Abstract
Background
Characterization of the Mycobacterium leprae genome has made possible the development of Polymerase Chain Reaction (PCR) systems that can amplify different genomic regions. Increased reliability and technical efficiency of quantitative PCR (qPCR) makes it a promising tool for early diagnosis of leprosy. Index cases that are multibacillary spread the bacillus silently, even before they are clinically diagnosed. Early detection and treatment could prevent transmission in endemic areas.
Methods
In this study, the qPCR technique is used to detect DNA of M. leprae in samples of slit skin smears (SSS) of the ear lobe and blood of leprosy patients and their asymptomatic household contacts residing in Governador Valadares, MG, Brazil, a hyperendemic area for leprosy. A total of 164 subjects participated in the study: 43 index cases, 113 household contacts, and, as negative controls, 8 individuals who reported no contact with patients nor history of leprosy in the family. The qPCR was performed to amplify 16S rRNA fragments and was specifically designed for M. leprae.
Results
Of asymptomatic household contacts, 23.89% showed bacillary DNA by qPCR in samples of SSS and blood. Also, 48.84% of patients diagnosed with leprosy were positive for qPCR while the bacillary load was positive in only 30.23% of patients. It is important to note that most patients were already receiving treatment when the collection of biological material for qPCR was performed. The level of bacillary DNA from household contacts was similar to the DNA levels detected in the group of paucibacillary patients.
Conclusion
Considering that household contacts comprise a recognizable group of individuals with a high risk of disease, as they live in close proximity to a source of infection, qPCR can be used to estimate the risk of progress towards leprosy among household contacts and as a routine screening method for a chemoprophylactic protocol.
Electronic supplementary material
The online version of this article (10.1186/s12879-018-3056-2) contains supplementary material, which is available to authorized users.
Keywords: Leprosy, Household contacts, qPCR
Background
Leprosy is a chronic disease caused by Mycobacterium leprae that results in neurological and skin damage [1]. Despite advances toward the elimination of leprosy over the last two decades, new case detection rates have stabilized over the last decade, and leprosy remains an important health problem in many regions [2]. Leprosy is considered endemic in several countries with low rates of social and economic development, especially in India and Brazil, which contain the largest absolute number of cases. Given delays in the diagnosis of multibacillary (MB) leprosy, transmission of M. leprae from infected individuals to their contacts continues, and in many cases, irreversible nerve damage occurs before those infected are registered as patients [3, 4]. It is remarkable that DNA amplification methods have been used for genomic studies and resistance-associated gene mutations [5].
The early diagnosis and prompt initiation of treatment is essential to the rapid interruption of the disease transmission chain. In this sense, the development of a sensitive test for the diagnosis of leprosy has been one of the main objectives of research related to the disease [6]. The M. leprae genome sequence has made it possible to target specific sequences of the bacillus. PCR is also a sensitive technique capable of detecting 25 fg (10− 15 g) of M. leprae DNA [7, 8]. The assays have been developed for regions such as the 36-kDa [9], 18-kDa [10] or 65-kDa [11] antigens as well as repeat sequences (RLEP) and the gene encoding the 16S rRNA of M. leprae [12, 13]. The analysis of sensitivity and specificity of real-time quantitative PCR (qPCR) amplification of the sodA gene, 16S rRNA, RLEP, and 85BAg for the differential diagnosis of leprosy showed that the RLEP gene confers greater sensitivity (although a lower specificity) to the technique. The assay 16S rRNA, albeit less sensitive, was highly specifically suited for M. leprae [12]. Previous data [14] confirmed the suitability of the 16S rRNA primer for M. leprae, comparing it with nine other Mycobacterium species, including M. tuberculosis and M. bovis as well as bacteria of other genera, such as Staphylococcus, Streptococcus, and Escherichia. We screened and followed household contacts (HHCs) along with patients, testing the 16S rRNA qPCR assay to evaluate the presence of M. leprae DNA in patients and their asymptomatic HHCs.
Methods
Study group
The study was conducted in the city of Governador Valadares in eastern Minas Gerais State, Brazil. This city is considered hyperendemic for leprosy: the new case detection rate () was 1.9/10,000 people in 2015. In Minas Gerais, the rate was 0.5/10,000 in 2016, and in Brazil as a whole, it is currently around 1.2/10,000. The study’s participants included leprosy patients and HHCs who came to the Reference Center for Endemic Diseases and Special Programs (CREDEN-PES) at the Department of Public Health of Governador Valadares municipality. The index cases were diagnosed and biological samples were collected for up to three months after treatment began. All contacts were subjected to careful clinical evaluation before being considered asymptomatic. One hundred sixty-four individuals participated in the study: 43 index cases and 113 HHCs. Eight individuals who reported no contact with patients or family history of leprosy were included as a negative control (NC) group. According to the classification used in CREDEN-PES, the index cases were identified as paucibacillary (PB) or MB based on the Guidelines of the Brazilian Health Ministry [15]. PB individuals showed Tuberculoid-Tuberculoid (2), Borderline Tuberculoid (18) in its clinical form (PB, n = 20), and were negative in bacillary load. MB individuals showed Borderline–Borderline (3), Borderline Lepromatous (7) and Lepromatous Lepromatous (13). HHCs were grouped and assigned as follows: contacts of PB patients (HHC-PB, n = 52) and contacts of MB patients (HHC-MB, n = 61).
Ethical approval
The study was approved by the Ethics Committee of the Universidade Vale do Rio Doce—Univale, filed under N° PQ 022/09–009. All participants signed an informed consent (IC) at the first evaluation.
Sample collection and DNA extraction
The bacillary load technique was performed only in index cases, according to the guidelines to technical procedures: smear microscopy in leprosy [16]. The bacteriological index (BI) was calculated according to the work of Ridley and Jopling (1966) [17]. Slit skin smears (SSS) from the right earlobe and blood samples were collected for DNA extraction using the DNeasy kit (QIAGEN®). Samples of SSS stored in ethanol at 70% were thawed and centrifuged at 2000 rpm for 10 min. The extraction was performed according to protocols described by the manufacturer. The concentration of DNA in the eluate was determined by spectrophotometer (NanoDrop 1000 Spectrophotometer—Thermo Scientific). DNA extraction from blood samples followed the same procedure as mentioned above, using 50 μL of blood.
Real time PCR assay—qPCR
The qPCR was performed using the TaqMan qPCR amplification system. The amplification target was the gene 16S rRNA specific for M. leprae as previously described [14]. The threshold values to define positive samples were used as described (Martinez et al., 2011; Barbieri et al., 2014), and the number of genomes was calculated by interpolating the Ct values in a dilution curve with the known number of M. leprae genomes using an Excel spreadsheet (Martinez et al., 2011). All qPCR reactions were run in triplicates and in the same thermocycler under calibrated conditions where positive controls with known numbers of genomes were tested (Step One, Applied Biosystems).
Statistical analysis
Statistical analysis was performed using the GraphPad Prism version 5.0 software. The bacterial DNA levels among groups were evaluated by Mann-Whitney test and Kruskal-Wallis test (Dunn’s Multiple Comparison Test).
Results
Efficiency of bacillary loads and qPCR for M. leprae detection
Among the index cases, 25% of patients in the PB group showed M. leprae DNA in any sample (blood or SSS). It is important to note that all patients from the PB group had a negative BI. With respect to the MB group, it was found that 69.56% of patients were positive in both SSS or blood in qPCR (Any-Sample). On the other hand, in the MB group, the BI was positive in 56.52% of patients. It was observed that 30% of MB individuals that showed negative BI were qPCR positive. In addition, 100% of MB patients with positive BI showed a qPCR positive result. In summary, it was possible to identify M. leprae DNA in 48.84% of all index cases investigated while the BI was positive in only 30.23% (Table 1).
Table 1.
Efficiency of bacilloscopy and qPCR for M. leprae detection
| Study Group | N | Bacilloscopy | qPCR blood | qPCR SSS | blood or SSS |
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | N (%) | ||
| PB | 20 | 0 (0.00) | 2 (10.00) | 4 (20.00) | 5 (25.00) |
| MB | 23 | 13 (56.52) | 4 (17.39) | 14(60.87) | 16 (69.56) |
| Total | 43 | 13 (30.23) | 6 (13.95) | 18 (41.86) | 21 (48.84) |
N number of patients, qPCR Slit Skin Smear = qPCR SSS
The Spearman test was used to correlate the genome numbers of M. leprae against the bacillary load. No association was observed (p = 0.3894, r = 0.3571) (Fig. 1). In this analysis were included only patients from the MB group who were positive for qPCR and bacillary load.
Fig. 1.
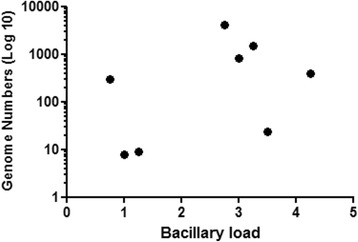
Spearman correlation between genome numbers and bacillary load for MB patients with positive bacilloscopy. (N = 12*; p = 0.3894; r = 0.3571) * Only individuals with bacillary DNA in SSS and positive bacilloscopy were selected
A comparison of bacterial genome numbers between the groups of index cases was performed using interpolation of the Ct values as described above, obtained from the qPCR blood or SSS. It was observed that the genome number of M. leprae was significantly higher in the MB group than in the PB group (Fig. 2).
Fig. 2.
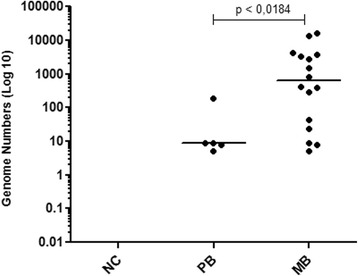
Comparison of M. leprae genome numbers between the groups of index cases. NC = Negative control (n = 8); PB = Paucibacillary (n = 5); MB = Multibacillary (n = 16). Each point represents the individual value of a genome number. The median is represented by the horizontal line. Mann-Whitney test was applied
M. leprae DNA detection in household contacts
The 113 HHCs were evaluated by qPCR using blood samples and SSS of the right earlobe, and it was found that 19.23% of HHC-PB were positive for M. leprae DNA in blood or SSS. Likewise, in relation to the HHC of MB cases (HHC-MB), 27.87% were positive. Among 113 asymptomatic contacts assessed by qPCR, 23.89% had positive M. leprae DNA results (Table 2).
Table 2.
Detection of M. leprae DNA in household contacts of index cases
| Study Group | N | qPCR blood | qPCR SSS | blood or SSS |
|---|---|---|---|---|
| N (%) | N (%) | N (%) | ||
| HHC-PB | 52 | 4 (7.69) | 7 (13.46) | 10 (19.23) |
| HHC-MB | 61 | 7 (11.48) | 11 (18.03) | 17 (27.87) |
| Total | 113 | 11 (9.73) | 18(15.93) | 27(23.89) |
N Number of household contacts, HHC-PB House hold contacts Pauciballary, HHC-MB House hold contacts Multibacillary, qPCR Slit Skin Smear qPCR SSS
Comparison of DNA levels of M. leprae between household contacts and groups of index cases
Analysis of genome numbers obtained in qPCR blood or SSS of HHCs showed no statistical difference between the HHC-PB and HHC-MB groups (Fig. 3). Interestingly, it was found that the median of the values of genome counts of all HHCs (HHC-PB and HHC-MB) was similar to the median of the index cases of PB. However, as expected, the median value of genome counts in MB cases was significantly higher than the median of the contact group (p < 0.0001). Note that two contacts showed similar genome numbers as the MB group (Fig. 4). After a one-year follow-up, three individuals were diagnosed with leprosy. But only one of those three infected individuals was contacted; the others moved and were not found.
Fig. 3.
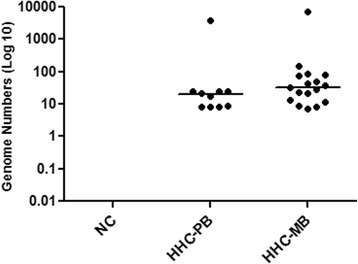
Comparison of M. leprae genome number among household contacts groups. NC = Negative control (n = 8); HHC-PB = Contact of paucibacillary (n = 10); HHC-MB = Contact of multibacillary (n = 17). Each point represents the individual value of a genome number. The median is represented by the horizontal line. Mann-Whitney test
Fig. 4.
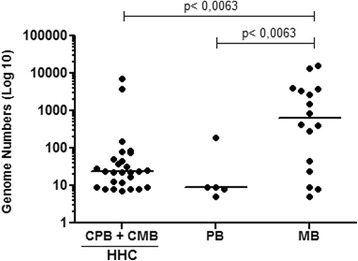
Comparison of the M. leprae genome number of all household contacts and index cases PB and MB. Household contacts (HHC) (n = 27); PB = paucibacillary (n = 5); and MB = multibacillary (n = 16). Each point represents the individual value of 1/Ct. The median is represented by the horizontal line. Kruskal-Wallis statistic and Dunn’s Multiple Comparison Test were applied
Discussion
It is clear that effective control of leprosy requires, together with multidrug therapy, new diagnostic tools that can detect M. leprae infection at an early stage. The PCR technique has been evaluated for DNA detection of M. leprae in different systems [18–20].
The high sensitivity of qPCR in relation to the BI makes this an extremely important technique in supporting the clinical diagnosis, as reported by other authors [12, 13, 20]. In our study, we confirmed the significant potential of qPCR for M. leprae DNA detection in leprosy cases with negative BI as well as in asymptomatic HHCs. It was observed that 25% of PB and 30% of MB individuals who showed negative BI (dimorphous 3/10) were qPCR positive. Considering that, 100% of MB patients with positive BI (4 dimorphous and 9 virchowian) were also qPCR positive (Tables 3 and 1).
Table 3.
Characterization of the study groups for clinical form and bacilloscopy
| Operational classification | Clinical form | Bacillary load | N |
|---|---|---|---|
| PB | TT | Negative | 02 |
| BT | Negative | 18 | |
| MB | BB | Negative | 03 |
| BL | Positive | 07 | |
| LL | Positive | 13 | |
| Total | 43 |
N Number of patients, PB Paucibacillary, MB Multibacillary, TT Tuberculoid-Tuberculoid, BT Borderline-Tuberculoid, BB Borderline-Borderline, BL Borderline-Lepromatous, LL Lepromatous-Lepromatous
Although our study did not present data about the sensitivity and specificity of the qPCR test, our assay was based in a series of previous works wherein the extensive evaluation of qPCR used here is 100% specific. The sensitivity is around 50% in patients with PB forms to near 100% in patients with MB forms of the disease [21].
Because collecting SSS is an invasive procedure, it was limited to one specific site (the right earlobe). Therefore, we believe that a higher frequency of positivity for qPCR could be achieved if other collection sites could be used, as is standard for smear microscopy [16].
Importantly, some patients were already receiving treatment at the time of the collection of biological material, which may have reduced the positivity rate for qPCR, in accordance with Banerjee et al. (2010) [18].
Interestingly, we noted a moderate positive correlation (p = 0.047; r = 0.5823) between the values of 1/Ct (DNA levels) and the bacterial index (BI) in MB patients, reinforcing the association between the M. leprae DNA level detected by qPCR and infection. Therefore, it was found that, the higher the BI, the higher the bacterial DNA level (Fig. 1) as demonstrated [22]. Evidence of a positive correlation between qPCR and BI led us to compare the M. leprae DNA levels among patients in an attempt to distinguish the groups. We found a significant difference in the level of bacterial DNA between the MB and PB groups (Fig. 2). Studies on leprosy transmission have demonstrated that people living with index cases are exposed to a greater risk of progressing towards the disease [23, 24]. An effective strategy for reducing the incidence of leprosy is contact tracing and diagnosis in the early stages of the disease. As shown by Banerjee et al. (2010) [18], MB contacts have increased the frequency of positivity in the multiplex PCR (M-PCR) compared to PB contacts; we also found a higher frequency of qPCR positive for HHC-MB (27.87%) in comparison with HHC-PB (19.23%). We consider that contacts of MB are more exposed to a higher bacterial load, possibly showing increased frequency of qPCR, positive in relation to HHC-PB contacts [25]. However, we did not detect significant differences in the number of genomes obtained from qPCR among HHC (HHC-PB + HHC-MB) groups.
It is important to note that all HHCs were considered asymptomatic for clinical evaluation. However, 23.89% of them had bacillary DNA in SSS of the earlobe. This high detection rate indicates a high dynamic transmission of leprosy in that study group. Knowing that leprosy has a long incubation period and that symptoms are difficult to detect in the early stages of the disease, we emphasize that the contacts’ surveillance with positive results for qPCR is extremely relevant. Six to 8% of HHCs develop clinical symptoms of leprosy within two years of diagnosis of the index case [26]. The fact that all contacts showed levels of bacterial DNA similar to the PB group can be explained by the slow progress of the disease; some of these contacts may develop leprosy in the future. In our study, with only one year of follow-up, we detected three new cases of leprosy among the examined contacts. Of these, two were CMB and one was positive for qPCR before the onset of clinical symptoms. The employment of strategy for early detection and/or identification of subclinical infection linked to chemoprophylaxis will certainly contribute to the effective control of leprosy [27]. More recently, the importance of serological and DNA-based techniques for the assessment and confirmation of diagnoses in suspected and early cases of leprosy was emphasized [28]. According Reis et al. 2014 [29], all HHC positive for qPCR should be addressed to control strategies to provide both chemoprophylaxis and immunoprophylaxis by vaccination to generate immediate protection that can be sustained in the long term.
Finally, we consider that the high sensitivity of qPCR allows the identification of a large number of asymptomatic contacts having M. leprae DNA. More qPCR tests need to be evaluated further as they could serve as a better diagnostic tool for early case detection and treatment to achieve faster control of leprosy.
Conclusion
The presence of bacterial DNA in the dermal scraping earlobe suggests subclinical infection, and therefore, contacts with positive qPCR should be monitored for disease development in the future. Early detection of leprosy cases and effective chemotherapy are the best strategies to reduce the incidence of new cases of leprosy and prevent transmission. Considering that HHCs comprise a recognizable group of individuals with a high risk of disease, as they live in close proximity to a source of infection, we suggest that, as a prevention strategy, qPCR should be used to follow-up with leprosy HHCs to confirm or rule out subclinical infection.
Additional file
Bank of data. Description of data: this file contains all raw data of the study. (PDF 219 kb)
Acknowledgments
We are very thankful to Maria de Fatima Silva, Marlucy Rodrigues Lima, Lilia Cardoso Pires, and Wallace Olimpio for their technical support. We are also thankful to all members of CREDEN-PES, especially Dr. Alexandre Castelo Branco for the diagnostics of the patients and Regina L.B. Cipriano for the administrative support.
Funding
This study received financial support from the Conselho de Desenvolvimento Tecnológico e Científico/CNPq/BRAZIL, DECIT 2008, DECIT 2012, FAPEMIG, TC 304/2013/FNS/MS. These fundings sources had no role in the design of the study and collection, analysis, implementation, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files (Additional file 1).
Abbreviations
- Any-Sample
Blood and/or dermal scraping
- BI
Bacilloscopy index
- CMB
Contacts of multibacillary
- CPB
Contacts of paucibacillary
- CREDEN-PES
Center for Endemic Diseases and Special Programs
- Ct
Cycle threshold
- DNA
Deoxyribonucleic acid
- HHC
Household contact
- IC
Informed consent
- M. bovis
Mycobacterium bovis
- M. leprae
Mycobacterium leprae
- M. tuberculosis
Mycobacterium tuberculosis
- MB
Multibacillary
- M-PCR
Multiplex polymerase chain reaction
- N
Number of patients
- PB
Paucibacillary
- PCR
Polymerase chain reaction
- PGL-1
Phenolic glycolipid I
- PNL
Pure neural leprosy
- qPCR BL
qPCR blood
- qPCR DS
qPCR dermal scraped
- qPCR
quantitative polymerase chain reaction
- RLEP
Repeat sequences
- rRNA
Ribosomal ribonucleic acid
- μL
Microliter
Authors’ contributions
RSG, LAOF, and MOM developed and standardized the protocol and helped to write the manuscript. TARG, CFMG, LBPO, and PHFM collected the samples. ENS, MOM, and RMGG contributed reagents/materials/analysis tools. SJMM performed the qPCR and helped to write the manuscript. FSNM analyzed the data and helped to write the manuscript. All of the authors read and approved the manuscript.
Ethics approval and consent to participate
We hereby declare that this study was approved by the Ethics Committee of the Universidade Vale do Rio Doce—Univale, filed under N° PQ 022/09–009. All participants signed a free and informed consent at the first evaluation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12879-018-3056-2) contains supplementary material, which is available to authorized users.
Contributor Information
Rafael Silva Gama, Email: rsilvagama@hotmail.com.
Thalisson Artur Ribeiro Gomides, Email: thalissonartur@hotmail.com.
Chaiana Fróes Magalhães Gama, Email: chaianafroes@hotmail.com.
Suelen Justo Maria Moreira, Email: sukamoreira@gmail.com.
Fernanda Saloum de Neves Manta, Email: femanta@yahoo.com.br.
Lorena Bruna P. de Oliveira, Email: lorena_bruninha@hotmail.com
Pedro Henrique Ferreira Marçal, Email: pedro.marcal@univale.br.
Euzenir Nunes Sarno, Email: euzenir@fiocruz.br.
Milton Ozório Moraes, Email: mmoraes@fiocruz.br.
Raúl Marcel González Garcia, Email: raul.garcia@ufjf.edu.br.
Lucia Alves de Oliveira Fraga, Phone: 55-33-32795983, Email: luciaalvesfraga@yahoo.com.br, Email: luciaalves.fraga@ufjf.edu.br.
References
- 1.Lyon S, Moura ACL, Grossi MAF, Silva RC. Dermatologia Tropical. 1a ed. Belo Horizonte: Medbook; 2016.
- 2.WHO Global leprosy update, 2016: accelerating reduction of disease burden. Wkly Epid Rec. 2017;92:501–520. [PubMed] [Google Scholar]
- 3.Sales AM, Leon AP, Duppre NC, Hacker MAJ, Nery AC, Sarno EN, et al. Leprosy among patient contacts: a multilevel study of risk factors. PLoS Negl Trop Dis. 2011;5(3):e1013. doi: 10.1371/journal.pntd.0001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos SD, Penna GO, Costa MD, Natividade MS, Teixeira MG. Leprosy in children and adolescents under 15 years old in an urban Centre in Brazil. Mem Inst Oswaldo Cruz. 2016;111(6):359–364. doi: 10.1590/0074-02760160002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De A, Reja AHH, Biswas S, Bhattacharya B, Chatterjee G, Basu K, et al. Unique TTC repeat base pair loss mutation in cases of pure neural leprosy: a survival strategy of mycobacterium leprae. Indian J Dermatol. 2015;60(4):351–355. doi: 10.4103/0019-5154.160478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett BH, Parker DL, Robson M. Leprosy: steps along the journey of eradication. Public Health Rep. 2008;123:198–205. doi: 10.1177/003335490812300212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang TJ, Kim SK, Lee SB, Chae GT, Kim JP. Comparision of two different PCR amplification products (the 18-kDa protein gene MBs. RLEP repetitive sequence) in diagnosis of Mycobacterim leprae. Exp Dermatol. 2003;28:420–424. doi: 10.1046/j.1365-2230.2003.01300.x. [DOI] [PubMed] [Google Scholar]
- 8.Goulart IMB, Goulart LR. Leprosy: diagnostic and control challenges for a worldwide disease. Arch Dermatol Res. 2008;300:269–290. doi: 10.1007/s00403-008-0857-y. [DOI] [PubMed] [Google Scholar]
- 9.Kampirapap K, Singtham N, Klatser PR, Wirimbawipart SDNA. Amplification for detection of leprosy and assessment of efficacy of leprosy chemotherapy. Int J Lepr Other Mycobact Dis. 1998;66:16–21. [PubMed] [Google Scholar]
- 10.Scollard DM, Gillis TP, Williams DL. Polymerase chain reaction assay for the detection and identification of Mycobacterium leprae in patients in the United States. Am J Clin Pathol. 1998;109(5):642–646. doi: 10.1093/ajcp/109.5.642. [DOI] [PubMed] [Google Scholar]
- 11.Plikaytis BB, Gelber RH, Shinnick TM. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J Clin Microbiol. 1990;8(9):1913–1917. doi: 10.1128/jcm.28.9.1913-1917.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez AN, Ribeiro-Alves M, Sarno EN, Moraes MO. Evaluation of qPCR-based assays for leprosy diagnosis directly in clinical specimens. PLoS Negl Trop Dis. 2011;5(10):1354. doi: 10.1371/journal.pntd.0001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turankar RP, Pandey S, Lavania M, Singh I, Nigam A, Darlong J, et al. Comparative evaluation of PCR amplification of RLEP, 16S rRNA, rpoT and sod a gene targets for detection of M. Leprae DNA from clinical and environmental samples. Int J Mycobacteriol. 2015;4(1):54–59. doi: 10.1016/j.ijmyco.2014.11.062. [DOI] [PubMed] [Google Scholar]
- 14.Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, Moraes MO, et al. Molecular detection of Mycobaterium leprae viability by use of real-time PCR. J Clin Microbiol. 2009;47(7):2124–2130. doi: 10.1128/JCM.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brasil Ministério da Saúde, Secretaria de Vigilância em Saúde, Departamento de Vigilância das Doenças Transmissíveis . Diretrizes para vigilância, atenção e eliminação da Hanseníase como problema de saúde pública: manual técnico-operacional. 2016. [Google Scholar]
- 16.Brasil Ministério da Saúde, Secretaria de Vigilância em Saúde . Guia de procedimentos técnicos: baciloscopia em hanseníse. 2010. [Google Scholar]
- 17.Ridley DS, Jopling WH. Classification of leprosy according to immunity: a five group system. Int J Lepr. 1966;34:255–273. [PubMed] [Google Scholar]
- 18.Banerjee S, Sarkar K, Gupta S, Mahapatra PS, Gupta S, Guha S, et al. Multiplex PCR technique could be an alternative approach for early detection of leprosy among close contacts: a pilot study from India. BMC Infect Dis. 2010;10:252. doi: 10.1186/1471-2334-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez NA, Britto CFPC, Nery JAC, Scollard D, Truman R, Moraes MO, et al. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J Clin Microbiol. 2006;44:3154–3159. doi: 10.1128/JCM.02250-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caleffi KR, Hirata RDC, Hirata MH, Caleffi ER, Siqueira VLD, Cardoso RF. Use of the polymerase chain reaction to detect Mycobacterium leprae in urine. Braz J Med Biol Res. 2012;45(2):153–157. doi: 10.1590/S0100-879X2012007500011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez AN, Talhari C, Moraes MO, Talhari S. PCR-based techniques for leprosy diagnosis: from the laboratory to the clinic. PLoS Negl Trop Dis. 2014;8(4):e2655. doi: 10.1371/journal.pntd.0002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truman RW, Andrews PK, Robbins NY, Adams LB, Krahenbuhl JL, Gillis TP. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2(11):e328. doi: 10.1371/journal.pntd.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarno EN, Duppre NC, Sales AM, Hacker MA, Nery JA, de Matos HJ. Leprosy exposure, infection and disease: a 25-year surveillance study of leprosy patient contacts. Mem Inst Oswaldo Cruz. 2012;107(8):1054–1059. doi: 10.1590/S0074-02762012000800015. [DOI] [PubMed] [Google Scholar]
- 24.Barreto JG, Bisanzio D, Frade MAC, Moraes TMP, Gobbo AR, Guimarães LS, et al. Spatial epidemiology and serologic cohorts increase the early detection of leprosy. BMC Infect Dis. 2015;15:527. doi: 10.1186/s12879-015-1254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Job CK, Jayakumar J, Kearney M, Gillis TP. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am J Trop Med Hyg. 2008;78:518–521. [PubMed] [Google Scholar]
- 26.Ottenhoff T. Immunology of leprosy: lessons from and for leprosy. Int J Lepr. 1994;62:108–121. [PubMed] [Google Scholar]
- 27.Fischer EAJ, De Vlas SJ, Habbema JDF, Richardus JH. The long term effect of current and new interventions on the new case detection of leprosy: a modeling study. PLoS Negl Trop Dis. 2011;5(9):e1330. doi: 10.1371/journal.pntd.0001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulart IMB, Araujo S, Botelho AF, De Paiva PHR, Goulart LR. Asymptomatic leprosy infection among blood donors may predict disease development and suggests a potential mode of transmission. J Clin Microbiol. 2015;53:3345–3348. doi: 10.1128/JCM.01305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reis EM, Araujo S, Lobato J, Neves AF, Costa AV, Gonçalves MA, et al. Mycobacterium leprae DNA in peripheral blood may indicate a bacilli migration route and high-risk for leprosy onset. Clin Microbiol Infect. 2014;20(5):447–452. doi: 10.1111/1469-0691.12349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bank of data. Description of data: this file contains all raw data of the study. (PDF 219 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files (Additional file 1).


