Significance
T helper type (Th) 9 cells demonstrate both pro- and antiinflammatory properties, pointing to a functional heterogeneity not examined so far. Applying single cell gene expression analysis of alloreactive Th9 cells, we revealed the existence of two major subsets, CD96high and CD96low Th9 cells, with strongly opposing inflammatory and, especially, colitis-inducing potential. Mechanistically, we found that CD96 controls cytokine and colitis-inducing potential of Th9 cells, providing strong evidence for an inhibitory role of CD96 in controlling CD4+ T-cell effector functions. Thus, interfering with CD96-mediated immune inhibition would be a promising approach in preventing Th9-mediated diseases, such as ulcerative colitis, or reinforcement of Th9-mediated immune control of tumors and infections.
Keywords: Th9 cells, IL-9, CD96, single-cell gene expression, colitis
Abstract
Recent findings demonstrated proinflammatory functions of interleukin (IL)-9–producing T helper type (Th) 9 cells in the pathogenesis of intestinal bowel diseases (IBDs). However, also antiinflammatory properties have been ascribed to Th9 cells, pointing to a functional heterogeneity. To dissect the specific expression pattern and, especially, diversity of murine antigen-specific Th9 cells, we applied single cell transcription profiling. Th9 cells displayed reduced expression of typical activation markers, such as Cd40 ligand and Cd96, whereas expression of Cd25 and Cd83 was increased compared with other Th subsets. Importantly, we identified two subsets of Th9 cells differing above all in their CD96 expression. The heterogeneous CD96 expression was specific for Th9 cells and not observed for other Th subtypes, such as Th1 cells. Lower CD96 expression was also observed in human IL-9+ compared with IFN-γ+ T cells. Although Il9 was highly transcribed by all Th9 cells, IL-9 mRNA and protein expression was increased in CD96low cells. Transfer of CD96low Th9 cells into recombination activating gene 1-deficient (Rag1−/−) mice caused severe weight loss, intestinal and colonic inflammation, and destruction of allogeneic skin grafts and thus showed high inflammatory potential. This was associated with their expansion and tissue accumulation. Contrastingly, CD96high Th9 cells did not cause colitis and showed reduced expansion and migratory potential. Blockade of CD96 completely restored the expansion and inflammatory properties of CD96high Th9 cells. Collectively, our data suggest an inhibitory role for the cosignaling receptor CD96 in Th9 cells, raising new opportunities in the treatment of IL-9–associated inflammations such as IBD.
More than 20 y ago, murine CD4+ T cells were identified as potent producers of IL-9 when activated in the presence of IL-2, IL-4, and transforming growth factor beta (TGF-β) (1). In 2008, IL-9–producing T cells gained new attention when they were found to lack expression of both known lineage-specific transcription factors and cytokines (2, 3). Hence, IL-9–producing T cells were considered to represent a distinct lineage, termed Th9 cell. Th9 differentiation requires a complex transcription factor network, including signal transducer and activator of transcription 6 (STAT6), IFN regulatory factor 4 (IRF4), transcription factor PU.1, and basic leucine zipper transcription factor, ATF-like (BATF) (reviewed in ref. 4), but no lineage-defining transcription factor comparable with t-box transcription factor (T-bet) being linked to Th1 differentiation has been identified yet (4).
Therefore, the classification of Th9 cells as a distinct and homogenous lineage is still under debate. However, the concept of a single “master regulator” controlling CD4+ T-cell differentiation has changed (5). Instead, Th cell fate is seen as an interplay of transcription factors providing the molecular explanation for lineage plasticity but, especially, functional diversity (5, 6).
Functionally, Th9 cells appear proinflammatory, driving host immune responses against worm infections (7) and tumors (8, 9), but also contribute to development of unwanted inflammatory responses, such as autoimmune diseases (10) and allergies (11, 12).
Moreover, studies in patients with intestinal bowel diseases (IBDs) revealed significantly enhanced Il9 mRNA expression levels in the mucosa, especially in active ulcerative colitis (UC) (13, 14). IL-9 colocalized with PU.1 and IRF4, demonstrating the presence of Th9 cells (14). Absence of IL-9 prevented development of experimental colitis in mice (13). Functionally, IL-9 was shown to target intestinal epithelial cells, resulting in impaired proliferation and mucosal wound healing (13, 14). Thus, Th9 cells play a crucial role in the pathogenesis of IBD and, especially, in UC.
Intriguingly, production of the signature cytokine IL-9 is not limited to proinflammatory properties but also includes antiinflammatory effects. For example, in transplantation immunology, IL-9 was found to be crucial for mast-cell recruitment and activation during allograft tolerance, which failed to develop in mast cell-deficient mice (15). At that time, IL-9–producing T cells as a separate Th subset were not defined yet, and IL-9 was proposed to be produced by CD4+CD25+Foxp3+ regulatory T cells. Recently, forkhead-box-protein P3 (Foxp3) was identified as a potent repressor of Il9 gene expression (16), strongly suggesting that Th9 cells, and not regulatory T cells, were the source of IL-9.
Taken together, these previous observations indicate a functional heterogeneity of Th9 cells—an aspect that has not been investigated yet.
Here, we utilized single cell profiling of murine alloantigen-reactive Th9 cells to elucidate potential phenotypic and functional heterogeneity. Thereby, we identified two main subsets of Th9 cells with strongly opposing inflammatory potential.
Results
Graft Rejection and Colitis in Rag1−/− Mice After Transfer of Th9 Cells.
Antigen-specific Th9 cells were generated by stimulation of naive WT BALB/c CD4+ T cells with allogeneic C57BL/6 dendritic cells (DCs). Addition of the Th9-polarizing cytokines IL-4 and TGF-β induced IL-9 production in 62.3% (median) of activated, i.e., CD44+ T cells (Fig. S1A). Comparable levels were achieved using the reversed strain combination (Fig. S1B).
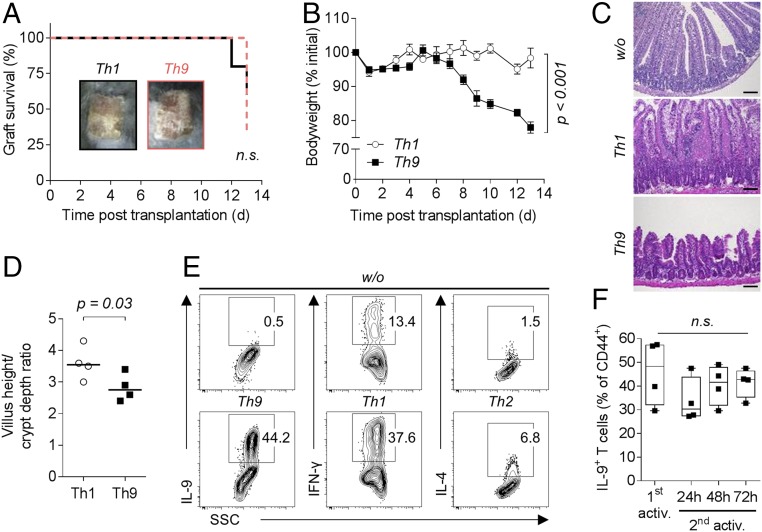
To analyze the inflammatory properties of in vitro generated alloreactive Th9 cells, we injected the cells into recombination activating gene 1-deficient (Rag1−/−) mice (17) and performed allogeneic skin transplantation. Since cytokine production is restricted to activated T cells (Fig. S1A), we enriched the cells for CD44 expression before injection (see Fig. S2 for cell-sorting strategies). Contradictory to previous findings, transfer of CD44+ IL-9–producing Th9 cells induced graft rejection within 13 d comparable with Th1-mediated rejection (Fig. 1A). Furthermore, mice reconstituted with CD44+ Th9 cells suffered from severe weight loss starting ∼7 d after cell injection (Fig. 1B). Neutralization of IL-9 nearly completely prevented Th9 cell-induced weight changes (Fig. S3). This was already described for polyclonally activated Th9 cells (2, 13). Contrary to our expectation, mice receiving CD44+ Th1 cells did not lose weight.
Fig. 1.
Transfer of Th9 cells into B cell- and T cell-deficient Rag1−/− mice causes graft rejection, weight loss, and intestinal inflammation. (A–D) C57BL/6 Rag1−/− mice received 1 or 2 × 105 activated (CD44+) T cells sorted from in vitro differentiated alloreactive C57BL/6 Th1 (n = 4–5) or Th9 (n = 4–7) cultures and a BALB/c skin graft on the following day. (A) Survival of BALB/c donor skin. A Log-rank (Mantel–Cox) test was applied. (B) Body weight of mice depicted as mean ± SEM. P value of the interaction term (group with time) was calculated using an ANOVA type III test after fitting a linear mixed-effect model to the body weight data. (C) Representative hematoxylin and eosin stained ileum sections collected on day 12 (w/o) or 13 (Th1, Th9) post skin transplantation. (Scale bar: 100 µm.) (D) Villus height-to-crypt depth ratio in ileal mucosa with median value. For statistical analysis, a one-tailed Mann–Whitney test was applied. (E) Staining of in vitro differentiated alloreactive (CD44+) BALB/c T cells for intracellular expression of signature cytokines. Cells were cultured for 3 d in the absence (w/o) or presence of Th9, Th1, or Th2 polarizing cytokines. (F) Intracellular staining for IL-9 in activated (1st activ.) and (with allogeneic dendritic cells) reactivated (2nd activ.) in vitro differentiated alloreactive C57BL/6 Th9 cells (n = 4). Statistical analysis by Kruskal–Wallis test. n.s., not significant (P > 0.05).
Weight loss was associated with development of severe intestinal inflammation, manifested by shortened and broadened villi in the ileal mucosa of mice injected with CD44+ Th9 cells (Fig. 1C). In contrast, mucosal morphology of animals that received CD44+ Th1 cells was comparable with control mice receiving no T cells. Changes in ileal morphology led to reduced villus height-to-crypt depth ratio in specimens from mice receiving Th9 cells in comparison with Th1 cells (Fig. 1D).
Alloreactivity is supposed to be a Th1-driven process (18, 19). Consequently, activation of CD4+ T cells with allogeneic DCs in vitro without additional cytokines resulted in production of IFN-γ with almost no production of IL-9 or IL-4 (Fig. 1E). However, we could obtain as many IL-9+ T cells as IFN-γ+ T cells when cultured in Th9 or Th1 skewing conditions, respectively. IL-9 production was stable and also detectable after reactivation of total Th9 cells (Fig. 1F).
Taken together, Th9 cells display IL-9–dependent inflammatory effector functions in vivo.
Unique and Heterogeneous Activation Phenotype of Th9 Cells.
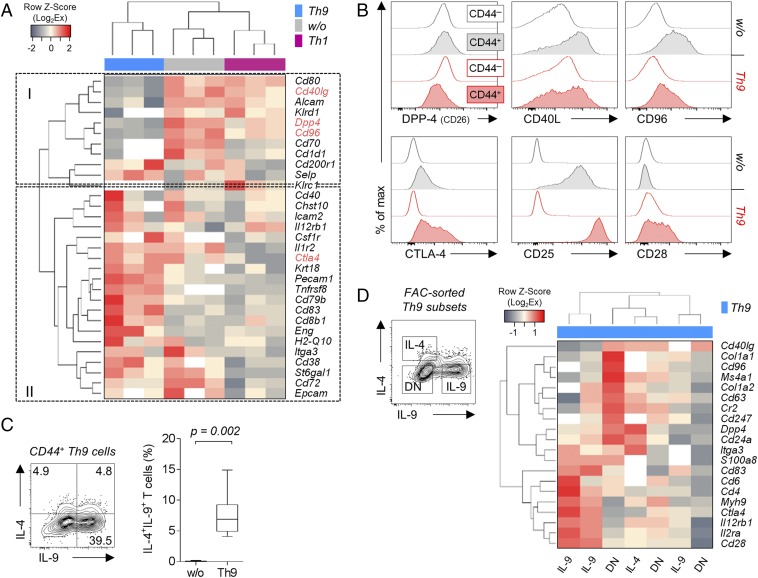
Enhanced colitis-inducing potential implies selective homing potential of Th9 cells. To investigate this, we measured gene expression of 84 cell surface proteins by PCR array and compared expression in CD44+ Th9 cells, CD44+ nonpolarized cells, and CD44+ Th1-polarized cells.
Clustering analysis of differentially expressed genes resulted in clear separation of Th9 samples, with identification of down-regulated (cluster I) and up-regulated (cluster II) genes (Fig. 2A). Many of the differentially expressed genes were associated with T-cell activation: e.g., Cd40 ligand (CD40lg), dipeptidyl peptidase 4 (Dpp4), Cd96, cytotoxic T lymphocyte-associated protein 4 (Ctla4), Cd83, and Cd38. Interestingly, expression of Cd40lg, Dpp4, and CD96 was down-regulated whereas that of Ctla4, Cd83, and Cd38 was up-regulated in bulk Th9 cells. Expression of multiple activation markers was verified on protein level (Fig. 2B). Additionally, CD44+ Th9 cells showed high expression of CD25 and did not lose CD28 expression compared with CD44+ nonpolarized T cells. Thus, Th9 cells exhibited a unique activation phenotype.
Fig. 2.
In vitro differentiated Th9 cells display a unique activation phenotype with heterogeneous cytokine and gene expression. (A) Nonsupervised hierarchical clustering analysis of differentially expressed genes encoding for cell surface expressed molecules in nonpolarized (w/o), Th1, or Th9 polarized alloreactive C57BL/6 T cells. Gene expression of three biological replicates (enriched for CD44+ cells) was determined by PCR array and normalized to B2m expression. White areas indicate undetermined (Ct > 35) values, and genes written in red were stained for protein expression as shown in B. (B) Histograms comparing protein expression of surface molecules associated with T-cell activation in nonactivated (CD44−) and activated (CD44+) alloreactive BALB/c T cells from PMA/ionomycin-stimulated cocultures. (C) Staining of intracellular cytokines in alloreactive BALB/c CD44+ Th9 cells. The P value of nine independent experiments was determined by one-tailed Wilcoxon matched-pairs signed rank test. (D) Nonsupervised hierarchical clustering analysis of differentially expressed genes encoding for cell surface relevant molecules in fluorescence activated cell (FAC)-sorted IL-9+, IL-4+, and double-negative (DN) cells. Cells were sorted from PMA/ionomycin-restimulated alloreactive Th9 cocultures with total BALB/c CD4+ T cells. The results of three biological replicates are shown (IL-4+ samples had to be pooled due to limited cell number).
As described above, bulk CD44+ alloantigen-reactive Th9 cells are inflammatory. However, it is conceivable that Th9 cells as a whole consist of subsets with contrasting inflammatory potential, with one subset causing colitis. To reveal Th9-specific mediators that contribute to such functional heterogeneity, we first analyzed their cytokine production. Notably, a fraction of cells coexpressed IL-4 and IL-9 (Fig. 2C). This was unexpected as Th9 cells have initially been reported to not express cytokines attributed to other Th cells (2). Moreover, this indicated a functional heterogeneity within CD44+ Th9 cells. To investigate that further, we sorted in vitro generated Th9 cells into IL-4+, IL-9+, and double negative (DN) cells and performed again the PCR array. Unfortunately, due to the low recovered cell number of pure (>90%) IL-9/IL-4 double producers, we could not perform the PCR array with those. Two of three samples of solely IL-9–producing cells clustered separately from the remaining samples (Fig. 2D). In contrast, IL-4+ cells did not separate from DN cells. Genes contributing to the separation were Cd6 and again Ctla4, Cd25, and Cd28 being higher expressed in IL-9+ cells. Cd96 and Cd40l were up-regulated in IL-4+ and DN cells. Interestingly, one IL-9+ and one DN cell sample clustered separately, indicating again heterogeneity of Th9 cells.
Two Main Subsets Differing in CD96 Expression Within Th9 Cells.
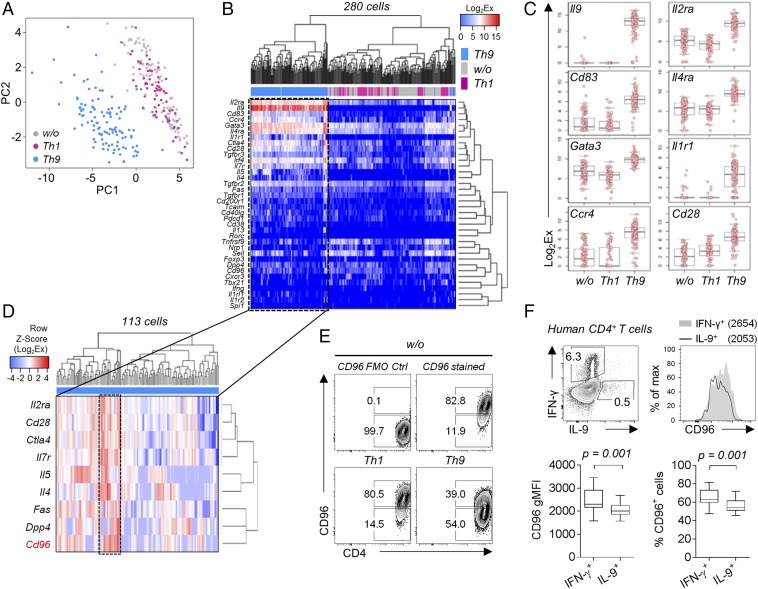
To reveal further differences between individual Th9 cells, we performed single cell gene expression analysis. We included 280 cells for data analysis. Application of principal component and clustering analyses resulted in formation of a separate Th9 cluster (Fig. 3 A and B). In contrast, the expression profiles of Th1 and nonpolarized T cells were comparable.
Fig. 3.
Single cell profiling identifies CD96high and CD96low subsets of Th9 cells. (A–D) Single cell gene expression analysis of CD44+ alloreactive T cells sorted from nonpolarized (w/o; n = 100), Th1 (n = 67), and Th9 (n = 113) polarized BALB/c cocultures. See Table S2 for a list of all genes measured. (A) Principal component analysis of gene expression in indicated Th subsets. (B) Nonsupervised hierarchical clustering analysis (Pearson’s correlation) comparing gene expression in Th subsets. Analysis includes all genes with detectable expression. (C) Box plots of most differentially expressed genes. Bonferroni-corrected ANOVA P values: Il9 (P = 1.4 × 10−126), Il2ra (P = 1.4 × 10−65), Cd83 (P = 7.1 × 10−64), Il4ra (P = 1.2 × 10−52), Gata3 (P = 3.6 × 10−47), Il1r1 (P = 5.5 × 10−47), Ccr4 (P = 2.2 × 10−46), and Cd28 (P = 2.3 × 10−43). (D) Nonsupervised hierarchical clustering analysis (complete linkage method) of gene expression in CD44+ T cells sorted from alloreactive BALB/c Th9 cultures. Displayed genes contributed to clustering. Frame indicates a cluster of Th9 cells characterized by high Cd96 expression. (E) Contour plots of surface CD96 expression in CD44+ polarized alloreactive BALB/c T cells, including fluorescence minus one (FMO) staining control (Ctrl). (F) Comparison of CD96 expression levels in IL-9+ and IFN-γ+ human CD4+ T cells. PBMCs from blood of 10 healthy individuals were stimulated ex vivo with Staphylococcus enterotoxin B (SEB) for 24 h. A one-tailed Wilcoxon matched-pairs signed rank test was applied. gMFI, geometric mean fluorescence intensity.
Astonishingly, Il9 mRNA was expressed at high levels in almost all CD44+ Th9 cells (Fig. 3 B and C). Contrastingly, Il9 mRNA was not detectable at all in Th1 or nonpolarized T cells, turning Il9 into the most differentially expressed gene. Other genes strongly contributing to the separation of CD44+ Th9 cells were Il2 receptor alpha (Il2ra), Cd83, Il4ra, and GATA-binding protein 3 (Gata3) (Fig. 3C).
However, within CD44+ Th9 cells, we identified a subset of cells characterized by particularly high expression of T-cell activation genes: e.g., Il2ra, Cd28, Ctla4, and Il7 receptor (Il7r). This cluster was dominated by high Cd96 expression, a gene weakly transcribed by the remaining Th9 cells (Fig. 3D). Consistently, CD96 protein was only expressed by a fraction of Th9 cells (Fig. 3E). In contrast, CD44+ Th1 and nonpolarized T cells showed high CD96 expression in almost all cells. Importantly, differential CD96 expression was similar between murine and human helper T cells as IFN-γ–producing T cells from blood of healthy donors also displayed high CD96 expression whereas IL-9–producing cells had reduced expression levels (Fig. 3F).
Thus, we identified two subsets of Th9 cells most distinguishable by the amount of CD96 expression and revealed similarities in differential CD96 expression across species.
Increased Cytokine Expression of CD96low Th9 Cells.
CD96 has inhibitory (20) but also activating (21) natural killer (NK) cell regulating functions. However, its role for controlling T-cell and, especially, CD4+ T-cell responses is less well investigated and understood (22, 23).
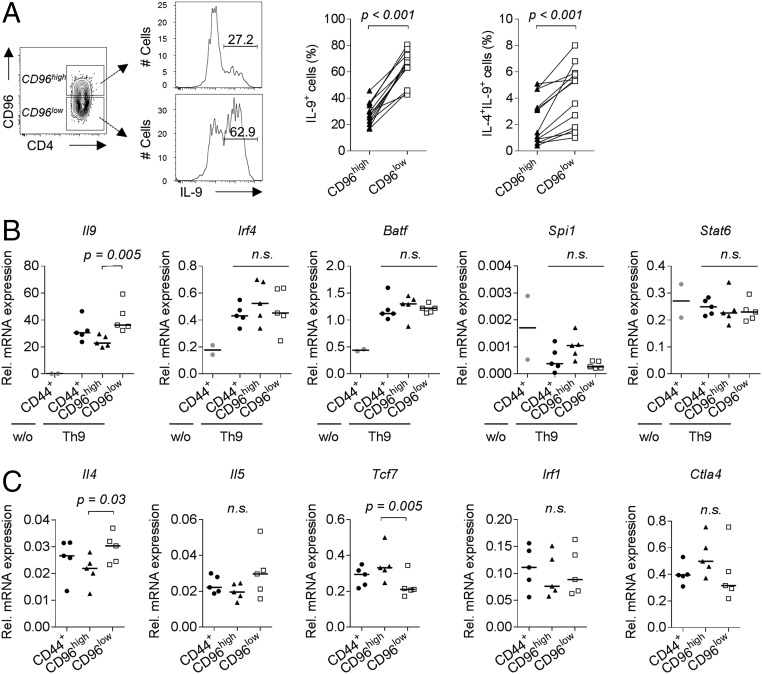
We observed that low CD96 expression was associated with both high frequencies of IL-9 single-positive and IL-9/IL-4 double-positive cells (Fig. 4A). This did not reflect differences in Th9 lineage commitment as total CD44+, CD96low, and CD96high Th9 cells, in contrast to nonpolarized CD44+ cells, showed high expression of Il9 mRNA and equally high expression of Th9-associated transcription factors Irf4 and Batf (Fig. 4B). Notably, CD96low Th9 cells expressed significantly more Il9 mRNA compared with CD96high Th9 cells. No significant differences were observed in expression of Spi1 and Stat6 between total CD44+, CD96low, and CD96high Th9 cells (Fig. 4B).
Fig. 4.
Molecular characterization reveals higher cytokine expression potential of CD96low Th9 cells. (A) Intracellular production of IL-9 and IL-4 in CD96high and CD96low in vitro differentiated alloreactive C57BL/6 Th9 cells (n = 13). P values were calculated with a one-tailed Wilcoxon matched-pairs signed rank test. (B) mRNA expression levels of Il9 and transcription factors related to Th9 differentiation and (C) mRNA expression levels of additional genes of interest in CD96high and CD96low cells sorted from in vitro differentiated alloreactive C57BL/6 Th9 cultures analyzed by real-time qRT-PCR. Results were compared with expression in total CD44+ Th9 cells and, if indicated, with total CD44+ unpolarized cells (w/o, n = 2). Statistical analyses of Th9 datasets (n = 5) were performed by applying Friedman’s test and Dunn’s multiple comparisons test. qRT-PCR, quantitative reverse transcription–PCR. n.s., not significant (P > 0.05).
Additionally, we analyzed mRNA expression of transcription factor 7 (Tcf7) and IFN regulatory factor 1 (Irf1), which are known to influence T helper cell differentiation, as well as of cytokines (Il4, Il5) and inhibitory molecules (Ctla4). We observed higher Tcf7 expression in CD96high Th9 cells but significantly more Il4 and a tendency toward higher Il5 expression in the CD96low subset (Fig. 4C). Taken together, both, CD96low and CD96high cells represent committed Th9 cells, but CD96low cells display increased cytokine expression.
High Inflammatory Potential of CD96low Th9 Cells.
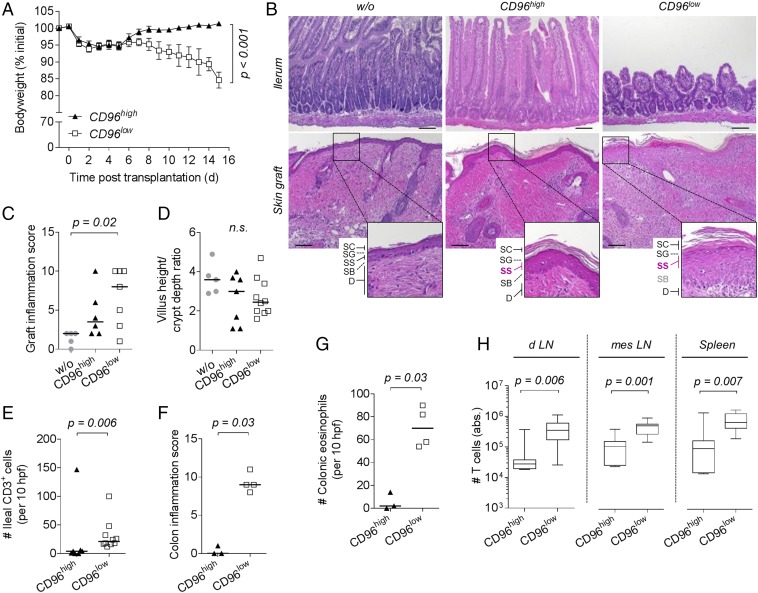
Purified CD96high and CD96low Th9 cells maintained their differential expression pattern for several days upon reactivation (Fig. S4A). To assess whether this translates into differential inflammatory potential in vivo, we reconstituted Rag1−/− mice with either CD96low or CD96high Th9 cells and performed allogeneic skin transplantation. Surprisingly, mice reconstituted with CD96high Th9 cells reached their initial weight within 8 d and constantly gained weight thereafter (Fig. 5A). In contrast, animals injected with CD96low Th9 cells lost body weight as rapidly as observed upon transfer of CD44+ Th9 cells (Fig. 1B).
Fig. 5.
CD96low Th9 cells have a high inflammatory and colitogenic potential. (A–H) C57BL/6 Rag1−/− mice received no cells (w/o, n = 5), 1 × 105 CD96high (n = 3–7), or CD96low (n = 4–10) Th9 cells sorted from in vitro differentiated alloreactive C57BL/6 cocultures and were grafted with BALB/c skin on the next day. Samples were collected between day 9 and 15 posttransplantation. Due to a sample collection error, colonic specimens could only be analyzed from three to four animals of three independent experiments. (A) Body weight of mice shown as mean ± SEM (CD96high, n = 7; CD96low, n = 10). P value of the interaction term (group with time) was calculated using an ANOVA type III test after fitting a linear mixed-effect model to the data. (B) Representative hematoxylin and eosin stained sections of ileum (Upper row) and skin graft (Lower row) harvested 9 to 13 d posttransplantation. Magnifications display the epidermis composed of stratum corneum (SC), stratum granulosum (SG), stratum spinosum (SS), stratum basale (SB) and part of the dermis (D). (Scale bar: 100 µm.) (C) Skin graft inflammation score with median. P value was calculated by Kruskal–Wallis test and Dunn’s test. (D) Villus height-to-crypt depth ratio in ileal mucosa with median. Kruskal–Wallis test was applied. (E) Number of CD3+ cells per 10 high power fields (hpfs) in immunohistochemically stained ileum sections. (E–G) Median is depicted, and one-tailed Mann–Whitney test was applied. (F) Colon inflammation score. (G) Number of eosinophils per 10 high power fields (hpfs) in immunohistochemically stained colon sections. (H) Absolute number of T cells isolated from graft draining lymph nodes (d LN), mesenteric lymph nodes (mes LN), and spleen of n = 7 (CD96high) or n = 10 (CD96low) mice. P values were determined by Mann–Whitney test. n.s., not significant (P > 0.05).
Since grafts were not completely rejected when mice injected with CD96low Th9 cells experienced 20% weight loss and had to be killed, we performed comparative histological analysis at this time point. The epidermis of skin grafts in mice receiving CD96high Th9 cells remained unharmed, with intact stratum corneum (SC), stratum granulosum (SG), stratum spinosum (SS), and stratum basale (SB), and thereby appeared like accepted grafts from Rag1−/− mice receiving no cells (Fig. 5 B and C). In contrast, separation of SS and SB was disrupted, and skin graft was severely infiltrated in animals reconstituted with CD96low Th9 cells, leading to a significantly increased inflammation score compared with that of grafts from nonreconstituted mice (Fig. 5 B and C).
Transfer of CD96low Th9 cells was associated with a trend toward a more altered ileal mucosal morphology, such as shortening and broadening of villi (Fig. 5 B and D). They also caused a significantly increased T-cell infiltration into the ileal mucosa (Fig. 5E). Importantly, CD96low Th9 cells induced a significantly more severe colonic inflammation (Fig. 5F and Fig. S4 B and C) and eosinophilic infiltration (Fig. 5G; see Fig. S4D for representative staining of eosinophils).
Differences between subsets were not due to early death of CD96high Th9 cells as T cells were still detectable at high numbers in lymphoid organs of Rag1−/− mice 9 to 15 d after transfer. However, T-cell numbers were significantly enhanced in graft draining and mesenteric lymph nodes and in the spleen of mice reconstituted with CD96low Th9 cells (Fig. 5H). This indicates a higher expansion capacity of CD96low Th9 cells compared with CD96high Th9 cells.
Next, we investigated whether CD96low Th9 cells would also show higher colitogenic potential in an immunocompetent setting: i.e., without homeostatic proliferation. Alloreactive CD96low, CD96high, or total CD44+ Th9 cells were transferred into OT-I-Rag1−/− mice containing ovalbumin-specific CD8+ T cells before allogeneic skin transplantation. OT-I-Rag1−/− mice were not susceptible to early weight loss upon cell transfer, but long-term effect was consistent with data obtained from transfer into Rag1−/− mice. Transfer of total CD44+ and, especially, CD96low Th9 cells induced weight loss in nonlymphopenic mice whereas transfer of CD96high Th9 cells did not (Fig. S5). Graft survival in OT-I-Rag1−/− mice was slightly prolonged in animals receiving CD96high Th9 cells compared with animals reconstituted with CD96low Th9 cells (Table S3). Collectively, these data indicate a high inflammatory potential of CD96low Th9 cells.
Restoration of Inflammatory Properties in CD96high Th9 Cells by CD96 Blockade.
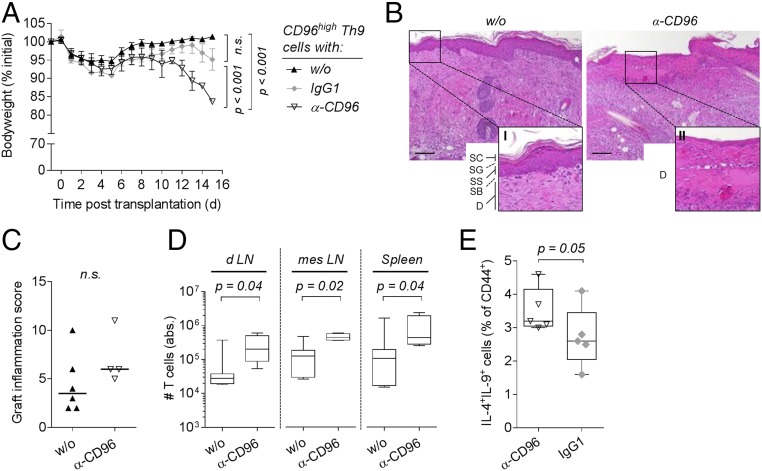
Our findings pointed to an inhibitory role of CD96 expression in Th9 cells. To prove this, we blocked CD96 expression in Rag1−/− mice reconstituted with CD96high Th9 cells and performed allogeneic skin transplantation again. In contrast to application of an isotype control antibody, i.p. injection of a single dose (250 µg) of anti-CD96 antibody (α-CD96 Ab) restored weight loss (Fig. 6A).
Fig. 6.
Blockade of CD96 in CD96high Th9 cells restores their inflammatory capacity. (A–D) C57BL/6 Rag1−/− mice were reconstituted with 1 × 105 CD96high cells sorted from alloreactive C57BL/6 Th9 cultures and received i.p. injection of 250 µg of anti-CD96 (α-CD96, n = 4) or isotype control (IgG1, n = 3) antibody or were left untreated (w/o, n = 6–7). All animals received a BALB/c skin graft on the next day. (A) Body weight of mice shown as mean ± SEM. P value of the interaction term (group with time) between two groups was calculated using an ANOVA type III test after fitting a linear mixed-effect model to the data. (B) Representative hematoxylin and eosin stained sections of skin grafts harvested 10 d post skin transplantation. Higher magnification of w/o photograph (I) displays the epidermis composed of the stratum corneum (SC), stratum granulosum (SG), stratum spinosum (SS), stratum basale (SB), and part of the dermis (D). Magnification of α-CD96 photograph (II) shows the altered dermis without epidermis. (C) Inflammation score of skin grafts harvested between day 9 and 15 posttransplantation. Median is shown, and a one-tailed Mann–Whitney test was applied. (D) Absolute number of T cells isolated from graft draining lymph nodes (d LN), mesenteric lymph nodes (mes LN), and spleen harvested 9 to 15 d posttransplantation. P values were calculated with a two-tailed Mann–Whitney test. (E) Frequency of IL-4+IL-9+ double-producing cells in alloreactive C57BL/6 Th9 cultures following treatment with 5 µg/mL anti-CD96 (α-CD96, n = 5) or isotype control antibody (IgG1, n = 5). Antibodies were added after 24 h of cocultivation, and intracellular cytokine expression was analyzed following an additional 48 h of culture. A one-tailed Mann–Whitney test was applied. n.s., not significant (P > 0.05).
Moreover, CD96 blockade in mice reconstituted with CD96low Th9 cells did not enhance but rather prevented weight loss (median of body weight at the end of the test: 89.3%, without α-CD96 Ab; 100.6%, with α-CD96 Ab). Thus, deterioration observed upon α-CD96 Ab treatment of mice injected with CD96high cells was not caused by interfering with other endogenous CD96-expressing cells: e.g., NK cells.
Treatment of CD96high reconstituted mice with α-CD96 Ab was associated with increased destruction of all four epidermal layers (Fig. 6B) and a tendency toward enhanced skin graft inflammation (Fig. 6C), but this did not reach statistical significance. Furthermore, colon samples of α-CD96–treated mice displayed more severe destruction and T-cell infiltration compared with untreated or isotype control antibody-treated mice (Fig. S6). Blocking CD96 also restored the expansion capacity of transferred CD96high Th9 cells as we detected significantly increased T-cell numbers in draining and mesenteric lymph nodes and in the spleen upon CD96 blockade (Fig. 6D).
Mechanistically, we found that blockade of CD96 increased the proportion of IL-9/IL-4 double-producing cells when added to Th9 cell-inducing cocultures (Fig. 6E). Furthermore, addition of a CD96 blocking antibody during reactivation of CD96high Th9 cells led to increased Il9 mRNA expression compared with isotype antibody treatment (Fig. S6B).
Thus, our data confirm an inhibitory role of CD96 expression for Th9 cell cytokine expression potential and inflammatory effector functions.
Discussion
This report describes a phenotypic and functional heterogeneity of Th9 cells. Using single cell profiling, we identified two subsets of Th9 cells differing mainly in CD96 expression. Transfer of CD96low in vitro differentiated alloreactive Th9 cells into immunocompromised mice caused severe weight loss, intestinal and colonic inflammation, eosinophil infiltration, and destruction of allogeneic skin grafts. In contrast, transfer of CD96high Th9 cells did neither cause weight loss nor changed epidermal organization of the allografts. Transcriptional profiling of Irf4, Batf, and Il9 revealed that both subsets were committed to Th9 differentiation but CD96low Th9 cells were characterized by higher IL-9 mRNA and protein levels, as well as IL-4 and IL-5 expression potential. Blockade of CD96 in vivo restored the inflammatory properties of CD96high Th9 cells, demonstrating that low expression of CD96 identifies a highly inflammatory subset. Thus, the predominance of either CD96high or CD96low Th9 cells during immune responses may account for the contradictory reports on the inflammatory potential of Th9 cells. Our hypothesis is supported by other findings reporting an association of differential CD96 expression with dissimilar effector functions for CD8+ T cells (24). There, absence of CD96 expression characterized highly cytotoxic CD8+ T cells, producing both IFN-γ and perforin, whereas CD96+ CD8+ T cells lacked perforin production.
Investigations by many groups have shown that T cell-deficient mice develop chronic intestinal inflammation and colitis 5 to 10 wk upon transfer of naive T cells, which differentiate into colitogenic (e.g., Th1) effector cells (25). Thus, we expected rapid weight loss in Rag1−/− mice by transferring activated IFN-γ–producing Th1 cells generated from naive CD25− CD4+ T cells. However, transfer of CD44+ Th9 cells and not CD44+ Th1 cells did induce acute weight loss in Rag1−/− mice, reflecting the high colitogenic potential of Th9 cells also described previously (2, 13). Interestingly, CD44+ Th9 cells showed decreased expression of T-cell activation markers, such as CD26, CD40L, and CD70, and therefore appeared less activated compared with nonpolarized and Th1-polarized cells. Moreover, CD44+ Th9 cells share phenotypic features with regulatory T cells: e.g., high expression of CD25 and CTLA-4. However, Foxp3 expression was not detectable in IL-9+ T cells (Fig. 3B and Fig. S7). Taken together, we observed a unique activation phenotype of Th9 cells resembling a noninflammatory cell, which is in accordance with a previous investigation showing also a unique transcriptional profile of Th9 cells compared with Th2 and regulatory T cells (26).
Our neutralization experiments revealed that the pathogenic potential of the transferred Th9 cells was mainly determined by expression of IL-9 (Fig. S3). This is in accordance with previous findings showing that Th9-driven intestinal inflammation is caused by IL-9 by impairing intestinal barrier function, resulting in translocation of bacteria into the mucosa (13).
CD96 is expressed on CD4+ T cells, CD8+ T cells, and NK cells in humans and mice and belongs to a family of molecules interacting with nectins and nectin-like proteins (27). Members of the family are known to provide costimulatory and coinhibitory signals during T-cell activation. For example, CD226 [DNAX accessory molecule-1 (DNAM1)] acts as a positive regulator of T-cell activation whereas T-cell immunoreceptor with Ig and ITIM domains (TIGIT) provides a negative signal (28). CD155, the main ligand of both, is also bound by CD96 (27), but the role of CD96 for CD4+ T-cell function is less clear (reviewed in refs. 22 and 23). Apart from a higher IL-9 production, CD96low Th9 cells were characterized by an increased IL-4 and IL-5 expression potential. The Th2-associated cytokines have also been implicated in the pathogenesis of colitis (reviewed in ref. 29) (30). Together with IL-9, they can also promote accumulation and activation of eosinophils (reviewed in refs. 31 and 32), which have been described to mediate colon destruction and inflammation during colitis (33). Therefore, increased IL-4 and IL-5 expression potential of CD96low Th9 cells might explain high numbers of infiltrating eosinophils in colon specimens, which could contribute to their colitogenic potential. Given the increased inflammatory and cytokine producing potential of CD96low Th9 cells, our study provides strong evidence for an inhibitory role of CD96 in controlling the effector functions of CD4+ cells as previously described for cytotoxic CD8+ T cells (24). This is further supported by our findings that CD96 blockade restored the inflammatory colitogenic potential of CD96high Th9 cells in vivo and Il9 mRNA expression in vitro. In mice, CD96 was shown to inhibit NK cell function (20), which is in accordance with our findings. Downstream signaling of CD96 is not well-studied yet, but the cytoplasmic tail of CD96 contains an immunoreceptor tyrosine-based inhibition motif (ITIM) likely to deliver an inhibitory signal (34).
Up-regulation of CD96 expression represents a late event of T-cell activation peaking 6 to 9 d after stimulation: e.g., in human CD3+ T cells (35). This is comparable with other activation-induced inhibitory receptors, such as programmed cell death protein 1 (PD-1), with an expression maximum between 4 and 7 d (36). Similarly, murine naive CD4+ T cells express low levels of CD96, which is up-regulated over a period of several days upon anti-CD3/anti-CD28 stimulation (37). However, gain of CD96 expression is rather a feature of differentiated than exhausted cells as Th1 cells mainly being positive for CD96 were functional and rejected an allograft. Additionally, we observed high IL-7R (CD127) expression in CD96high Th9 cells, a molecule known to be down-regulated in exhausted T cells (38). Furthermore, the differential expression pattern of CD96high and, especially, CD96low Th9 cells was kept for several days after reactivation. After prolonged reactivation, CD96high cells lose CD96 expression and adopt a similar phenotype as CD96low cells, arguing against an exhaustion-associated expression pattern. However, even though some of the purified CD96high cells down-regulate CD96 expression and adopt a CD96low phenotype, this is accompanied by an increase in their IL-9 production, meaning that the opposite expression pattern between low CD96 expression and high IL-9 production is stable and kept even upon reactivation.
As mentioned earlier, both CD96low and CD96high Th9 cells expressed high levels of Irf4 and Batf, as well as Il9, transcripts in comparison with nonpolarized or Th1-polarized cells, supporting their commitment toward Th9 differentiation (4). Moreover, there was a tendency of higher Spi1 (encoding PU.1) expression in CD96high Th9 cells. As Spi1 is reported to decrease Th2 cytokine expression (39), this might contribute to the diminished expression of Il4 and Il5 mRNA in CD96high Th9 cells and, thus, reduced inflammatory potential. However, we observed comparably low Spi1 expression levels in alloreactive Th9 and nonpolarized cells. This might be due to the late time point of analysis as Spi1 expression decreases over culture period reported for Th2 (39) and Th9 (40) culture conditions.
Recently, IRF1 was implicated in counteracting IRF4-driven Il9 promoter activity (40). We did not detect differences in Irf1 expression between CD96low and CD96high Th9 cells. This might explain why both subsets, CD96low and CD96high Th9 cells, express Il9 mRNA in contrast to activated nonpolarized T helper cells.
Interestingly, CD96high Th9 cells did express more Tcf7. Tcf7 encoding T-cell factor 1 (TCF1) has been implicated in the control of T helper cell differentiation and responses (41–44). Thus, higher Tcf7 expression in CD96high Th9 cells might contribute to their altered phenotype and function.
Taken together, our data indicate that expression of CD96 inhibits expansion and IL-9 production of Th9 cells, causing less pathology in vivo. Currently, the signaling events downstream of CD96 remain elusive, and also the regulation of CD96 expression needs further consideration. However, the clinical relevance of CD96-mediated immune inhibition was already shown in mice, where blockade of CD96 improved NK cell-dependent tumor control in multiple tumor models (20, 45).
Thus, manipulation of CD96-mediated immune inhibition may open new therapeutic opportunities: e.g., as a checkpoint inhibitor for reactivation of protective CD4+ T-cell memory in cancer or reinforcement of CD96 expression in the treatment of Th9-driven IBD, such as ulcerative colitis.
Materials and Methods
Mice.
Specific pathogen-free BALB/c and C57BL/6N mice were purchased from Charles River Laboratories. Rag1−/− mice on a C57BL/6 background were bred in the central animal facility of the Charité–Universitaetsmedizin Berlin. OT-I-Rag1−/− and Rag2−/− mice (both on a C57BL/6 background) were obtained from the animal facility of the Max Delbrück Center for Molecular Medicine, Berlin. Male mice aged between 2 and 3 (BALB/c, C57BL6N) or 4 and 6 mo (Rag1−/−, Rag2−/−, OT-I-Rag1−/−) were used for the experiments. Studies were performed in accordance with the recommendations of the Federation of European Laboratory Animal Science Association (FELASA) and approved by the relevant authority [Landesamt für Gesundheit und Soziales Berlin (LAGeSo)].
Human Samples.
Human blood donors gave their written consent to take part in this study approved by the local ethics committee (Ethikkommission der Charité–Universitaetsmedizin Berlin, EA1/293/16).
Cell Culture.
Dendritic cells (DCs) were generated by culturing isolated bone marrow cells for 8 d in the presence of 6 ng/mL granulocyte-macrophage colony stimulating factor (GM-CSF) (Miltenyi Biotec) in complete medium [RPMI 1640 with glutamine containing 10% (vol/vol) FCS, 100 U/mL penicillin, and 100 mg/mL streptomycin; all Biochrom AG]. Medium was changed every other day. On day 7, maturation was induced by adding 1 µg/mL lipopolysaccharide (LPS) (Sigma-Aldrich). Mature DCs were cocultured in a 1:12.5 ratio with naive CD4+ T cells isolated from a single cell suspension of spleen and lymph nodes using a Naive CD4+ T Cell Isolation Kit, mouse (Miltenyi) in complete medium supplemented with 50 µM β-mercaptoethanol (Sigma-Aldrich) for 3 d at 37 °C and 5% CO2 (= primary coculture). Cytokines for Th cell differentiation were added as follows: Th9: 50 ng/mL IL-4 (PeproTech), 5 ng/mL porcine TGF-β1 (R&D Systems); Th1: 10 ng/mL IL-12, 5 ng/mL IL-2 (both PeproTech), 10 µg/mL anti–IL-4 [produced in the Deutsches Rheuma-Forschungszentrum (DRFZ)]; and Th2: 2 ng/mL IL-2, 50 ng/mL IL-4, 10 µg/mL anti–IFN-γ (DRFZ), 10 µg/mL anti–IL-12 (Biolegend). If indicated, 5 µg/mL anti-CD96 (3.3) or IgG1 isotype control antibody (RTK2071; both Biolegend) were added to 24-h primary Th9 coculture for additional 48 h of culture.
For reactivation, cells were harvested from 3-d primary Th9 cocultures, and T cells were separated from DCs using a CD4+ T Cell Isolation Kit, mouse (Miltenyi) or were sorted into CD3+CD4+TCRβ+CD25+CD96high or -CD96low cells (see Fig. S2C for cell-sorting strategy). After 24 h resting, T cells were reactivated (second activ.) for an additional 3 d with mature allogeneic DCs in the presence of Th9-polarizing cytokines and, if indicated, 50 µg/mL anti-CD96 antibody or IgG1 isotype control antibody.
Flow Cytometry.
See Table S1 for all antibodies used. Cell staining was performed at 4 °C in the dark. Surface antigens were stained in PBS containing 2% (vol/vol) FCS and 0.1% sodium azide (Serva).
For cytokine detection in murine T cells, cells were treated with phorbol 12-myristate 13-acetate (PMA) (10 ng/mL; Sigma-Aldrich) and ionomycin (1 µg/mL; Biotrend) for 4 h at 37 °C, and Brefeldin A (2 µg/mL; Sigma-Aldrich) was added for the last 2 h. Cells were incubated with Mouse BD Fc Block (1:1,000; BD Biosciences) and Fixable Viability Dye (eBioscience) in PBS (Gibco/Life Technologies). For staining of intracellular antigens, cells were fixed and permeabilized using Fixation and Permeabilization Solution (BD Biosciences) and Intracellular Staining Permeabilization Wash Buffer (Biolegend) according to the manufacturer’s protocol. Data were acquired using an LSRFortessa or LSR II (both BD Biosciences) flow cytometer with FACSDiva software V6.2 (BD Biosciences) and analyzed with FlowJo v9 software (FlowJo, LLC).
For quantification of intracellular cytokines in human CD4+ T cells, peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll (Biochrom) gradient centrifugation and restimulated with 1 µg/mL staphylococcal enterotoxin B (SEB) (Sigma-Aldrich) for 24 h at 37 °C. After 20 h, 10 µg/mL Brefeldin A (Sigma-Aldrich) was added. Dead cells were stained using Zombie Aqua (Biolegend), and Fc receptors were blocked with 1 mg/mL Beriglobin (CSL Behring GmbH). Intracellular antigens were stained with Foxp3 Staining Buffer Set (Miltenyi). Data were acquired on a CytoFlex S with CytExpert 1.2 software (Beckman Coulter).
Cell Sorting.
Cells were stained in PBS containing 0.5% BSA and 2 mM EDTA and sorted on a FACSAria II (BD Biosciences).
T Cell Transfer and Skin Transplantation.
CD3+CD4+TCRβ+CD44+, CD3+CD4+TCRβ+CD25+CD96high, or CD3+CD4+TCRβ+CD25+CD96low cells were sorted from primary cocultures (see Fig. S2 for cell-sorting strategies). Then, 1 or 2 × 105 cells in PBS were injected into the tail vein of Rag1−/− recipients 1 d before skin transplantation. If indicated, animals were additionally injected with 250 µg of anti-CD96 antibody (3.3; Biolegend) intraperitoneally. For allogeneic skin transplantation, full-thickness BALB/c tail skin grafts with areas of 0.8 mm × 1.1 mm were prepared and engrafted onto the recipients’ backs. Weight changes and graft survival were monitored daily within the first 7 d and then at least every second day until fast weight loss started. Due to interexperimental variability, body weight was not recorded for each animal on each day but uniformly between all animals of one experiment. Rejection was defined as necrosis of more than 80% of the graft tissue. Body weight 1 d before skin transplantation was set as initial (= 100%), and experiments ended if one animal lost 20% of initial weight. Graft draining and mesenteric lymph nodes, spleen, ileum, colon, and skin graft were harvested. Total CD4+ T-cell number was determined with flow cytometry after restimulation with PMA and ionomycin.
RT2 Profiler PCR Array.
CD3+CD4+TCRβ+CD44+ cells were sorted from primary cocultures. Total RNA was isolated using a NucleoSpin RNA Kit (Macherey-Nagel) according to the manufacturer’s information. RNA was reverse transcribed into cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer’s protocol.
Extra- and intracellular stained Th9 cells were sorted into CD44+IL-4−IL-9+ (IL-9)/IL-4+IL-9− (IL-4), or IL-4−IL-9− (DN) cells (see Fig. S2B for gating strategy), and RNA was extracted using the RNeasy FFPE Kit (Qiagen). Deparaffinization of samples was not required, and cells were immediately resuspended in proteinase K digestion buffer, followed by the remaining steps of the manufacturer’s instructions. Up to 1 µg of RNA was reverse transcribed into cDNA as described before. Each cDNA sample was used to examine gene expression in one coculture, except for IL-4, where cDNA from three different cocultures had to be pooled due to limited cell number recovered. cDNA was diluted with RNase-free water and added to SYBR Green PCR Master Mix (Thermo Fisher). Then, 25 µL of the mixture were added to each well of the RT2 Profiler PCR Array Mouse Cell Surface Markers (Qiagen), and real-time qRT-PCR was performed on a 7500 Real Time PCR System (Applied Biosystems) using the following cycling conditions: 10 min at 95 °C and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Data analysis was conducted in R (46). Therefore, a cutoff at Ct 35 was set, and beta-2 microglobulin (B2m) was used as a reference gene. Variances (v) over all samples were calculated, and ΔCt values of genes with v > 0.5 (Fig. 2A) or v > 1 (Fig. 2D) were selected for hierarchical clustering analysis using default settings (Euclidian distances, complete linkage) of heatmap.2 function of R package “gplots” (47). Data were scaled for analysis and displayed as Log2Ex = −[Ct (Gene) − Ct (B2m)].
Single Cell Gene Expression Analysis.
CD3+CD4+TCRβ+CD44+ cells were harvested from primary cocultures. cDNA of single cells was prepared using the C1 Single-Cell Auto Prep System (Fluidigm). Freshly sorted cells at a concentration of 750 cells per microliter were loaded on a 10- to 17-µm C1 Single-Cell PreAmp Integrated Fluidic Circuit (IFC). Cell capture efficiency was controlled by visual inspection of each capture site under a microscope. To monitor technical accuracy, RNA Spike 1 (750 nucleotides; Thermo Fisher) was added to the Lysis Final Mix. Real-time qRT-PCR was performed using the BioMark Gene Expression 48.48 IFC and TaqMan gene expression assays (Thermo Fisher; see Table S2 for a list of all assays used). Data were obtained with the Biomark HD system (Fluidigm), and Ct values were calculated from Fluidigm Real-time PCR Analysis v4.1.3 software. Limit of detection (LOD) was set at Ct 24. Ct values were converted to expression levels using Log2Ex = LOD Ct − Ct [Gene]. If the value was negative, the expression was set to ND (not detectable). The following samples were excluded from data analysis in R: debris and samples with multiple cells, samples without detectable expression of B2m and/or RNA Spike 1, and genes that were expressed by less than 10 cells in total. Reference genes [B2m and hypoxanthine guanine phosphoribosyl transferase (Hprt)] were removed from principal component analysis (PCA) and hierarchical clustering analysis. Default settings of heatmap.2 function of R package “gplots” (47) were used.
Real-Time Quantitative Reverse Transcription–PCR.
Total RNA was isolated with a NucleoSpin RNA Kit (Macherey–Nagel), and cDNA was synthesized as described in RT2 Profiler PCR Array. Gene expression was measured using TaqMan gene expression assays (Thermo Fisher; see Table S2 for a list of all assays used) and TaqMan Universal PCR Master Mix (Thermo Fisher) on a 7500 Real Time PCR System (Applied Biosystems). Thermal cycling started with 2 min at 50 °C, followed by 10 min at 95 °C and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. All reactions were assessed as duplicates. Data were analyzed with 7500 Systems SDS Software v1.4 or v2.3. Relative gene expression was calculated using the 2-ΔΔCt method, with Hprt serving as reference gene.
Immunohistology.
Ileum, colon (both rinsed), and skin grafts were fixed in 10% formalin and embedded in paraffin. Paraffin sections of 1- to 2-µm thickness were cut, dewaxed, and stained histochemically with hematoxylin and eosin. Sections were coverslipped with corbit balsam (Hecht). For calculation of crypt height/villus depth ratio, length of 10 crypts and villi per sample was determined. Skin inflammation was scored according to the summary of epidermal and dermal infiltration: 1 = minimal, 2 = mild, 3 = moderate, 4 = severe. One additional point was given for complete loss of epidermis and one for sloughing (4 + 4 + 1 + 1 = maximum 10 points). Colon inflammation was scored based on mucosal and submucosal infiltration, epithelial hyperplasia, goblet cell loss, and crypt abscesses as described (48). See Fig. S4B for description of a moderately inflamed specimen.
For immunohistochemical detection of T cells, dewaxed paraffin sections were subjected to a heat-induced epitope retrieval step before incubation with anti-CD3 antibody (M-20; Santa Cruz Biotechnology), followed by incubation with biotinylated secondary antibody (Dianova). For detection, alkaline phosphatase-labeled streptavidin and chromogen RED (both Dako) were used. Nuclei were counterstained with hematoxylin (Merck). Eosinophils were detected with modified Sirius Red staining as described before (49). Cells were quantified in 10 high power fields (hpfs) (0.237 mm2) per section. Images were acquired using the AxioImager Z1 microscope (Carl Zeiss MicroImaging, Inc.). All evaluations were performed in a blinded manner.
Statistical Analysis.
Datasets are shown with median, as box plots with median and whiskers displaying minimum and maximum values or as mean with SEM indicated in the respective figure legend. Kaplan–Meier plots were analyzed using a Log-rank (Mantel–Cox) test. Statistical analyses of body weight data were performed with R package “lmerTest” (50). Therefore, a linear mixed-effect model was fitted to the longitudinal body weight data, and P values of the interaction term (group:time) were calculated using an ANOVA type III test.
Datasets of two matched samples were tested with a nonparametric Wilcoxon matched-pairs signed rank test. For unpaired datasets, a nonparametric Mann–Whitney test was used. If a preformed hypothesis was tested, one-tailed tests were applied and as such specified; otherwise, two-tailed tests were used. More than two samples were analyzed with a nonparametric Friedman test (paired samples) or nonparametric Kruskal–Wallis test (unpaired samples). P values were calculated using post hoc Dunn’s multiple comparison test. P < 0.05 was considered statistically significant. Statistical analyses were calculated with GraphPad Prism 6.00 or R v3.3.1 (46).
Supplementary Material
Acknowledgments
We thank Dr. Désirée Kunkel, Dr. Sarah Warth, and Jens Hartwig [Berlin–Brandenburg Center for Regenerative Therapies Flow Cytometry Laboratory (BCRT-FCL)] for assistance with cell sorting; and Dr. Eva J. Koziolek [Berlin Experimental Radionuclide Imaging Center (BERIC)] for great support with i.v. cell application. This work was supported by the Deutsche Forschungsgemeinschaft (Grant SFB650) and Bundesministerium für Bildung und Forschung (Grant e:Kid).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. O.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708329115/-/DCSupplemental.
References
- 1.Schmitt E, et al. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 2.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan MH. The transcription factor network in Th9 cells. Semin Immunopathol. 2017;39:11–20. doi: 10.1007/s00281-016-0600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci USA. 2015;112:7061–7066. doi: 10.1073/pnas.1415675112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licona-Limón P, et al. Th9 cells drive host immunity against gastrointestinal worm infection. Immunity. 2013;39:744–757. doi: 10.1016/j.immuni.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y, et al. Th9 cells promote antitumor immune responses in vivo. J Clin Invest. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Végran F, et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat Immunol. 2014;15:758–766. doi: 10.1038/ni.2925. [DOI] [PubMed] [Google Scholar]
- 10.Jäger A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staudt V, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Jones CP, Gregory LG, Causton B, Campbell GA, Lloyd CM. Activin A and TGF-β promote T(H)9 cell-mediated pulmonary allergic pathology. J Allergy Clin Immunol. 2012;129:1000–1010.e3. doi: 10.1016/j.jaci.2011.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlach K, et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol. 2014;15:676–686. doi: 10.1038/ni.2920. [DOI] [PubMed] [Google Scholar]
- 14.Nalleweg N, et al. IL-9 and its receptor are predominantly involved in the pathogenesis of UC. Gut. 2015;64:743–755. doi: 10.1136/gutjnl-2013-305947. [DOI] [PubMed] [Google Scholar]
- 15.Lu LF, et al. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 2006;442:997–1002. doi: 10.1038/nature05010. [DOI] [PubMed] [Google Scholar]
- 16.Xiao X, et al. GITR subverts Foxp3(+) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat Commun. 2015;6:8266. doi: 10.1038/ncomms9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 18.Stinn JL, et al. Interferon-gamma-secreting T-cell populations in rejecting murine cardiac allografts: Assessment by flow cytometry. Am J Pathol. 1998;153:1383–1392. doi: 10.1016/s0002-9440(10)65725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Elios MM, et al. Predominant Th1 cell infiltration in acute rejection episodes of human kidney grafts. Kidney Int. 1997;51:1876–1884. doi: 10.1038/ki.1997.256. [DOI] [PubMed] [Google Scholar]
- 20.Chan CJ, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol. 2014;15:431–438. doi: 10.1038/ni.2850. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 22.Blake SJ, Dougall WC, Miles JJ, Teng MW, Smyth MJ. Molecular pathways: Targeting CD96 and TIGIT for cancer immunotherapy. Clin Cancer Res. 2016;22:5183–5188. doi: 10.1158/1078-0432.CCR-16-0933. [DOI] [PubMed] [Google Scholar]
- 23.Dougall WC, Kurtulus S, Smyth MJ, Anderson AC. TIGIT and CD96: New checkpoint receptor targets for cancer immunotherapy. Immunol Rev. 2017;276:112–120. doi: 10.1111/imr.12518. [DOI] [PubMed] [Google Scholar]
- 24.Eriksson EM, et al. Differential expression of CD96 surface molecule represents CD8+ T cells with dissimilar effector function during HIV-1 infection. PLoS One. 2012;7:e51696. doi: 10.1371/journal.pone.0051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiesler P, Fuss IJ, Strober W. Experimental models of inflammatory bowel diseases. Cell Mol Gastroenterol Hepatol. 2015;1:154–170. doi: 10.1016/j.jcmgh.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabeen R, et al. Th9 cell development requires a BATF-regulated transcriptional network. J Clin Invest. 2013;123:4641–4653. doi: 10.1172/JCI69489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bamias G, Cominelli F. Role of type 2 immunity in intestinal inflammation. Curr Opin Gastroenterol. 2015;31:471–476. doi: 10.1097/MOG.0000000000000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen MJ, et al. STAT6 deficiency ameliorates severity of oxazolone colitis by decreasing expression of claudin-2 and Th2-inducing cytokines. J Immunol. 2013;190:1849–1858. doi: 10.4049/jimmunol.1201373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uppal V, Kreiger P, Kutsch E. Eosinophilic gastroenteritis and colitis: A comprehensive review. Clin Rev Allergy Immunol. 2016;50:175–188. doi: 10.1007/s12016-015-8489-4. [DOI] [PubMed] [Google Scholar]
- 32.Wynn TA. Type 2 cytokines: Mechanisms and therapeutic strategies. Nat Rev Immunol. 2015;15:271–282. doi: 10.1038/nri3831. [DOI] [PubMed] [Google Scholar]
- 33.Moshkovits I, et al. A key requirement for CD300f in innate immune responses of eosinophils in colitis. Mucosal Immunol. 2017;10:172–183. doi: 10.1038/mi.2016.37. [DOI] [PubMed] [Google Scholar]
- 34.Meyer D, et al. CD96 interaction with CD155 via its first Ig-like domain is modulated by alternative splicing or mutations in distal Ig-like domains. J Biol Chem. 2009;284:2235–2244. doi: 10.1074/jbc.M807698200. [DOI] [PubMed] [Google Scholar]
- 35.Wang PL, O’Farrell S, Clayberger C, Krensky AM. Identification and molecular cloning of tactile. A novel human T cell activation antigen that is a member of the Ig gene superfamily. J Immunol. 1992;148:2600–2608. [PubMed] [Google Scholar]
- 36.Bennett F, et al. Program death-1 engagement upon TCR activation has distinct effects on costimulation and cytokine-driven proliferation: Attenuation of ICOS, IL-4, and IL-21, but not CD28, IL-7, and IL-15 responses. J Immunol. 2003;170:711–718. doi: 10.4049/jimmunol.170.2.711. [DOI] [PubMed] [Google Scholar]
- 37.Seth S, et al. The murine pan T cell marker CD96 is an adhesion receptor for CD155 and nectin-1. Biochem Biophys Res Commun. 2007;364:959–965. doi: 10.1016/j.bbrc.2007.10.102. [DOI] [PubMed] [Google Scholar]
- 38.Yi JS, Cox MA, Zajac AJ. T-cell exhaustion: Characteristics, causes and conversion. Immunology. 2010;129:474–481. doi: 10.1111/j.1365-2567.2010.03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang HC, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Campos Carrascosa L, et al. Reciprocal regulation of the Il9 locus by counteracting activities of transcription factors IRF1 and IRF4. Nat Commun. 2017;8:15366. doi: 10.1038/ncomms15366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Yu D. TCF-1 at the Tfh and Th1 divergence. Trends Immunol. 2015;36:758–760. doi: 10.1016/j.it.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Choi YS, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muranski P, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blake SJ, et al. Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discov. 2016;6:446–459. doi: 10.1158/2159-8290.CD-15-0944. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team 2015. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.3.1.
- 47.Warnes GR, et al. 2016. gplots: Various R Programming Tools for Plotting Data, Version 3.0.1. Available at https://CRAN.R-project.org/package=gplots. Accessed November 17, 2016.
- 48.Zimmermann J, et al. T-bet expression by Th cells promotes type 1 inflammation but is dispensable for colitis. Mucosal Immunol. 2016;9:1487–1499. doi: 10.1038/mi.2016.5. [DOI] [PubMed] [Google Scholar]
- 49.Peine M, et al. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 2013;11:e1001633. doi: 10.1371/journal.pbio.1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuznetsova A, Brockhoff PB, Christensen RHB. 2016. lmerTest: Tests in Linear Mixed Effects Models, Version 2.0-33. Available at https://CRAN.R-project.org/package=lmerTest. Accessed December 18, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.