and CMTM4 in the regulation of PD-L1. The CMTM6 gene belongs to the CMTM family, which contains eight members (CMTM1-4 on 16q21-22 chromosomal region, CMTM5 on 14q11 region, CMTM6-8 on 3p22 region) (29). All CMTM members belong to the chemokine-like factor gene superfamily, a superfamily similar to the chemokine and transmembrane 4 superfamilies. One obvious question was to determine whether CMTM6 directly interacts with PD-L1. Reciprocal co-immunoprecipitation of CMTM6 or PD-L1 showed that CMTM6 was readily detected in association with PD-L1. Mass spectrometry analysis of CMTM6 immunoprecipitates revealed only a small number of other high-confidence interacting proteins [Lymphocyte function-associated antigen 3 (CD58), Arginase-1 (ARG1), Alpha-Enolase (ENO1), Lamina-associated polypeptide 2, alpha (TMPO)]. PD-L1 was one of the top-ranked proteins, attesting, once again, of the specificity of the CMTM6-PD-L1 association. Such co-localization of PD-L1 and CMTM6 was confirmed by IHC in human tumors.
CMTM6, like the other members of this family, is a type-3 transmembrane protein with a MARVEL domain and comprising at least three transmembrane helices. MARVEL domain proteins have been implicated in regulating the trafficking of transmembrane and secretory proteins. The other exact functions of CMTM6 were unknown until now. Interestingly, CMTM6 is widely expressed in all tested tissues and cell types and in many cancers (https://www.proteinatlas.org/ENSG00000091317-CMTM6, and TCGA database), suggesting a likely role outside the immune system. In addition to its predicted localization at the plasma membrane, CMTM6 expression was mostly observed in the cytosol and/or localized at the intermediate filaments (https://www.proteinatlas.org/ENSG00000091317-CMTM6/cell#human). This was confirmed by the two Nature papers, which refined CMTM6 cellular localization to recycling endosomes, together with TFRC and RAB11, two molecules that define the endocytic recycling compartment. This observation suggested that CMTM6 might have a role in protein stabilization and recycling, which is coherent with its predicted function due to the presence of the MARVEL domain.
To shed light on the functional role of CMTM6, IFNG-stimulated CMTM6 parental and KO cells were pulse-labelled with 35S-cysteine/methionine to study the maturation and the trafficking of newly synthesized PD-L1 molecules. CMTM6 loss did not impair PD-L1 export from the endoplasmic reticulum and trafficking beyond the medial Golgi. However, in CMTM6-KO cells, PD-L1 was rapidly degraded (visible after 6 hours), leading to fewer PD-L1 molecules expressed at the cell surface. The reverse experiment in wild-type cells showed that blocking endocytic recycling induced rapid loss of PD-L1 from the surface of wild-type cells, suggesting that a large proportion of surface PD-L1 is continuously internalized and recycled (every 15 minutes), whereas endocytosed PD-L1 is not effectively recycled in CMTM6-deficient cells and may instead be rerouted for degradation in the lysosome. The dissection of this pathway revealed that CMTM6 protects PD-L1 from ubiquitination, thus resulting in the increase of PD-L1 protein half-life at the cell membrane and in recycling endosomes, where it prevents PD-L1 from being targeted for lysosome-mediated degradation.
Collectively, these data shed light on a novel mechanism of regulation of PD-L1 and open potential new avenues to block this pathway. However, this is just the beginning of a new story: the mechanism(s) of regulation of CMTM6 itself is (are) still unknown, as well as whether CMTM6 is dysregulated in human cancers. Interestingly, RNA expression levels of CMTM6 and PD-L1/CD274 are weakly or not correlated in most tumor types. Furthermore, CMTM6 expression is not subject to the IFN-pathway regulation, whereas CMTM6 KO leads to repression of IFNG-induced PD-L1 expression, without compromising antigen presentation via MHC-class I. This means that the silencing of efficient anti-tumor response through PD-L1 upregulation (as part of the IFN-pathway retro-control loop) could potentially be reverted with the inhibition of CMTM6. In this line, the two Nature studies demonstrated, both in vitro and in vivo, that knocking-down CMTM6 favored tumor cells clearance by specific T-cells and enhanced T-cells cytotoxic functions (perforin, granzymes, granulysin, and IL2), via the decreased expression of PD-L1. This makes CMTM6 a very interesting alternative therapeutic target to enhance anti-tumor immunity, either as a novel immunotherapy or in a combination strategy, and notably in patients that may have developed mechanisms of resistance to PD-1/PD-L1 inhibitors.
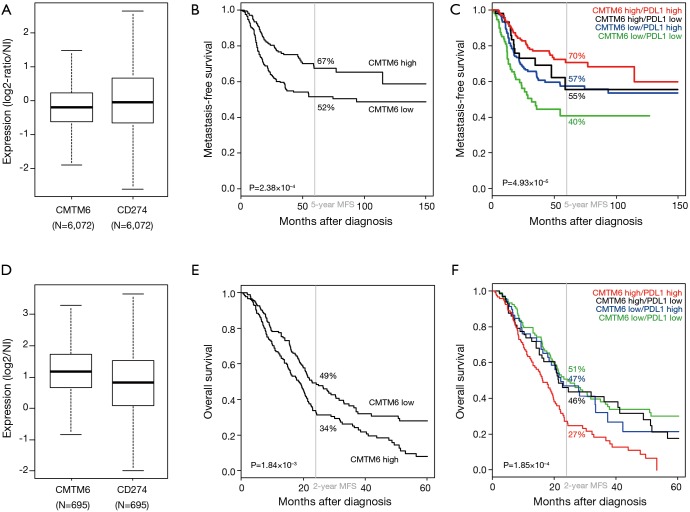
To further explore the link between CMTM6 and PD-L1 expression in human cancers, we studied how CMTM6 expression, alone and in combination with PD-L1 expression, could impact patients’ survival in our two gene expression databases of 509 operated triple-negative (TN) primary breast cancers and of 403 operated primary pancreatic ductal adenocarcinomas (PDAC) for which we had previously reported a prognostic value for PD-L1 mRNA expression (20,30). In TN breast cancers, CMTM6 mRNA expression levels varied among samples, as did PD-L1 expression (Figure 1A), suggesting heterogeneous expression, which allowed a search for eventual correlations with survival. High CMTM6 expression was associated with longer metastasis-free survival (MFS) than low expression (67% vs. 52%, P=2.38×10–4, log-rank test; Figure 1B), as did high PD-L1 expression. Interestingly, CMTM6 expression status enhanced the prognostic value of PD-L1 expression by separating the patients in three groups with different MFS (Figure 1C): PD-L1high CMTM6high group (“High:High” phenotype) with 70% 5-year MFS (good-prognosis), PD-L1high CMTM6low or PD-L1low CMTM6high (“Mixed” phenotype) with similar respective 5-year MFS of 57% and 55% (intermediate-prognosis), and PD-L1low CMTM6low (“Low:Low” phenotype) with 40% 5-year MFS (poor-prognosis). Such prognostic complementarity was confirmed by multivariate analysis in which both CMTM6 and PD-L1 expression statutes remained associated with longer MFS with respective hazard ratios (HR) for metastatic relapse equal to 0.56 (P=8.19×10–4, Wald test) and 0.63 (P=6.78×10–3, Wald test). Thus, in TN breast cancers, CMTM6 expression increased the known favorable prognostic value of PD-L1 expression. In pancreatic adenocarcinomas (PDAC), similar results were observed in terms of heterogeneous expression of CMTM6 (Figure 1D) and of prognostic value. However, and in contrast to breast cancer, high CMTM6 expression, like high PD-L1 expression, was associated with shorter overall survival (OS) with 34% 2-year OS vs. 49% in case of low expression (P=1.84×10–3, log-rank test; Figure 1E). And here too, CMTM6 expression enhanced the prognostic value of PD-L1 expression (Figure 1F): PD-L1high CMTM6high group displayed 27% 2-year OS vs. 47% for the PD-L1high CMTM6low group. In multivariate analysis, both CMTM6 and PD-L1 expression statutes were associated with shorter OS with respective HR for death equal to 1.38 (P=1.90×10–2, Wald test) and 1.46 (P=7.47×10–3, Wald test). Thus, in both cancer types and by stabilizing PD-L1 expression, CMTM6 expression added prognostic information to that of PD-L1 expression, suggesting cooperative effect in disease progression, and nicely complementing the two Nature papers.
Figure 1.
CMTM6 expression and prognostic value in triple negative breast cancers and in pancreatic ductal adenocarcinoma. (A) Box-plot of mRNA expression levels of CMTM6 and CD274/PD-L1 in our own database of 6,072 primary breast cancers. (B) Kaplan-Meier metastasis-free survival according to CMTM6 mRNA expression in 509 patients with triple negative breast cancer. High and low expressions were defined by reference to the median expression level in the whole breast cancer database including 6,072 samples. The P value if for the log-rank test. (C) Similar to B, but according to both CMTM6 and CD274/PD-L1 mRNA expression. The survival curves of the four groups are color-coded as follows: red for PD-L1high CMTM6high group, green for PD-L1low CMTM6low group, black for PD-L1low CMTM6high group, and blue for PD-L1high CMTM6low group. (D), (E), and (F) Similar to (A), (B), and (C), but in our own database of 695 primary pancreatic ductal adenocarcinomas and for overall survival (n=403).
Of course, all those data are preliminary and numerous questions remain open. For example, whether and how CMTM6 is deregulated in cancers; or what is the predictive value of CMTM6 expression for the response to PD-1/PD-L1 inhibitors. But yet, these findings identify a previously unknown regulator of the PD-1/PD-L1 immune checkpoint, which could provide a new therapeutic target to block this critical pathway and to help overcoming immune evasion by tumor cells.
Acknowledgements
Our work was supported by Institut Paoli-Calmettes, Institut National de la Santé et de la Recherche Médicale, Institut National du Cancer, Site de Recherche Intégrée sur le Cancer Marseille (INCa-DGOS-Inserm 6038 grant), and la Ligue contre le cancer (Label Ligue). None of them had any role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Provenance: This is a Guest Editorial commissioned by Section Editor Dr. Xue-Feng Leng, MD, PhD (Department of Thoracic Surgery, West China Hospital, Sichuan University; Department of Cardiothoracic Surgery, the Affiliated Hospital of Chengdu University, Chengdu, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ishida Y, Agata Y, Shibahara K, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J 1992;11:3887-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000;192:1027-34. 10.1084/jem.192.7.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2:261-8. 10.1038/85330 [DOI] [PubMed] [Google Scholar]
- 4.Eggermont A, Robert C, Soria JC, et al. Harnessing the immune system to provide long-term survival in patients with melanoma and other solid tumors. Oncoimmunology 2014;3:e27560. 10.4161/onci.27560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56-61. 10.1126/science.aaa8172 [DOI] [PubMed] [Google Scholar]
- 6.Restifo NP, Smyth MJ, Snyder A. Acquired resistance to immunotherapy and future challenges. Nat Rev Cancer 2016;16:121-6. 10.1038/nrc.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. 10.1038/nrc3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kataoka K, Shiraishi Y, Takeda Y, et al. Aberrant PD-L1 expression through 3'-UTR disruption in multiple cancers. Nature 2016;534:402-6. 10.1038/nature18294 [DOI] [PubMed] [Google Scholar]
- 9.Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227-31. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolle MA, Calin HN, Pichler M, et al. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J 2017;284:1952-66. 10.1111/febs.14030 [DOI] [PubMed] [Google Scholar]
- 11.Lim SO, Li CW, Xia W, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016;30:925-39. 10.1016/j.ccell.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Diaz A, Shin DS, Moreno BH, et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep 2017;19:1189-201. 10.1016/j.celrep.2017.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SJ, Jang BC, Lee SW, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett 2006;580:755-62. 10.1016/j.febslet.2005.12.093 [DOI] [PubMed] [Google Scholar]
- 14.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005;5:375-86. 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Garcia M, Porichis F, de Jong OG, et al. Expression of PD-L1 and PD-L2 on human macrophages is up-regulated by HIV-1 and differentially modulated by IL-10. J Leukoc Biol 2011;89:507-15. 10.1189/jlb.0610327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gowrishankar K, Gunatilake D, Gallagher SJ, et al. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-kappaB. PLoS One 2015;10:e0123410. 10.1371/journal.pone.0123410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang G, Wen Q, Zhao Y, et al. NF-kappaB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS One 2013;8:e61602. 10.1371/journal.pone.0061602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng J, Hamanishi J, Matsumura N, et al. Chemotherapy Induces Programmed Cell Death-Ligand 1 Overexpression via the Nuclear Factor-kappaB to Foster an Immunosuppressive Tumor Microenvironment in Ovarian Cancer. Cancer Res 2015;75:5034-45. 10.1158/0008-5472.CAN-14-3098 [DOI] [PubMed] [Google Scholar]
- 19.Noman MZ, Janji B, Abdou A, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 2017;6:e1263412. 10.1080/2162402X.2016.1263412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorand RD, Nthale J, Myers JT, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 2016;353:399-403. 10.1126/science.aae0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burr ML, Sparbier CE, Chan YC, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017;549:101-5. 10.1038/nature23643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mezzadra R, Sun C, Jae LT, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017;549:106-10. 10.1038/nature23669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Strome SE, Salomao DR, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 2002;8:793-800. 10.1038/nm730 [DOI] [PubMed] [Google Scholar]
- 24.Brown JA, Dorfman DM, Ma FR, et al. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol 2003;170:1257-66. 10.4049/jimmunol.170.3.1257 [DOI] [PubMed] [Google Scholar]
- 25.Bertucci F, Finetti P, Colpaert C, et al. PDL1 expression in inflammatory breast cancer is frequent and predicts for the pathological response to chemotherapy. Oncotarget 2015;6:13506-19. 10.18632/oncotarget.3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertucci F, Finetti P, Mamessier E, et al. PDL1 expression is an independent prognostic factor in localized GIST. Oncoimmunology 2015;4:e1002729. 10.1080/2162402X.2014.1002729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatier R, Finetti P, Mamessier E, et al. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 2015;6:5449-64. 10.18632/oncotarget.3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li CW, Lim SO, Xia W, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun 2016;7:12632. 10.1038/ncomms12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han W, Ding P, Xu M, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 2003;81:609-17. 10.1016/S0888-7543(03)00095-8 [DOI] [PubMed] [Google Scholar]
- 30.Birnbaum DJ, Finetti P, Lopresti A, et al. Prognostic value of PDL1 expression in pancreatic cancer. Oncotarget 2016;7:71198-210. 10.18632/oncotarget.11685 [DOI] [PMC free article] [PubMed] [Google Scholar]