Significance
Because auxin herbicides selectively control broadleaf weeds, their use is extremely valuable in crops, such as wheat and corn. Although auxin-resistant weeds have appeared rarely over the past 60 years of herbicide use, they pose a major challenge in these cropping systems. Several groups have investigated the mechanisms of resistance for several of these weed species; this paper reports the identification of the underlying genetic mechanism of auxin resistance in a field-derived weed species. This mutation sits within a highly conserved region previously identified in Arabidopsis studies as vital for auxin signaling and points to the importance of studies in model systems to predict resistance mechanisms.
Keywords: herbicide, weed resistance, auxin, KsIAA16, KsTIR1
Abstract
The understanding and mitigation of the appearance of herbicide-resistant weeds have come to the forefront of study in the past decade, as the number of weed species that are resistant to one or more herbicide modes of action is on the increase. Historically, weed resistance to auxin herbicides has been rare, but examples, such as Kochia scoparia L. Schrad (kochia), have appeared, posing a challenge to conventional agricultural practices. Reports of dicamba-resistant kochia populations began in the early 1990s in areas where auxin herbicides were heavily utilized for weed control in corn and wheat cropping systems, and some biotypes are resistant to other auxin herbicides as well. We have further characterized the auxin responses of one previously reported dicamba-resistant biotype isolated from western Nebraska and found that it is additionally cross-resistant to other auxin herbicides, including 2,4-dichlorophenoxyacetic acid (2,4-D) and fluroxypyr. We have utilized transcriptome sequencing and comparison to identify a 2-nt base change in this biotype, which results in a glycine to asparagine amino acid change within a highly conserved region of an AUX/indole-3-acetic acid (IAA) protein, KsIAA16. Through yeast two-hybrid analysis, characterization of F2 segregation, and heterologous expression and characterization of the gene in Arabidopsis thaliana, we show that that the single dominant KsIAA16R resistance allele is the causal basis for dicamba resistance in this population. Furthermore, we report the development of a molecular marker to identify this allele in populations and facilitate inheritance studies. We also report that the resistance allele confers a fitness penalty in greenhouse studies.
Auxin is a central regulator in plant growth and development and has been implicated in numerous developmental and response pathways (1). Because this plant hormone is key to so many critical plant pathways, perturbation of auxin levels or response often leads to abnormal development and/or plant death, which has led to the development of herbicides that mimic auxin action. Whereas the predominant naturally occurring auxin is indole-3-acetic acid (IAA), many other compounds have been found to confer auxin-like activity when applied to plants (2). Auxin herbicides include 2,4-dichlorophenoxyacetic acid (2,4-D), which was the first widely used commercial herbicide; 3,6-dichloro-2-methoxybenzoic acid (dicamba); [(4-amino-3,5-dichloro-6-fluoro-2-pyridinyl)oxy]acetic acid (fluroxypyr); 4-amino-3,5,6-trichloro-2-pyridinecarboxylic acid (picloram); [(3,5,6-trichloro-2-pyridinyl)oxy]acetic acid (triclopyr); 3,7-dichloro-8-quinolinecarboxylic acid (quinclorac); and many others. Weed resistance to auxin herbicides has been selected only rarely, despite widespread use of the herbicides for more than 60 y. Worldwide, there are a total of 36 weeds classified as resistant to auxin herbicides (3), with only 7 of these weeds being resistant to dicamba. Dicamba-resistant species include Amaranthus hybridus (smooth pigweed), Sinapis arvensis L. (wild mustard), Chenopodium album L. (common lambsquarters), Lactuca serriola L. (prickly lettuce), Centaurea cyanus L. (cornflower), Galeopsis tetrahit L. (common hempnettle), and Kochia scoparia L. Schrad (kochia) (3).
Kochia is a prolific seed producer, has protogynous flowers that are wind pollinated, and at maturity, becomes a tumbleweed that can disperse seed over great distances as it is blown across fields. These characteristics have given rise to great genetic diversity and rapid spread of favorable alleles, including those for herbicide resistance (4). In kochia, populations with resistance to auxin herbicides, acetolactate synthase (ALS) inhibitors, photosystem II inhibitors, and glyphosate have been reported (3), and recently, a kochia biotype from Kansas has been reported with resistance to all four of these herbicide classes (5).
Dicamba resistance was first documented in kochia in 1994 in populations isolated from fields in western Nebraska and Montana (6). Since the initial occurrence, multiple reports of dicamba-resistant kochia biotypes from Nebraska, Montana, and North Dakota have been published (4, 6–10). Since many of these studies have utilized newly isolated field-harvested samples, it is not yet clear how many distinct loci/alleles may be responsible for the observed auxin resistance. In some cases, the resistance has been postulated to be due to quantitative traits (6), whereas in other cases, the resistance has been shown to be inherited from a single dominant allele (4). For some biotypes, cross-resistance to other auxin herbicides has also been observed (8, 9), and based on reports by Goss and Dyer (8), Cranston et al. (6), and Preston et al. (4), there seem to be at least two to three biotypes with distinct resistance profiles. Only genetic studies or sequence analysis, however, will determine if these different biotypes contain mutations in the same gene(s).
Although auxin function has been investigated for more than a century, it is only recently that the molecular transport and signaling mechanisms have been uncovered for this plant hormone. The natural auxin IAA has been shown to be transported in a polar fashion throughout the plant via the function of influx carrier AUX1/LAX family members and efflux carrier PIN and ABCB families (Fig. S1) (reviewed in ref. 11). This transport can be further regulated by the function of additional interacting proteins and by flavonoids, which modulate the activity of ABCB transporters (12). In two recent reports, reduced translocation of auxins has been shown to be associated with resistance (13, 14). Furthermore, this reduction in auxin transport was recently associated with chalcone synthase up-regulation in the same kochia line used in this study (14). In addition to our understanding of auxin transport, F-box proteins in the TIR1/AFB family have been shown to be receptors for auxin as illustrated in Fig. S1 (15, 16), and they bind the auxin molecule in concert with AUX/IAA family proteins. The association of auxins with TIR/AFB proteins fills a hydrophobic pocket within the binding domain of the protein and facilitates stronger interaction with the AUX/IAA proteins (17, 18). The F-box proteins are subunits of the SCF E3 ubiquitin ligase complex, and this auxin-enhanced interaction results in the ability of the larger SCF complex to efficiently ligate ubiquitin onto the AUX/IAA proteins, which targets them for degradation via the 26S proteasome, thus releasing ARF transcriptional regulators from their effects (15–17). Mutations that disrupt the ability of these AUX/IAA proteins to bind auxin within the context of the TIR/AFB coreceptor complex result in the loss of TIR/AFB-mediated ubiquitination (19) and thus, protect these proteins from degradation, resulting in dominant auxin-resistant phenotypes.
Despite its rarity to date, weed resistance to auxin herbicides still poses a serious threat to agriculture, and diligence must be practiced to control these resistant weeds to prevent their spread. One tool for combating weed resistance to any herbicide is an understanding of the molecular mechanisms giving rise to the resistance. This knowledge can be used to inform cropping practices as well as to drive future innovation. To this end, we have investigated the genetic basis of dicamba resistance in kochia, which has become problematic in the Northern Great Plains of the United States. We have based our studies on the observed phenotypes of resistant biotype 9425R (R) from western Nebraska (4) as well as on information garnered from the literature on auxin-resistant mutants from Arabidopsis thaliana. We report here the identification of the genetic basis for the dicamba-resistant phenotype of this kochia biotype, which results from a mutation in an AUX/IAA gene, as well as the characterization of the function and expression of wild-type and mutant alleles responsible for this resistance. We also report the development of a molecular marker to rapidly identify this allele in kochia populations as well as greenhouse-based fitness studies that indicate that the presence of the resistance allele conveys a fitness penalty.
Results
The Dicamba-Resistant Kochia Biotype Is also Resistant to Other Auxin Herbicides.
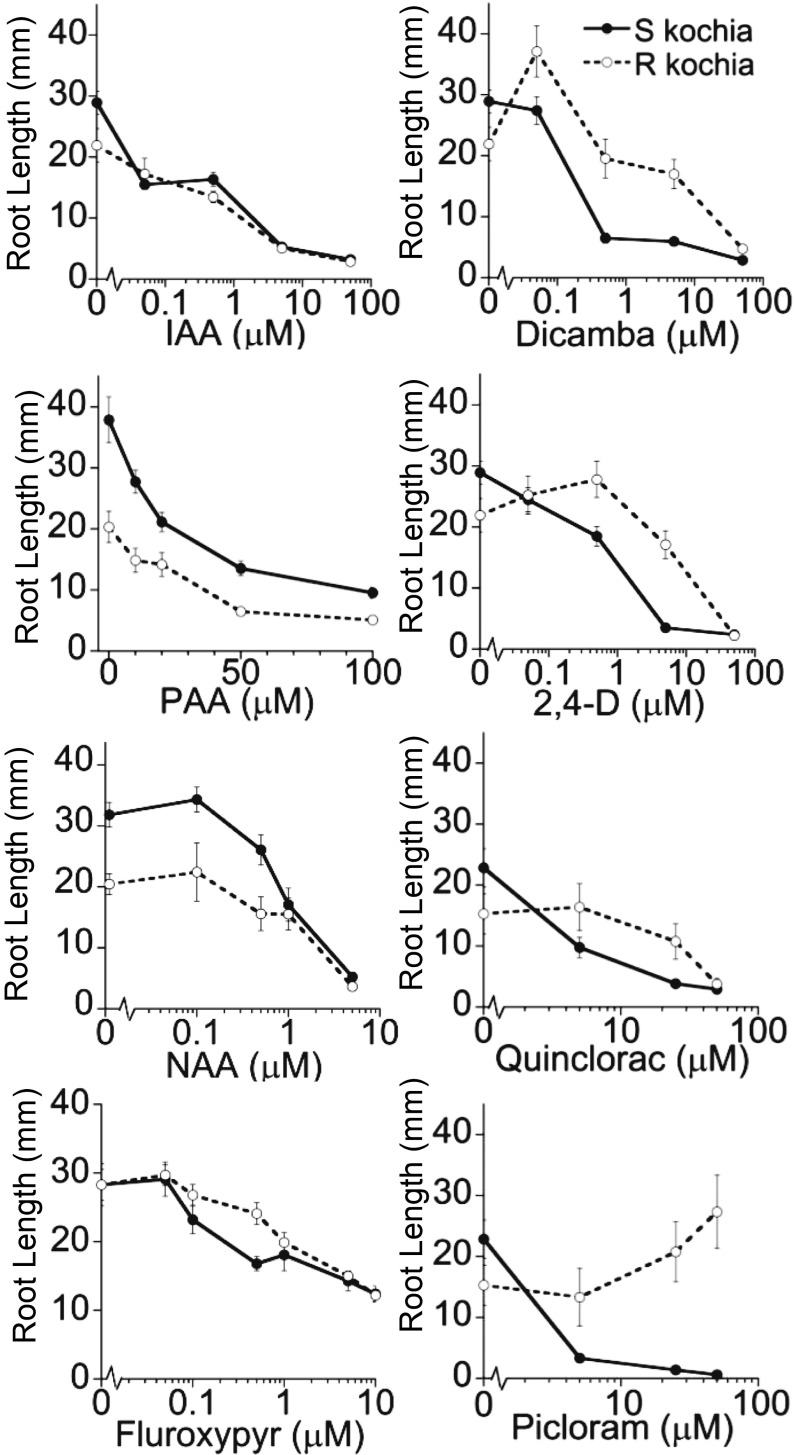
The dicamba-resistant biotype used in these studies was described in detail previously, and it was estimated to convey a nearly 30-fold increase in dicamba tolerance and to result from a dominant or semidominant single genetic locus (4); however, to our knowledge, the resistance of this biotype to other synthetic and natural auxins has not been examined in detail. We, therefore, tested the root elongation response of seedlings from this R biotype and a sensitive (S) kochia biotype in the presence of the natural auxins IAA, and phenyl acetic acid (PAA) and the synthetic auxins naphthalene acetic acid (NAA), dicamba, 2,4-D, picloram, quinclorac, and fluroxypyr to determine if the R biotype displayed cross-resistance to other forms of auxin. Results are shown in Fig. 1. Auxins are known to inhibit root elongation; thus, longer root length indicates a resistance to this inhibition. One observation that we made is that the R biotype displays a short root phenotype in the absence of auxin; thus, we tested several concentrations of each auxin to get a clearer picture of response. We noted that the R kochia biotype remains sensitive to root elongation inhibition by IAA, PAA, and NAA but is resistant to inhibition by dicamba, 2,4-D, picloram, and some concentrations of fluroxypyr and quinclorac, indicating that the mechanism of resistance in this biotype is not specific to dicamba alone.
Fig. 1.
The kochia R biotype is resistant to inhibition by several auxin herbicides. Kochia-sensitive (S) and dicamba-resistant (R) biotype root elongation inhibition by various auxins. Seeds were germinated and grown on plant nutrient media containing 0.5% sucrose in the presence of various auxin concentration as described in Materials and Methods. Root lengths were measured 8 d after plating. Each data point represents the mean root length of 12 seedlings, and error bars represent SEMs.
Because the R biotype showed decreased sensitivity to root inhibition by 2,4-D and fluroxypyr, both of which are also commonly used to control kochia in the field, we performed greenhouse dose–response studies with these two herbicides to determine the levels of resistance to these two additional auxin herbicides. We also included dicamba to serve as a comparator between our results and previous studies. Visible control ratings and photos were taken at both 14 and 21 d after treatment (DAT), and fresh weights were measured at the end of 21 d. Data were analyzed in R software, and calculations are described in SI Materials and Methods. GR10, GR50, and GR90 values represent the herbicide doses required to provide 10%, 50%, and 90% reduction in fresh weight, respectively, compared with untreated control and are reported in Table 1. Photos taken 14 DAT and R curves from 21 DAT fresh weights, expressed as the percentage of untreated control, are shown in Fig. S2. These data show that, in addition to having a greater than 30-fold decrease in dicamba sensitivity, which agrees with previous reports (4), this biotype is also resistant to 2,4-D and fluroxypyr, with R/S ratios showing about a 12-fold increase in GR50 values.
Table 1.
Growth reduction values and resistance ratios on dicamba, 2,4-D, and fluroxypyr
| Herbicide | GR10* (±SE) | GR50† (±SE) | GR90‡ (±SE) | R/S§ (ED50) |
| Dicamba | ||||
| R line | 1,596 (323) | 6,784 (439) | 28,827 (5,752) | 38 |
| S line | 14 (6) | 179 (27) | 2,230 (612) | |
| 2,4-D | ||||
| R line | 702 (122) | 2,619 (272) | 9,770 (2,227) | 12 |
| S line | 29 (10) | 220 (29) | 1,655 (430) | |
| Fluroxypyr | ||||
| R line | 72 (19) | 1,057 (139) | 15,583 (5,706) | 13 |
| S line | 19 (5) | 83 (7) | 368 (73) |
GR10 represents the effective dose in grams per hectare required to achieve 10% reduction in fresh weight compared with untreated control.
GR50 represents the effective dose in grams per hectare required to achieve 50% reduction in fresh weight compared with untreated control.
GR90 represents the effective dose in grams per hectare required to achieve 90% reduction in fresh weight compared with untreated control.
R/S is the resistance ratio, and it is calculated by dividing the GR50 of the R line by the GR50 of the S line to determine the relative resistance in fold.
Sequence Analysis of the K. scoparia Transcriptome Reveals a Mutation in an AUX/IAA Gene.
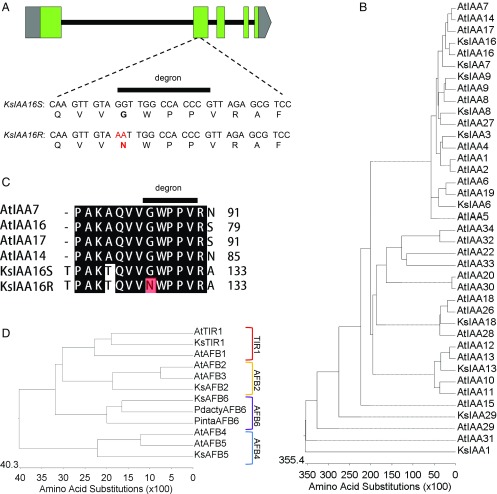
Since the R biotype is resistant to foliar auxin herbicide sprays, we chose to focus our sequencing efforts on aerial tissues, where we hypothesized that the resistance factor would be expressed and operational. Transcriptome sequencing of cDNA samples derived from kochia leaf tissues of greenhouse-grown R and S biotypes allowed comparative analysis (BLAST) (20) of putatively orthologous candidate genes known to be involved in auxin synthesis, transport, and regulatory response in other species, such as Arabidopsis. Comparators included one AUX1/LAX transporter, one ABP homolog, several multidrug resistance-like proteins, four TIR/AFB coreceptors (Fig. 2D), and 10 AUX/IAA proteins (phylogeny is shown in Fig. 2B, and accession numbers are listed in Table S1). Using this approach, we discovered only one seemingly significant difference between the kochia R and S biotypes in any of the transcripts examined. The identified difference is a GG to AA nucleotide change, which results in a glycine (G) to asparagine (N) amino acid substitution within the highly conserved degron region (GWPPV/I) of a homolog of IAA16 (Fig. 2 A and C). This degron region has been shown to be indispensable for auxin binding and interaction between the TIR/F-box proteins and AUX/IAA proteins (17, 21). This interaction is required for E3 ligase complex-mediated ubiquitination of the AUX/IAA proteins and their subsequent degradation via the 26S proteasome as shown in Fig. S1 (17). Changes within this 5-aa region prevent this interaction, lead to increased stability of the AUX/IAA proteins, and thus, have been reported to result in dominant auxin resistance in a number of Arabidopsis mutants, including axr5 (IAA1) (22), shy2 (IAA3) (23), axr2 (IAA7) (24), slr (IAA14) (25), axr3 (IAA17) (26), crane (IAA18) (27), bodenlos (IAA12) (28), msg2 (IAA19) (29), iaa16 (30), and iaa28 (31) as shown in Table S2. We, therefore, hypothesized that this KsIAA16 mutation is likely the basis of the observed dicamba resistance in this kochia biotype and sought to test this hypothesis.
Fig. 2.
KsIAA16 gene structure, IAA degron domain mutations, phylogeny, and expression. A illustrates the gene structure of KsIAA16 spanning 4,059 bp. Boxes indicate exons, green coloration indicates the translated region, and gray coloration indicates the 5′ and 3′ UTRs, whereas lines show intron positioning. Nucleotide and amino acid differences between the sensitive (S) and resistant (R) alleles of KsIAA16 (degron domain indicated) are shown. B is a phylogenetic tree containing all of the Arabidopsis AUX/IAA proteins as well as all of the putatively orthologous kochia AUX/IAA proteins identified in the transcriptome sequences. C shows the region of interest in an alignment with the most closely related Arabidopsis homologs, with the mutation in the KsIAA16R highlighted in red. D shows the phylogenetic tree of the Arabidopsis and kochia TIR/AFB family of F-box proteins identified in the transcriptome sequences.
Sequence-Based Analysis Identifies Four TIR/AFB Homologs from the Kochia Leaf Transcriptome.
Because AUX/IAA proteins are known to act as auxin coreceptors in concert with TIR/AFB proteins (Fig. S1), we also sought to express putative homologs of TIR/AFB from kochia to allow us to determine if the KsIAA16 protein could interact with these proteins in the presence of auxins. Such an interaction would indicate that the proteins are auxin coreceptors as expected based on sequence homology. As mentioned above, we identified four TIR/AFB homologs in kochia using BLAST analysis. To gain a clearer understanding of the phylogenetic relation of these four homologs, proteins were aligned with TIR1, AFB1, AFB2, AFB3, AFB4, and AFB5 from Arabidopsis as well as AFB6 homologs from Phoenix dactylifera L. (date palm) and Pinus taeda L. (loblolly pine) (32). Alignments were performed using the Clustal W method in MegAlign software (DNASTAR), and a phylogenetic tree was generated as shown in Fig. 2D. From this phylogeny, it seems that kochia contains at least one homolog from each known clade of the TIR/AFB family. Since our transcriptome dataset was generated from only aerial tissues of vegetative-stage kochia plants, we cannot rule out the possibility of additional homologs that are not expressed in these tissues at the time of sampling and are, therefore, not represented in our dataset.
Yeast Two-Hybrid Assays Show That KsIAA16 Is an Auxin Coreceptor.
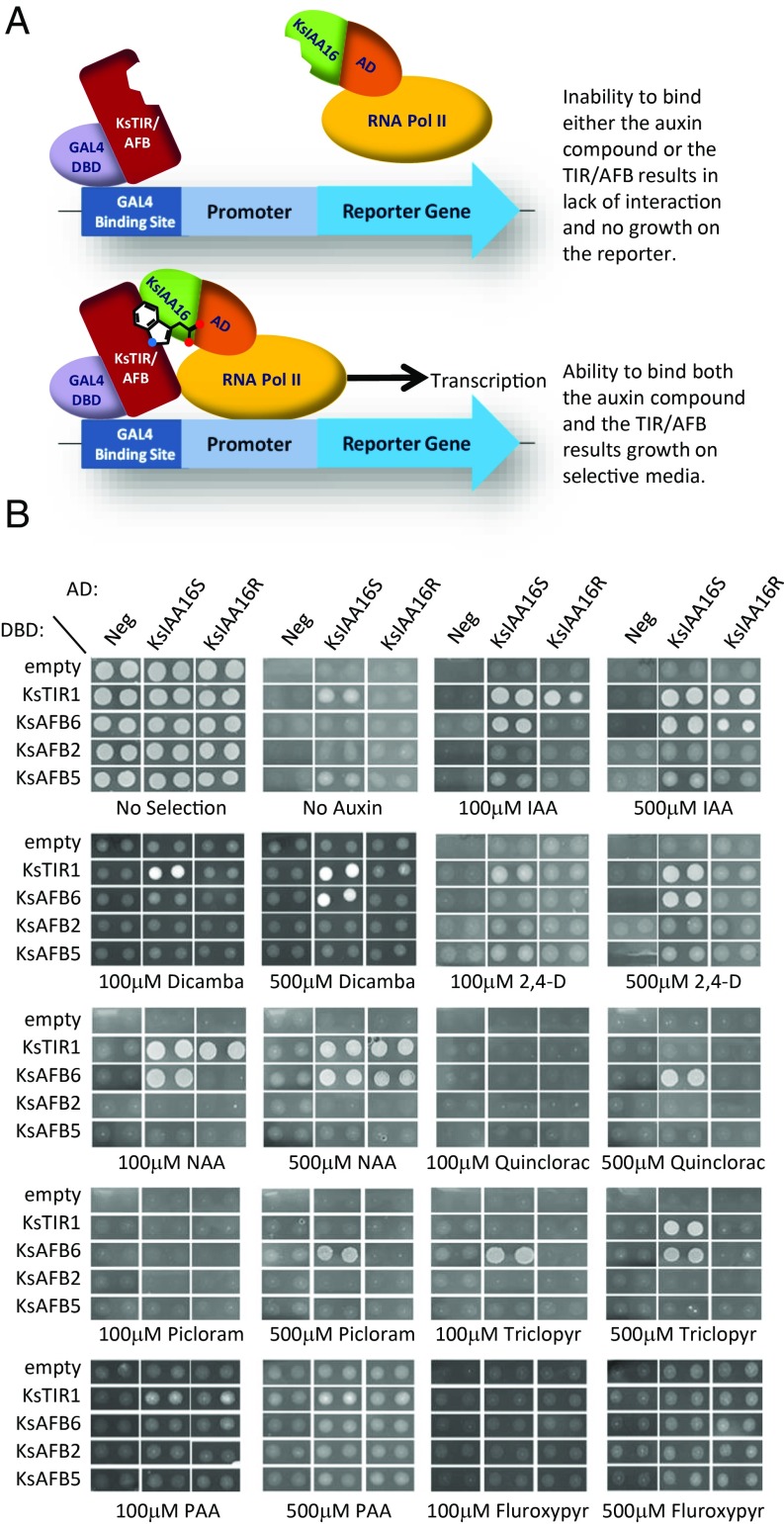
To further understand the role of KsIAA16 in auxin perception as well as to determine whether the identified mutation alters the ability of the protein to interact with TIR/AFB homologs in the presence of auxins (33), we performed yeast two-hybrid (Y2H) analysis. This method allowed us to query binding/interaction of the kochia R and S forms of KsIAA16 with each of the four identified kochia TIR/AFB homologs KsTIR1, KsAFB6, KsAFB2, and KsAFB5 in the absence or presence of the natural auxins IAA and PAA and the synthetic auxins NAA, dicamba, 2,4-D, picloram, triclopyr, fluroxypyr, and quinclorac. Based on previously published work (33), we utilized concentrations of 100 and 500 μM exogenous auxin for these tests. While these levels are one to two orders of magnitude higher than that used in our kochia root elongation assays, we would expect that compartmentalization of the auxin in the root studies could result in subcellular concentrations higher than the exogenous dose. Furthermore, it is likely that yeast differs from plants in its uptake and compartmentalization of these auxins; thus, these experiments are not meant to predict effective in planta concentrations. Results from the Y2H studies are shown in Fig. 3. We showed that all combinations of bait and prey vectors displayed good growth in the absence of selection, indicating that none of the transcript expression had inherent negative effects on yeast growth. In the absence of auxin, little to no growth is observed for either the R or S allele of KsIAA16 with any tested KsTIR/AFB homolog, indicating that the proteins do not interact in the absence of auxin, which is the expected result based on known interactions in Arabidopsis (17, 33). We did not observe interaction of either allele of KsIAA16 with either KsAFB2 or KsAFB5 in our system. Both KsIAA16S and KsIAA16R exhibited interaction with KsTIR1 in the presence of 100 and 500 μM IAA and NAA; however, KsIAA16R did not show interaction with KsTIR1 in the presence of 100 or 500 μM dicamba or 500 μM 2,4-D or triclopyr, whereas KsIAA16S did. Furthermore, we observed that KsIAA16S could interact with KsAFB6 in the presence of 100 and 500 μM IAA and NAA as well as 500 μM dicamba, 2,4-D, picloram, quinclorac, and triclopyr, but KsIAA16R could only interact with KsAFB6 in the presence of 500 μM IAA and NAA. Surprisingly, we did not observe any interaction in the presence of PAA or fluroxypyr with any of the protein combinations tested. These findings support the hypothesis that KsIAA16 is an auxin coreceptor and that it is capable of binding with either KsTIR1 or KsAFB6 in the presence of several different auxin compounds. Furthermore, the mutant form KsIAA16R has a decreased ability to interact with KsTIR1 and/or KsAFB6 in the presence of several tested auxin herbicides, although it still has at least some capacity to interact with these proteins in the presence of IAA and NAA. This is consistent with the observed resistance to these compounds in our root inhibition and greenhouse studies (Figs. 2 and 3). These observations in combination with the auxin herbicide resistance results provide further support for the hypothesis that the auxin resistance observed in this kochia biotype is due to the mutation in KsIAA16R.
Fig. 3.
Interactions of KsIAA16, KsTIR/AFB proteins, and auxins. A shows a cartoon representation of this type of Y2H assay and the possible results. B shows the results for various combinations of KsTIR/AFB homologs fused to the DNA-binding domain (DBD) as “bait” and KsIAA16S or R fused to the activation domain (AD) as “prey.” Five microliters of dilutions from each colony were replica pipetted onto –Trp –Leu –His media containing either 100 or 500 μM of the indicated auxin compound to test for expression of the HIS3 reporter as an indicator of protein–protein interaction. The “no selection” plate contained histidine and served as a positive control for colony viability. The noninteracting protein SV40 large T antigen fused to the activation domain served as a noninteracting control for the TIR/AFB proteins, and the empty DNA binding domain construct was a negative control for KsIAA16 interaction.
Segregation Analysis Shows That KsIAA16R Segregates with Dicamba Resistance.
To facilitate genetic inheritance and fitness studies as well as to allow the rapid identification of this auxin resistance allele in other kochia populations, we sought to develop a molecular maker to distinguish the identified KsIAA16R allele. Based on the identified base pair substitutions in KsIAA16 between the S and R kochia biotypes, we generated a TaqMan genotyping (34) assay (Fig. S3A). To determine if the identified KsIAA16R allele segregates with the dicamba resistance phenotype and therefore, represents the causal mutation leading to resistance, we performed crosses between S kochia and R kochia individuals by dusting pollen from the R line on the flowers of the S line. To allow easier determination of crossing success, we utilized the S biotype as the female and the R biotype as the male to allow dicamba sprays to be conducted to confirm individuals resulting from cross-pollination. Because of the anatomy of kochia reproductive organs, preventing some self-pollination is impractical; however, utilization of the TaqMan genotyping assay allowed us to distinguish which progeny were true crosses heterozygous for KsIAA16 (RS) vs. those resulting from self-pollination (SS). We planted 250 putative F1 seeds harvested from the crossed female plants and performed TaqMan assays on DNA from a leaf of each individual. We thus identified 24 heterozygous F1 plants resulting from successful cross-pollination. Several verified heterozygous F1 plants were selected for F2 seed production. In addition, a subset of 15 SS and 7 RS individuals from this generation was subjected to dicamba sprays to ensure that the marker could effectively identify the R allele (Fig. S3B). We observed perfect correlation in this small F1 spray test between those that were determined to be heterozygous for the R allele and those that showed resistance to dicamba, indicating that the marker is effective in detecting the KsIAA16R allele. This result also further supported the previous finding by Preston et al. (4) that the dicamba resistance trait in this kochia biotype has a high degree of dominance.
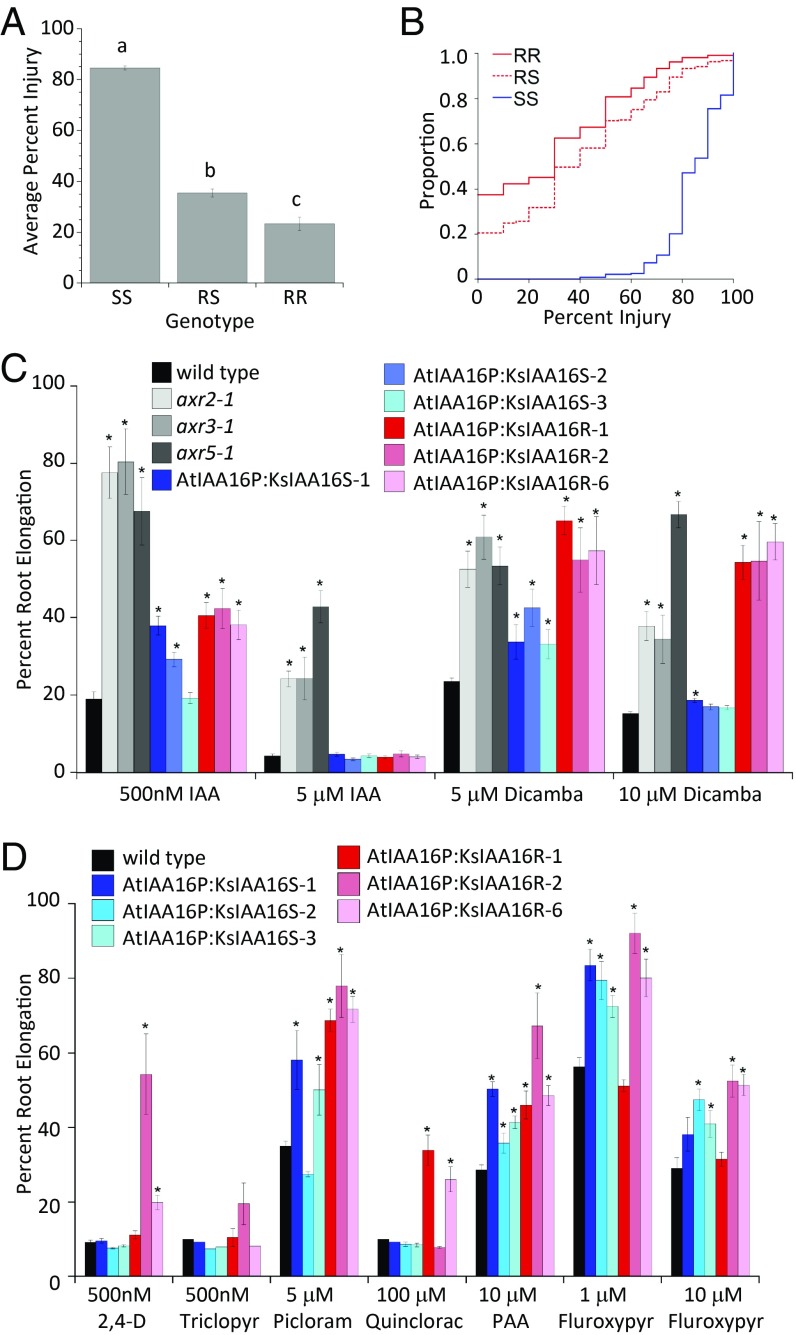
To determine definitively if the KsIAA16R allele cosegregates with the dicamba resistance phenotype, 720 of the F2 progeny from verified F1 parental plants were sampled, genotyped, and sprayed with dicamba at a 1,120 g ha−1 rate. Results are shown in Fig. 4 A and B. Based on the TaqMan genotyping assay, we identified 234 homozygous-sensitive plants (SS), 371 heterozygous plants (RS), and 105 homozygous-resistant plants (RR), with 14 plants returning an undetermined genotype and thus, representing a 2% PCR failure rate. These genotype numbers are significantly different (1:1.6:0.4) than the expected Mendelian 1:2:1 segregation pattern (P < 0.05); however, the pots were hand-thinned to one plant per plug before sampling. We have observed a height difference for the R biotype and suspect that this skewed numbers, as smaller plants are typically discarded when thinning. Plant identity was maintained throughout the experiment, and genotype and percentage of visible dicamba injury were determined for each of the F2 progeny. We found good correlation between our genotype results and the observed dicamba injury, with a mean injury rating of 82.3% for SS individuals, 35.9% for RS individuals, and 24.6% for R/R individuals. Statistical analysis of these data using ANOVA indicates with high likelihood that the resistance phenotype is associated with the presence of the KsIAA16R allele (P < 0.0001), and Cramér–von Mises statistical analysis shows the trait to be semidominant (Fig. 4B). These results further prove that the KsIAA16R allele is the causal basis for dicamba resistance in this biotype.
Fig. 4.
The KsIAA16 R allele confers dicamba resistance in kochia and Arabidopsis. A shows average injury by genotype of the segregating F2 kochia population and different letters a, b, and c indicate values that are significantly different from one another; 710 F2 progeny from three independently verified F1 parental plants were sampled, successfully genotyped, and sprayed with dicamba at a 1,120-g ha−1 rate. Based on the TaqMan genotyping assay, n = 234 for SS individuals, n = 371 for RS, and n = 105 for RR. B shows a Cramér–von Mises plot of injury by genotype for this same kochia experiment, which indicates that the R allele is semidominant. C shows the root elongation inhibition by 5 and 10 μM dicamba and 500 nM and 5 μM IAA in three independent transgenic Arabidopsis lines carrying either the KsIAA16S allele (blue shades) or the KsIAA16R (red shades) as well as the Arabidopsis mutants axr2-1, axr3-1, and axr5-1 (gray shades) for comparison. Each column represents the mean of 12 individuals, error bars represent SEs, and asterisks denote significant difference from the wild type. *P < 0.05. D shows the percentage of root elongation of these same transgenic Arabidopsis lines on other auxins, including 500 nM 2,4-D, 500 nM triclopyr, 5 μM picloram, 100 μM quinclorac, 10 μM PAA, and 1 and 10 μM fluroxypyr. Each column represents the mean of 12 individuals, error bars represent SEs, and asterisks denote significant difference from the wild type. *P < 0.05.
Heterologous Expression in A. thaliana Shows That the KsIAA16R Allele Is Sufficient to Cause Auxin Resistance.
To determine if the presence of the KsIAA16R allele was sufficient to cause dicamba resistance on its own, we generated Arabidopsis transgenic lines containing either the KsIAA16R or -S allele driven by the native Arabidopsis IAA16 promoter (AtIAA16). As shown in Fig. 4C, three independent transformant lines carrying AtIAA16P:KsIAA16R showed strong resistance to dicamba at both 5 and 10 μM and slight but significant resistance to IAA at 0.5 μM but not at 5 μM in a root elongation inhibition assay. For lines carrying AtIAA16P:KsIAA16S, all three tested lines showed slight resistance to 5 μM dicamba, and two separate lines showed slight resistance to 0.5 μM IAA. For comparison, we also tested resistance of previously reported Arabidopsis mutants axr2-1 (24, 35), axr3-1 (26, 36), and axr5-1 (22), which have mutations in the degron regions of IAA7, IAA17, and IAA1, respectively. Table S2 shows specific mutations. We found that these mutants had strong resistance to IAA as previously reported, and all also showed resistance to dicamba. This finding indicates that KsIAA16R is not unique in its ability to confer dicamba tolerance and shows that these similar gain-of-function mutations in Arabidopsis AUX/IAA genes also result in tolerance to dicamba. This result suggests that these proteins are also involved in dicamba perception.
To determine whether heterologous expression of KsIAA16 alleles was also sufficient to result in resistance to the other tested auxins, we also tested response to 2,4-D, triclopyr, picloram, quinclorac, fluroxypyr, and PAA as shown in Fig. 4D. We found that at least two of three KsIAA16R lines showed significant resistance to 2,4-D, picloram, quinclorac, fluroxypyr, and PAA, and two or more of the KsIAA16S lines were also resistant to picloram, fluroxypyr, and PAA. It thus seems that heterologous expression of KsIAA16R is sufficient to cause resistance to dicamba as well as all of the other auxins tested, with the exception of triclopyr, which was not statistically different. Heterologous expression of KsIAA16S can sometimes cause resistance to exogenous auxins, albeit to a somewhat lesser extent than that observed for the R allele. We hypothesize that the slight resistance observed for the S allele is due to an increased amount of total AUX/IAA protein present compared with that in a wild-type plant. Slightly increased resistance to IAA in root elongation assays has been previously reported to result from the heterologous expression of the wild-type AUX/IAA gene PtrIAA14 (37).
Examination of Transcript Levels and Reporter Gene Expression Indicates That KsIAA16 Is Expressed Throughout Development.
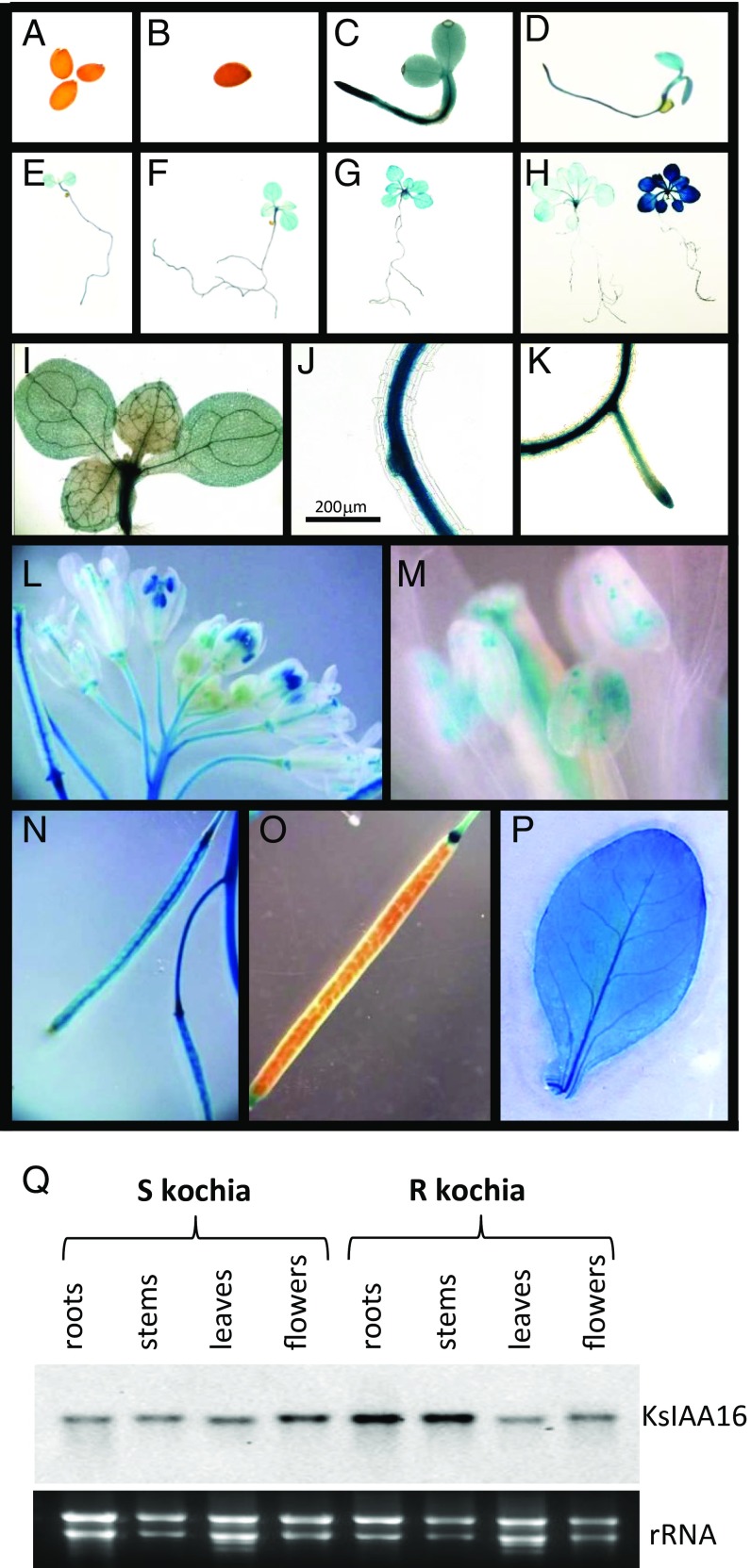
To gain a fuller understanding of expression of KsIAA16 during development as well as to determine if there may be growth stages or tissues in kochia that may be expected to be sensitive to dicamba, we introduced a KsIAA16 promoter:GUS reporter construct (KsIAA16P:GUS) into Arabidopsis. Results are shown in Fig. 5. Although we did not observe any expression in mature or imbibed seed (Fig. 5 A, B, and O), staining was observed ubiquitously in aerial tissues from the time of germination onward, which occurred at ∼3 d postimbibition (Fig. 5C). GUS expression remained apparent diffusely throughout leaves, with stronger staining in the vascular tissues of both roots and shoots throughout development (Fig. 5 D–K and P). In flowering plants, we also observed staining in the stigma and anthers as well as in the vascular tissues of young siliques (Fig. 5 L–N). In mature siliques, expression remained high in abscission zones (Fig. 5O). In addition, we performed a Northern blot on tissues from R and S kochia and observed that the KsIAA16 transcript is expressed in all kochia tissues tested as shown in Fig. 5Q. Given the widespread expression of KsIAA16 in kochia tissues as well as in the heterologous Arabidopsis reporter system, we would not predict that any vegetative growth stage of R kochia would remain sensitive to dicamba herbicide. Additionally, we also observed that expression of KsIAA16P:GUS was increased 24 h after treatment with 25 mM dicamba (equivalent to a 560-g ha−1 field application rate) (Fig. 5H). Auxin induction of AUX/IAA expression is a hallmark of this family of proteins (38, 39) and is thus not surprising; however, this positive feedback mechanism could make resistance even more difficult to mitigate.
Fig. 5.
Developmental expression of KsIAA16. Plants containing the KsIAA16 promoter:GUS reporter were harvested and stained at different stages of development. Seeds were surface sterilized and imbibed; then, seeds or seedlings were stained after 1 (A), 2 (B), 3 (C), 4 (D), 7 (E), 10 (F and I–K), or 15 d (G). After 20 d, seedlings were sprayed with either water (H, Left) or 25 mM dicamba (H, Right) and then stained 24 h later. Floral organs, leaves, and developing siliques of mature plants were also stained (L–P). Q, Upper shows the expression level by Northern blot of KsIAA16 in mature kochia tissues from the resistant and sensitive lines. Q, Lower shows the ethidium bromide-stained rRNA bands as a loading control.
KsIAA16R Imparts a Fitness Cost in Kochia.
To understand if the KsIAA16R allele had an effect on plant fitness, we performed greenhouse tests to measure various fitness parameters. Details are provided in SI Materials and Methods. Seed was sown on plant nutrient agar (40) without sucrose in 48-well dishes, and germination of each seed was scored daily for 14 d. Plants were numbered, and plant identity was tracked throughout the experiment. At the end of this stage, DNA was extracted from a leaf of each seedling or from any ungerminated seeds. Genotypes were aligned with germination data, and no differences were found in germination time between the genotypes or the independent lines (n = 1,440). Next seedlings were transplanted to 5-gallon pots either in monoculture (with eight plants of the same genotype per pot) or in competition [with four plants of either homozygous (RR) or heterozygous (RS) plus four plants that were homozygous sensitive (SS)]. Plant height was measured weekly, and flowering status was scored every 2 d. Results for fitness parameters are shown in Table 2. Reduced plant height associated with the R allele was observed throughout the experiment as shown in Table 2 and Fig. S4. Similarly, relative growth rate (RGR), which is a function of plant height, was also significantly different in both the presence and absence of competition. In addition, a significant reduction in biomass (dry weight) was observed for RR and RS genotypes when grown in competition with SS plants but not when grown in monoculture, indicating that they are less able to compete with SS plants for nutrients and/or moisture. Although we did not observe any differences in flowering time between the genotypes, the RR genotype produced significantly less seed than the SS genotype when grown in competition but not when grown in monoculture. No differences in 1,000 seed weights were observed between any of the genotypes, regardless of competition. In a separate experiment, 96 F2 seeds as well as 24 of the R parental line and 24 of the S parental line were sown on nutrient agar in 48-well plates; roots were measured after 7 d, and genotypes were determined by TaqMan assay. Results are shown in Fig. S2. Whereas we did observe a significant reduction in root length for the R parental line similar to what we reported above, in the F2, no root length difference was observed between the genotypes, indicating that the root length defect is not linked to the R allele. Furthermore, we also did not observe any deviation from expected Mendelian inheritance patterns in the large-scale germination study; thus, the deviation observed in the previous F2 segregation studies was an artifact of manual thinning.
Table 2.
Comparison of the fitness traits of different genotypes with and without competition
| Fitness traits | With competition (RR/SS or RS/SS = 4:4) | Monoculture (eight plants per pot) | ||||||
| RR | RS | SS | P value | RR | RS | SS | P value | |
| Germination* | ||||||||
| Germination time, d | 1.1 ± 0.1(a) | 1.4 ± 0.2(a) | 1.2 ± 0.2(a) | ND | 1.2 ± 0.1(a) | 1.0 ± 0.0(a) | 1.2 ± 0.1(a) | ND |
| Vegetative growth | ||||||||
| Plant height, cm† | 37.1 ± 2.4(a) | 50.1 ± 4.1(a) | 70.1 ± 3(b) | <0.001 | 40.9 ± 1.7(a) | 50.8 ± 3.6(a) | 70.6 ± 2.4(b) | <0.001 |
| RGR, fold‡ | 7.3 ± 0.8(a) | 7.5 ± 0.8(a) | 12.9 ± 0.9(b) | <0.001 | 9.4 ± 0.7(a) | 9.4 ± 0.9(a) | 15.9 ± 0.9(b) | <0.001 |
| Biomass, g | 4.2 ± 1.1(a) | 9.9 ± 4.3(a) | 26.7 ± 4.0(b) | <0.001 | 13.5 ± 2.8(a) | 18.3 ± 5.1(a) | 18.0 ± 2.2(a) | ND |
| Reproductive growth | ||||||||
| Flowering time, d | 50 ± 5(a) | 38 ± 6(a) | 53 ± 4(a) | ND | 47.6 ± 3.3(a) | 42.6 ± 5.6(a) | 55.0 ± 2.9(a) | ND |
| Seed production, g | 0.9 ± 0.3(a) | 1.9 ± 0.6(ab) | 3.7 ± 0.6(b) | 0.0013 | 3.0 ± 0.5(a) | 2.2 ± 0.7(a) | 2.0 ± 0.2(a) | ND |
| 1,000 Seed weight, g | 0.54 ± 0.03(a) | 0.59 ± 0.04(a) | 0.61 ± 0.03(a) | ND | 0.60 ± 0.02(a) | 0.62 ± 0.02(a) | 0.59 ± 0.02(a) | ND |
ND, no statistical difference. Letters (a) and (b) indicate statistically similar groups. Different letters indicate the groups are statistically different from one another.
Germination assays were performed on agar in 48-well plates before transplant. Individual plant identity was maintained throughout the experiment, and these values represent only the individuals represented here; however, no difference in germination time was observed between the genotypes in the entire experiment, which represented 1,440 seeds from three independent lines (480 seeds per line).
Six weeks after the transplanting time point.
RGR is calculated as (plant height at harvest − plant height at transplanting)/plant height at transplanting.
Discussion
Understanding the mechanism(s) of weed resistance to a herbicide is crucial in developing effective control measures. By utilizing a transcriptome sequencing approach, we were able to identify and compare transcripts of several candidate genes from both a sensitive and a dicamba-resistant kochia biotype, and thus, we were able to identify a mutation that changes a highly conserved residue in an AUX/IAA gene, which we designated KsIAA16 based on phylogeny (Fig. 2). We utilized this nucleotide difference as well as additional sequence that we obtained from the genomic DNA to develop a TaqMan genotype assay to distinguish the S and R alleles. By performing crosses between a resistant male and a sensitive female, we were able to utilize this marker to identify heterozygous F1 individuals that resulted from successful cross-pollination. F1 spray studies showed the heterozygotes were resistant to twice the field use rate of dicamba (1,120 g ha−1), which confirmed both the success of the marker at distinguishing the alleles and previous reports that indicated that the trait was partially dominant (4). Dicamba spray studies combined with segregation analysis on over 700 of the resulting F2 progeny showed with a very high level of confidence (P < 0.0001) that the R allele segregates with the resistance phenotype, indicating that it is the causal mutation.
Whereas the resistant biotype used in these studies had been previously reported to be strongly resistant to dicamba (4), its response to other auxins had never been reported. We conducted both greenhouse spray studies as well as root elongation studies to characterize the response of this line to several additional auxin herbicides as well as the natural auxin IAA. We found this kochia biotype to be resistant to all tested auxin herbicides, although it remained sensitive to the native auxins IAA and PAA as well as the synthetic analog NAA. Most significantly, this biotype also displays resistance to field use rates of both 2,4-D and fluroxypyr, thus making it an even more agriculturally challenging problem than previously recognized.
Although a great deal is known about the perception and transport of IAA in the model plant Arabidopsis (15, 32, 33, 41–43), relatively little has been published regarding the perception or transport of auxin herbicides other than 2,4-D (2). In addition to identifying the KsIAA16R allele and establishing that it is the causal basis of auxin herbicide resistance in this kochia biotype, we have also fully characterized its interaction with four identified TIR/AFB proteins identified from kochia. We show through Y2H analysis that the native KsIAA16S protein is capable of interacting with several auxin herbicides in addition to IAA and NAA in this system, whereas KsIAA16R has an impaired ability to interact in the presence of the herbicidal auxin compounds (Fig. 3). It is somewhat surprising that KsIAA16R retains the ability to interact with KsTIR1 and KsAFB6 in the presence of IAA and NAA, although the interaction with KsAFB6 does seem to be impaired at the lower concentration of both compounds. This finding that KsIAA16R has impaired interaction in the presence of several tested auxins is consistent with previous reports from Arabidopsis, which show decreased auxin binding affinity of the degron domain mutants with the TIR/AFB F-box proteins (33). With the exception of fluroxypyr, Y2H findings are also consistent with the observed auxin resistance profiles of the kochia R biotype in greenhouse spray tests as well as in the root inhibition results. The lack of observed Y2H interaction in the presence of fluroxypyr may be due to either a less stable interaction that prevents detection in the yeast system or a lack of metabolism of the fluroxypyr methylheptyl ester, which plants and bacteria are reported to cleave to release the free acid (44).
In addition to establishing through the Y2H studies that KsIAA16 functions as an auxin coreceptor with KsTIR1 and KsAFB6 in the presence of several auxin compounds, we also characterized expression in adult kochia tissues by Northern blot as shown in Fig. 5Q as well as by heterologous expression of a KsIAA16P:GUS construct in Arabidopsis (Fig. 5 A–P). We detected the KsIAA16 transcript in all tested tissues from reproductive-stage kochia plants in both R and S lines. Furthermore, we saw GUS staining ubiquitously in most tissues examined with the reporter system, with strongest staining in vascular tissue of both roots and leaves, which correspond to sites that generally have high levels of expression of auxin biosynthetic genes in Arabidopsis (45). The only tissues that did not show expression of the KsIAA16 reporter were mature seeds and imbibed seeds before germination. Based on these results, we would expect that this kochia biotype would be very difficult to control with auxin herbicide sprays at all stages postgermination, which is consistent with our phenotypic observations.
There has been much speculation in the literature about the low incidence and slow spread of auxin-resistant weeds reported to date. Redundancies in auxin perception pathways, complex inheritance patterns of quantitative traits, and fitness penalties associated with auxin resistance (4, 46, 47) have all been proposed to slow the appearance of auxin resistance; however, none of these theories could be tested without knowledge of a genetic mechanism. To address questions surrounding the relative fitness of this biotype and to further our understanding of the factors affecting evolution/selection of auxin-resistant weed biotypes, we conducted greenhouse-based fitness studies using our segregating F2 lines in conjunction with our molecular marker to compare RR, RS, and SS lines. As shown in Table 2, we observed a significant reduction in plant height associated with the KsIAA16R allele. Furthermore, we observed that plants containing the R allele were also less competitive, as they showed a significant reduction in both biomass and seed production when grown in competition with SS plants. As RS individuals were statistically similar to RR plants for biomass and were intermediate between RR and SS individuals for seed production, this fitness penalty seems to have a high degree of dominance. This finding is similar to the dominant fitness penalty identified for the Arabidopsis axr2-1 mutant (48), which has a similar dominant gain-of-function mutation in an AUX/IAA gene (Table S2) (35), and it suggests that a decrease in fitness is likely to be at least partially responsible for the rarity of auxin-resistant weeds. In addition, studies showing that this trait is dominant and that heterologous expression of KsIAA16R is sufficient to result in resistance to several auxin herbicides argue against the need for quantitative traits for auxin resistance.
As our understanding of auxin biosynthesis and signaling pathways grows, so too does our knowledge surrounding the types of genes that may be key leverage points for resistance development in weeds. A great deal of work has been done in the model plant Arabidopsis over the past two decades to identify and characterize mutants resistant to various forms of auxin. This information, especially the phenotypes associated with the mutants, may be informative when considering resistant weed populations and may help predict the mechanisms that have or potentially could appear in the field.
Identification of an AUX/IAA protein as the causal basis for dominant auxin resistance in kochia may facilitate the identification of other instances of dominant auxin weed resistance. It is noteworthy that most dominant auxin-resistant mutants from Arabidopsis also contain mutations within the degron domain of AUX/IAA family proteins (22–31) as illustrated in Table S2. (4-Chloro-2-methylphenoxy)acetic acid resistance in wild radish (Raphanus raphanistrum L.) has recently been shown to be due to a single incompletely dominant trait (49), and wild mustard, which is classified interchangeably as S. arvensis L. and Brassica kaber (DC.) L.C. Wheeler, has been reported to display dominant inheritance of resistance to several auxin herbicides (50–52). Similarly, another member of the mustard family from Australia, Sisymbrium orientale L., was recently shown to display 2,4-D resistance resulting from a single dominant allele (46); thus, we speculate that these auxin-resistant biotypes may also have similar mutations in AUX/IAA proteins given their reported dominant auxin resistance.
The findings reported here provide several lines of evidence that show that the mechanism of dicamba resistance in the kochia population from western Nebraska first reported by Westra and others (6) in 1994 is a mutation within a highly conserved amino acid region in KsIAA16. We have provided several lines of evidence, including sequence information, expression patterns, Y2H evidence that KsIAA16 functions as an auxin coreceptor for several auxin herbicides as well as the native auxin IAA, development of molecular markers to allow the identification of the allele and follow its inheritance, F2 segregation analysis showing that the KsIAA16R allele segregates with resistance to dicamba, and heterologous expression data showing that expression of KsIAA16R is sufficient to confer resistance to several auxins. Taken together, these data show that the KsIAA16R allele is both necessary and sufficient to cause auxin herbicide resistance.
Materials and Methods
Plant Materials, Growth, and Evaluation.
Arabidopsis seed for Columbia (wild type), axr2-1, axr3-1, and axr5-1 were obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH. Seed for a dicamba-sensitive (S) kochia biotype was purchased from Herbiseed, and generation of the dicamba-resistant (R) kochia biotype 9425R has been reported previously (4).
For root elongation studies, plant nutrient medium (40) containing 0.5% sucrose (PNS) was used. Auxin solutions were diluted from 100 mM stock solutions in ethanol to the appropriate final concentrations listed. Seeds were surface-sterilized in 1% sodium hypochlorite with 0.1% Triton-X100 for 30 min, then rinsed twice in sterile water, and resuspended in 0.1% agar for plating. All plates were grown under white light in a Percival growth chamber (Percival Scientific, Inc.) at 22 °C with 16-h day/8-h night light cycle and a light intensity of 200 μmol/min per 1 m2. Relative humidity was maintained at 50%.
Generation of a TaqMan Genotyping Assay for the KsIAA16R Allele.
Genomic DNA was isolated from leaf tissue of individual kochia plants using the ZR-96 genomic DNA tissue miniprep kit (Zymo Research Corporation) according to the manufacturer’s protocol. To determine the genomic DNA sequence and intron/exon boundaries, several forward and reverse primers were designed across the KsIAA16 coding region. PCR was performed using several different combinations of F and R primers, and the resulting PCR products were sequence and aligned with the known cDNA sequence using SeqMan from DNAstar. The GenBank accession number for the genomic region and the KsIAA16 cDNA is listed in Table S1.
The forward primer CGTAACTAATTCTTAATTGTTTGTTCTTCAG and the reverse primer TGAACGCTCTAACGGGTGG were designed to amplify the genomic DNA of both KsIAA16 alleles. The VIC-labeled probe CACAAGTTGTAAATTG was designed to detect the R allele, and the FAM-labeled probe CACAAGTTGTAGGTTGG was designed to detect the S allele. Assays were performed according to the manufacturer’s conditions on a ViiA7 System (Applied Biosystems) using standard TaqMan genotyping assay conditions. Allele calls were designated by the ViiA7 software, and known homozygous kochia R and S genomic DNA as well as no DNA controls were included in each experiment.
cDNA Cloning from K. scoparia.
Total RNA was isolated from leaf tissue of kochia R and S biotypes; 1 μg of total RNA from each biotype was used for first-strand cDNA synthesis with the ProtoScriptII kit (New England BioLabs). Coding regions of KsIAA16R, KsIAA16S, KsTIR1, KsAFB2, KsAFB5, and KsAFB6 were PCR-amplified and cloned into pENTR (Invitrogen) following the manufacturer’s recommendations. All cDNA clones were sequence verified by Sanger sequencing before use. Accession numbers are listed in Table S1.
Y2H Evaluations.
Y2H studies were conducted using the Matchmaker Gold Yeast Two-Hybrid System (Clontech Laboratories, Inc.). The coding sequences for KsIAA16R, KsIAA16S, KsTIR1, KsAFB6, KsAFB2, and KsAFB5 from clones listed above were PCR amplified and gel purified, and then, they were recombined into NdeI/NotI-digested pGADT7 or pGBKT7 vectors using the In-Fusion cloning system (Clontech Laboratories, Inc.). All vectors were sequence verified, and then, the listed vector combinations were cotransformed into the yeast strain, Y2HGold. Vectors combinations were pGADT7 –T + pGBKT7 empty (negative control), pGADT7 –T + pGBKT7 –Lam (weak interaction control), and pGADT7 –T + pGBKT7 –53 (strong interaction control); pGBKT7 empty was cotransformed with pGADT and KsIAA16 constructs as a negative control, and pGADT7 –T was cotransformed with each pGDKT7TIR/AFB construct as a negative control. All other combinations are as shown in the figures. Transformants were plated on SC-Trp-Leu plates to select for those containing both plasmids. Four healthy colonies were selected at random and resuspended into 300 μL of 0.1% agar in water. Five microliters of each dilution was replica spotted onto each plate using a multichannel pipette. Plates were incubated at 25 °C for 3 d before photographing. Plates were photographed, and figures were assembled showing the growth in two typical individual colonies for each combination.
Promoter:GUS and cDNA Expression Binary Vectors.
To isolate the promoter region of KsIAA16, genomic DNA was isolated from leaf tissue of R kochia as described above. Inverse PCR was performed by digesting with HindIII, SspI, SpeI, or PacI and self-ligating followed by several subsequent rounds of PCR; 2.5 kb of promoter was PCR amplified using primers KsIAA16P-F+R and fused to uidA (GUS) in a binary vector that also carried a Spectinomycin selection gene cassette within the same transfer DNA (T-DNA).
For heterologous expression of KsIAA16 alleles in Arabidopsis, constructs contained the native Arabidopsis IAA16 promoter (AtIAA16P) elements driving either the R or S allele of KsIAA16. Constructs contained a glufosinate resistance gene in the T-DNA region to serve as the selectable marker.
For Arabidopsis transformations, Agrobacterium tumefaciens was used to introduce the binary vectors in the plants using the floral dip method (53). For all constructs, T1 seed was harvested, surface sterilized as described above, and plated on the appropriate selective media to identify independent T1 transformants. Single-seed decent and segregation on selective media were used to estimate transgene insertion number and identify homozygous lines.
Supplementary Material
Acknowledgments
We thank Beiyan Zheng for consultation on statistical analysis, Cynthia Trembly for propagation and plant care in the greenhouse, Jenny Braun for performing herbicide sprays, and Steve Voss for performing visual injury ratings of segregating F2 plants.
Footnotes
Conflict of interest statement: S.L., C.W., and R.D.S. are employed by Monsanto.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MF376151, MF376153, MF376154, MF376152, MF376149, MF376150, MF465806, MF376158, MF376159, MF376162, MF376155, MF376157, MF376156, MF376160, MF376161, MF376163, AAC49048, CAA67308, NP_192207, and Q570C0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712372115/-/DCSupplemental.
References
- 1.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossmann K. Auxin herbicides: Current status of mechanism and mode of action. Pest Manag Sci. 2010;66:113–120. doi: 10.1002/ps.1860. [DOI] [PubMed] [Google Scholar]
- 3.Heap I. 2017. The International Survey of Herbicide Resistant Weeds. Available at www.weedscience.org. Accessed March 5, 2018.
- 4.Preston C, Belles DS, Westra PH, Nissen SJ, Ward SM. Inheritance of resistance to the auxinic herbicide dicamba in kochia (Kochia scoparia) Weed Sci. 2009;57:43–47. [Google Scholar]
- 5.Varanasi VK, et al. Field-evolved resistance to four modes of action of herbicides in a single kochia (Kochia scoparia L. Schrad.) population. Pest Manag Sci. 2015;71:1207–1212. doi: 10.1002/ps.4034. [DOI] [PubMed] [Google Scholar]
- 6.Cranston HJ, et al. Dicamba resistance in kochia. Weed Sci. 2001;49:164–170. [Google Scholar]
- 7.Crespo RJ, et al. Response of Nebraska kochia (Kochia scoparia) accessions to dicamba. Weed Technol. 2014;28:151–162. [Google Scholar]
- 8.Goss GA, Dyer WE. Physiological characterization of auxinic herbicide-resistant biotypes of kochia (Kochia scoparia) Weed Sci. 2003;51:839–844. [Google Scholar]
- 9.Jha P, Kumar V, Lim CA. Variable response of kochia [Kochia scoparia (L.) Schrad.] to auxinic herbicides dicamba and fluroxypyr in Montana. Can J Plant Sci. 2015;95:965–972. [Google Scholar]
- 10.Nandula VK, Manthey FA. Response of kochia (Kochia scoparia) inbreds to 2,4-D and dicamba. Weed Technol. 2002;16:50–54. [Google Scholar]
- 11.Vieten A, Sauer M, Brewer PB, Friml J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007;12:160–168. doi: 10.1016/j.tplants.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Teale W, Palme K. Naphthylphthalamic acid and the mechanism of polar auxin transport. J Exp Bot. 2018;69:303–312. doi: 10.1093/jxb/erx323. [DOI] [PubMed] [Google Scholar]
- 13.Goggin DE, Cawthray GR, Powles SB. 2,4-D resistance in wild radish: Reduced herbicide translocation via inhibition of cellular transport. J Exp Bot. 2016;67:3223–3235. doi: 10.1093/jxb/erw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettinga DJ, et al. Increased chalcone synthase (CHS) expression is associated with dicamba resistance in Kochia scoparia. Pest Manag Sci. October 30, 2017 doi: 10.1002/ps.4778. [DOI] [PubMed] [Google Scholar]
- 15.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 16.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 17.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 18.Uzunova VV, Quareshy M, Del Genio CI, Napier RM. Tomographic docking suggests the mechanism of auxin receptor TIR1 selectivity. Open Biol. 2016;6:160139. doi: 10.1098/rsob.160139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Estelle M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, et al. The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1) Plant J. 2004;40:772–782. doi: 10.1111/j.1365-313X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- 23.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 24.Nagpal P, et al. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 26.Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- 27.Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1025–1038. doi: 10.1093/pcp/pcn079. [DOI] [PubMed] [Google Scholar]
- 28.Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatematsu K, et al. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinaldi MA, Liu J, Enders TA, Bartel B, Strader LC. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol Biol. 2012;79:359–373. doi: 10.1007/s11103-012-9917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calderón Villalobos LIA, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′––3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leyser HMO, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, et al. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front Plant Sci. 2015;6:388. doi: 10.3389/fpls.2015.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guilfoyle TJ. Aux/IAA proteins and auxin signal transduction. Trends Plant Sci. 1998;3:205–207. [Google Scholar]
- 40.Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- 41.Prigge MJ, et al. The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 2016;6:1383–1390. doi: 10.1534/g3.115.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, et al. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, et al. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nat Plants. 2015;1:14030. doi: 10.1038/nplants.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bengal W. Analytical method, validation and degradation kinetics of Fluroxypyr-Meptyl in onion using gas chromatography-tandem mass spectrometry. J Crop Weed. 2014;10:472–479. [Google Scholar]
- 45.Cheng Y, Dai X, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20:1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston C, Malone JM. Inheritance of resistance to 2,4-D and chlorsulfuron in a multiple-resistant population of Sisymbrium orientale. Pest Manag Sci. 2015;71:1523–1528. doi: 10.1002/ps.3956. [DOI] [PubMed] [Google Scholar]
- 47.Mithila J, Hall JC, Johnson WG, Kelley KB, Riechers DE. Evolution of resistance to auxinic herbicides: Historical perspectives, mechanisms of resistance, and implications for broadleaf weed management in agronomic crops. Weed Sci. 2011;59:445–457. [Google Scholar]
- 48.Roux F, Gasquez J, Reboud X. The dominance of the herbicide resistance cost in several Arabidopsis thaliana mutant lines. Genetics. 2004;166:449–460. doi: 10.1534/genetics.166.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jugulam M, Dimeo N, Veldhuis LJ, Walsh M, Hall JC. Investigation of MCPA (4-chloro-2-ethylphenoxyacetate) resistance in wild radish (Raphanus raphanistrum L.) J Agric Food Chem. 2013;61:12516–12521. doi: 10.1021/jf404095h. [DOI] [PubMed] [Google Scholar]
- 50.Preston C, Mallory-Smith CA. Herbicide Resistance and World Grains. CRC; Boca Raton, FL: 2001. Biochemical mechanisms, inheritance, and molecular genetics of herbicide resistance in weeds; pp. 23–60. [Google Scholar]
- 51.Jugulam M, McLean MD, Hall JC. Inheritance of picloram and 2, 4-D resistance in wild mustard (Brassica kaber) Weed Sci. 2005;53:417–423. [Google Scholar]
- 52.Jasieniuk M, Morrison IN, Brûlé-Babel AL. Inheritance of dicamba resistance in wild mustard (Brassica kaber) Weed Sci. 1995;43:192–195. [Google Scholar]
- 53.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 54.Ritz C, Baty F, Streibig JC, Gerhard D. Dose-response analysis using R. PLoS One. 2016;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seefeldt SS, Jensen JE, Fuerst EP. Log-logistic analysis of herbicide dose-response relationships. Weed Technol. 1995;9:218–227. [Google Scholar]
- 56.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rice EA, et al. Expression of a truncated ATHB17 protein in maize increases ear weight at silking. PLoS One. 2014;9:e94238. doi: 10.1371/journal.pone.0094238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.