Significance
Transcriptome profiling quantitatively measures gene expression genome-wide. There is widespread interest in using transcriptomic profiles as phenotypes for epistasis analysis. Though epistasis measurements can be performed using individual transcripts, this results in many scores that must be interpreted independently. We developed a statistic that summarizes these measurements, simplifying analysis. Moreover, epistasis analysis has previously only been performed on cDNA extracted from single cells. We show that whole-organism RNA-sequencing (RNA-seq) can be used to characterize interactions between genes. With the advent of genome engineering, mutants can be created easily in many organisms. Thus, phenotyping is now the rate-limiting step toward reconstructing interaction networks. Our work potentially represents a solution to this problem because RNA-seq is sensitive to a variety of genetic perturbations.
Keywords: hypoxia, transcriptomics, epistasis, genetics, gene expression
Abstract
RNA-sequencing (RNA-seq) is commonly used to identify genetic modules that respond to perturbations. In single cells, transcriptomes have been used as phenotypes, but this concept has not been applied to whole-organism RNA-seq. Also, quantifying and interpreting epistatic effects using expression profiles remains a challenge. We developed a single coefficient to quantify transcriptome-wide epistasis that reflects the underlying interactions and which can be interpreted intuitively. To demonstrate our approach, we sequenced four single and two double mutants of Caenorhabditis elegans. From these mutants, we reconstructed the known hypoxia pathway. In addition, we uncovered a class of 56 genes with HIF-1–dependent expression that have opposite changes in expression in mutants of two genes that cooperate to negatively regulate HIF-1 abundance; however, the double mutant of these genes exhibits suppression epistasis. This class violates the classical model of HIF-1 regulation but can be explained by postulating a role of hydroxylated HIF-1 in transcriptional control.
Genetic analysis of molecular pathways has traditionally been performed through epistatic analysis. If the mutants of two distinct genes have a quantifiable phenotype and the double mutant has a phenotype that is not the sum of the phenotypes of the single mutants, this nonadditivity is referred to as generalized epistasis and indicates that these genes interact functionally. Such interactions can occur at the biochemical level between their products or as a consequence of their functions (1). Epistasis analysis remains a cornerstone of genetics today (2).
Recently, biological studies have shifted in focus from studying single genes to studying all genes in parallel. In particular, RNA-sequencing (RNA-seq) (3) enables biologists to identify genes that change expression in response to a perturbation. RNA-seq has been used to identify genetic modules involved in a variety of processes, such as in the Caenorhabditis elegans linker cell migration (4), planarian stem cell maintenance (5, 6). The role of transcriptional profiling has been restricted to target gene identification, and so far there are only a few examples where transcriptomes have been used to generate quantitative genetic models of any kind. In quantitative genetics, expression quantitative trait loci (eQTL) studies have established the power of transcriptomes for genetic mapping (7–10). Genetic pathway analysis via epistasis has been performed in Saccharomyces cerevisiae (11, 12) and in Dictyostelium discoideum (13). Recently, Dixit et al. described a protocol for epistasis analysis in dendritic and K562 cells using single-cell RNA-seq (14). Epistasis analysis of single cells or single-celled organisms is popular because of the concern that whole-organism sequencing will mix information from multiple cell types, preventing the accurate reconstruction of genetic interactions. Using whole-organism transcriptome profiling, we have recently identified a developmental state of C. elegans caused by loss of a single cell type (sperm cells) (15), which suggests that whole-organism transcriptome profiling contains sufficient information for epistatic analysis. To investigate the ability of whole-organism transcriptomes to serve as quantitative phenotypes for epistatic analysis in metazoans, we sequenced the transcriptomes of four well-characterized loss-of-function mutants in the C. elegans hypoxia pathway (16–19).
Metazoans depend on the presence of oxygen in sufficient concentrations to support aerobic metabolism. Hypoxia inducible factors (HIFs) are an important group of oxygen-responsive genes that are highly conserved in metazoans (20). A common mechanism for hypoxia-response induction is heterodimerization between a HIF and a HIF subunit; the heterodimer then initiates transcription of target genes (21). The number and complexity of HIFs varies throughout metazoans. In the roundworm, C. elegans, there is a single HIF gene, hif-1 (19), and a single HIF gene, ahr-1 (22).
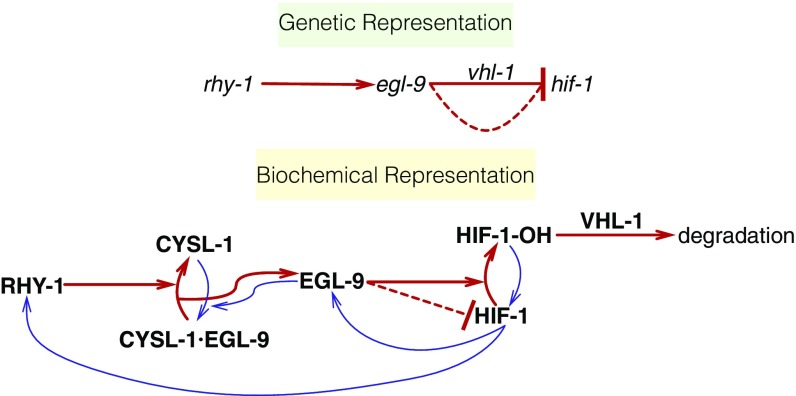
Levels of HIF proteins are tightly regulated. Under conditions of normoxia, HIF-1 exists in the cytoplasm and partakes in a futile cycle of protein production and rapid degradation (23). In C. elegans, HIF-1 is hydroxylated by a proline hydroxylase (EGL-9) (24). HIF-1 hydroxylation increases its binding affinity to Von Hippel–Lindau tumor suppressor 1 (VHL-1), which in turn allows ubiquitination of HIF-1, leading to its degradation. In C. elegans, EGL-9 activity is inhibited by binding of CYSL-1, a homolog of sulfhydrylases/cysteine synthases, and CYSL-1 activity is in turn inhibited by the putative transmembrane O-acyltransferase RHY-1, possibly by posttranslational modifications to CYSL-1 (25) (see Fig. 1).
Fig. 1.
Genetic and biochemical representation of the hypoxia pathway in C. elegans. Red arrows are arrows that lead to inhibition of HIF-1, and blue arrows are arrows that increase HIF-1 activity or are the result of HIF-1 activity. EGL-9 is known to exert VHL-1–dependent and –independent repression on HIF-1 as shown in the genetic diagram. The VHL-1–independent repression of HIF-1 by EGL-9 is denoted by a dashed line and is not dependent on the hydroxylating activity of EGL-9. RHY-1 inhibits CYSL-1, which in turn inhibits EGL-9, but this interaction was abbreviated in the genetic diagram for clarity.
Our reconstruction of the hypoxia pathway in C. elegans shows that whole-animal transcriptome profiles can be used as phenotypes for genetic analysis and that epistasis, a hallmark of genetic interaction observed in double mutants, holds at the molecular systems level. We demonstrate that transcriptomes can aid in ordering genes in a pathway using only single mutants. We were able to identify genes that appear to be downstream of vhl-1 but not downstream of hif-1. Using a single set of transcriptome-wide measurements, we observed most of the known transcriptional effects of hif-1 as well as effects not described before in C. elegans. Taken together, this analysis demonstrates that whole-animal RNA-seq is a fast and powerful method for genetic analyses in an area where phenotypic measurements are now the rate-limiting step.
Results
The Hypoxia Pathway Controls Thousands of Genes in C. elegans.
We selected four null single mutants within the hypoxia pathway for expression profiling: egl-9(sa307), rhy-1(ok1402), vhl-1(ok161), and hif-1(ia4). We also sequenced the transcriptomes of two double mutants, egl-9; vhl-1 and egl-9 hif-1 as well as wild type (N2). Each genotype was sequenced in triplicate using mRNA extracted from 30 worms at a depth of 15 million reads per sample. Of these 15 million reads, 50% of the reads mapped to the C. elegans genome on average. All samples were analyzed under normoxic conditions. We measured differential expression of 19,676 isoforms across all replicates and genotypes (70% of the protein coding isoforms in C. elegans; see SI Appendix, Basic Statistics). We included in our analysis a fog-2(q71) mutant we have previously studied (15), because fog-2 is not reported to interact with the hypoxia pathway. We analyzed our data using a general linear model (GLM) on logarithm-transformed counts. Changes in gene expression are reflected in the regression coefficient , which is specific to each isoform within a genotype (excluding wild type, which is used as baseline). Statistical significance is achieved when the q value of a coefficient ( values adjusted for multiple testing) are less than 0.1. Transcripts that are differentially expressed between the wild type and a given mutant have values that are statistically significantly different from 0 (i.e., greater than 0 or less than 0). coefficients are analogous to the logarithm of the fold-change between the mutant and the wild type. Larger magnitudes of correspond to larger perturbations (see Fig. 2). When we refer to coefficients and values, it will always be in reference to isoforms. However, we report the sizes of each gene set by the number of differentially expressed genes (DEGs), not isoforms, they contain. For the case of C. elegans, this difference is negligible since the great majority of protein-coding genes have a single isoform. We have opted for this method of referring to gene sets because it simplifies the language considerably. A complete version of the code used for this analysis with ample documentation is available at https://wormlabcaltech.github.io/mprsq.
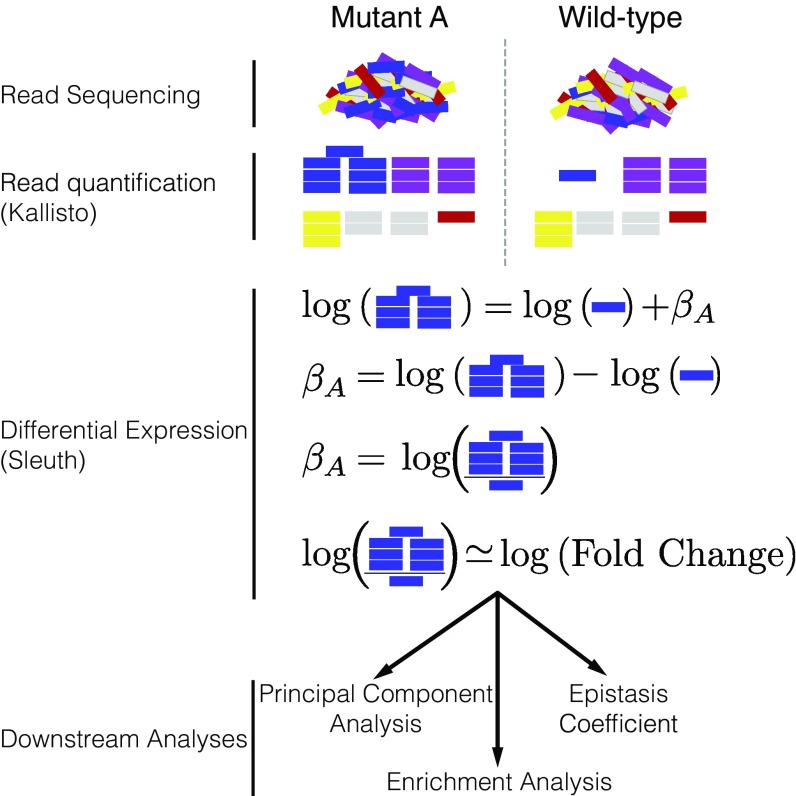
Fig. 2.
Analysis workflow. After sequencing, reads are quantified using Kallisto. Bars show estimated counts for each isoform. Differential expression is calculated using Sleuth, which outputs one coefficient per isoform per genotype. coefficients are analogous to the natural logarithm of the fold-change relative to a wild-type control. Downstream analyses are performed with coefficients that are statistically significantly different from 0. values less than 0.1 are considered statistically different from 0.
Transcriptome profiling of the hypoxia pathway revealed that this pathway controls thousands of genes in C. elegans (see Table 1; see Dataset S1 for a complete list of DEGs). The egl-9(lf) transcriptome showed differential expression of 2,549 genes. A total of 3,005 genes were differentially expressed in rhy-1(lf) mutants. The vhl-1(lf) transcriptome showed considerably fewer DEGs (1,275), possibly because vhl-1 is a weaker inhibitor of hif-1 than egl-9 (18). The egl-9(lf); vhl-1(lf) double mutant transcriptome showed 3,654 DEGs. The hif-1(lf) mutant showed a transcriptomic phenotype involving 1,075 genes. The egl-9(lf) hif-1(lf) double mutant showed a similar number of genes with altered expression (744 genes). We do not think that this transcriptional response is the due to transiently induced hypoxia during harvesting. If the wild-type strain had become hypoxic, then the hif-1(lf) genotype should show significantly lower levels of nhr-57, a marker that increases during hypoxia. We do not observe altered levels of nhr-57 when comparing the wild type and hif-1(lf) mutant, nor between the wild type and egl-9(lf) hif-1(lf) double mutant. Finally, the egl-9(lf), vhl-1(lf), rhy-1(lf) and egl-9(lf); vhl-1(lf) mutants did show altered nhr-57 transcript levels (see SI Appendix, Fig. S1). Of the DEGs in hif-1(lf) mutants, 161/1,075 were also differentially expressed in egl-9(lf) hif-1(lf) mutants, which suggests these transcripts are hif-1–dependent under normoxia. For the remaining genes, we cannot rule out cumulative effects from loss of hif-1, strain-specific eQTLs present in the strain background or that loss of egl-9 suppresses the mutant phenotype. We designed our experiments to probe the constitutive hypoxia response, and not the effects of hif-1 under normoxia, which we did not foresee. As a result, we have limited resolving power to explain the transcriptome of hif-1(lf) mutants.
Table 1.
Number of DEGs in each mutant strain with respect to the wild type (N2)
| Genotype | DEGs |
| egl-9(lf) | 2,549 |
| rhy-1(lf) | 3,005 |
| vhl-1(lf) | 1,275 |
| hif-1(lf) | 1,075 |
| egl-9(lf); vhl-1(lf) | 3,654 |
| egl-9(lf) hif-1(lf) | 744 |
| fog-2(lf) | 2,840 |
Principal Component Analysis Visualizes Epistatic Relationships Between Genotypes.
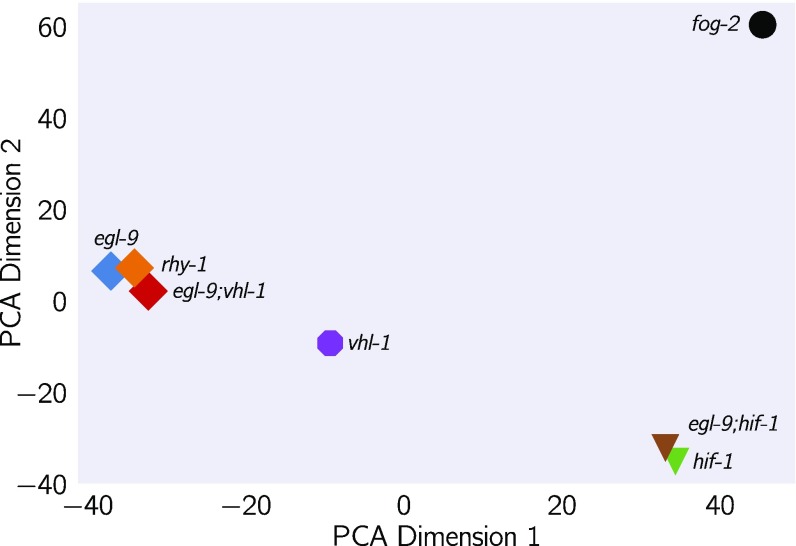
Principal component analysis (PCA) is used to identify relationships between high-dimensional data points (26). We used PCA to examine whether each genotype clustered in a biologically relevant manner. PCA identifies the vector that explains most of the variation in the data; this is called the first principal component. PCA can identify the first components that explain more than 95% of the data variance. Clustering in these dimensions can indicate biological relationships, although interpreting principal components can be difficult. In our analysis, the first principal component discriminated mutants that have constitutive high levels of HIF-1 from mutants that have no HIF-1, whereas the second component was able to discriminate between mutants within the hypoxia pathway and outside the hypoxia pathway (see Fig. 3; fog-2 is not reported to act in the hypoxia pathway and acts as a negative control; see SI Appendix).
Fig. 3.
PCA of various C. elegans mutants. Genotypes that have a constitutive hypoxia response [i.e., egl-9(lf)] cluster far from genotypes that do not have a hypoxic response [i.e., hif-1(lf)] along the first principal component. The second principal component separates genotypes that do not participate in the hypoxic response pathway.
Reconstruction of the Hypoxia Pathway from First Genetic Principles.
To reconstruct a genetic pathway, we must assess whether two genes act on the same phenotype. If they do not act on the same phenotype (two mutations do not cause the same genes to become differentially expressed relative to wild type), these mutants are independent. Otherwise, we must measure whether these genes act additively or epistatically on the phenotype of interest; if there is epistasis, we must measure whether it is positive or negative, to assess whether the epistatic relationship is a genetic suppression or a synthetic interaction. To allow coherent comparisons of different mutant transcriptomes (the phenotype we are studying here), we define the shared transcriptomic phenotype (STP) between two mutants as the shared set of genes or isoforms whose expression in both mutants are different from wild-type, regardless of the direction of change.
Genes in the Hypoxia Mutant Act on the Same Transcriptional Phenotype.
All of the hypoxia mutants had a significant STP: the fraction of DEGs that was shared between mutants ranged from a minimum of 10% between hif-1(lf) and egl-9(lf); vhl-1(lf) to a maximum of 32% between egl-9(lf) and egl-9(lf); vhl-1(lf) (see SI Appendix, Table S1 and https://wormlabcaltech.github.io/mprsq). For comparison, we also analyzed a previously published fog-2(lf) transcriptome (15). The fog-2 gene is involved in masculinization of the C. elegans germline, which enables sperm formation, and is not known to be involved in the hypoxia pathway. The hypoxia pathway mutants and the fog-2(lf) mutant also had STPs (8.8% to 14%).
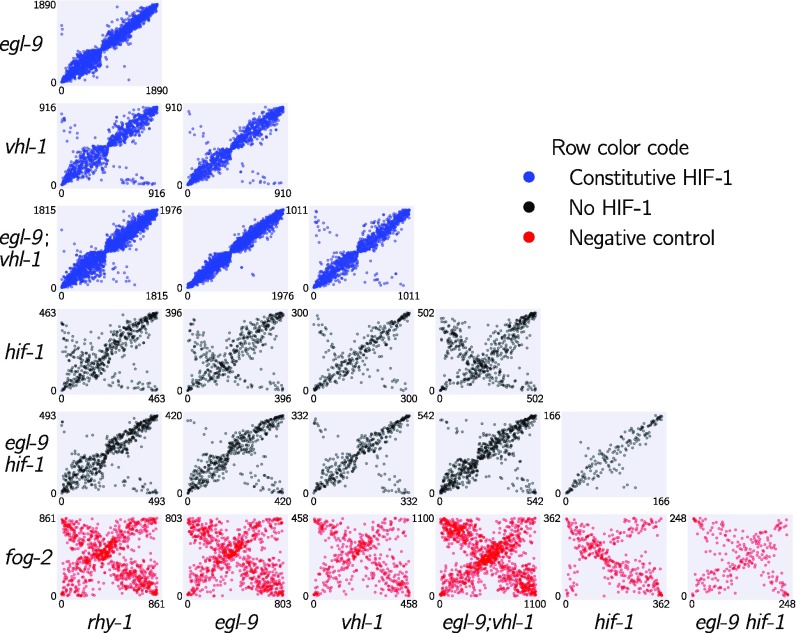
Next, we analyzed pairwise correlations between all mutant pairs. We rank-transformed the coefficients of each isoform between the STP of two mutants and plotted the transcript ranks between genotypes (see Fig. 4). Although hif-1 is known to be genetically repressed by egl-9, rhy-1, and vhl-1 (16, 17), all of the correlations between mutants of these genes and hif-1(lf) were positive (see SI Appendix). We reasoned that this apparent contradiction could be due to either strain-specific effects in our N2 background (an artifactual signal) or that it could reflect a previously unrecognized aspect of HIF-1 biology. This motivated us to look for genes that exhibited verifiable extreme patterns of anomalous behavior and led us to propose a new model of the hypoxia pathway (see Identification of Nonclassical Epistatic Interactions).
Fig. 4.
Interacting genes have correlated transcriptional signatures. The rank order of transcripts contained in the shared transcriptional phenotype is plotted for each pairwise combination of genotypes. Correlations between in-pathway genotypes are strong, whereas comparisons with a fog-2(lf) genotype are dominated by noise. Comparisons between some genotypes show populations of transcripts that are anticorrelated, possibly as a result of feedback loops. Plots are color-coded by row. Comparisons with genotypes with a constitutive hypoxia response are in blue, comparisons with genotypes negative for hif-1(lf) are in black, and comparisons involving fog-2(lf) are in red. The x and y axes show the rank of each transcript within each genotype.
Transcriptome-Wide Epistasis.
Ideally, any measurement of transcriptome-wide epistasis should conform to certain expectations. First, it should make use of the regression coefficients of as many genes as possible. Second, it should be summarizable in a single, well-defined number. Third, it should have an intuitive behavior, such that special values of the statistic have an unambiguous interpretation.
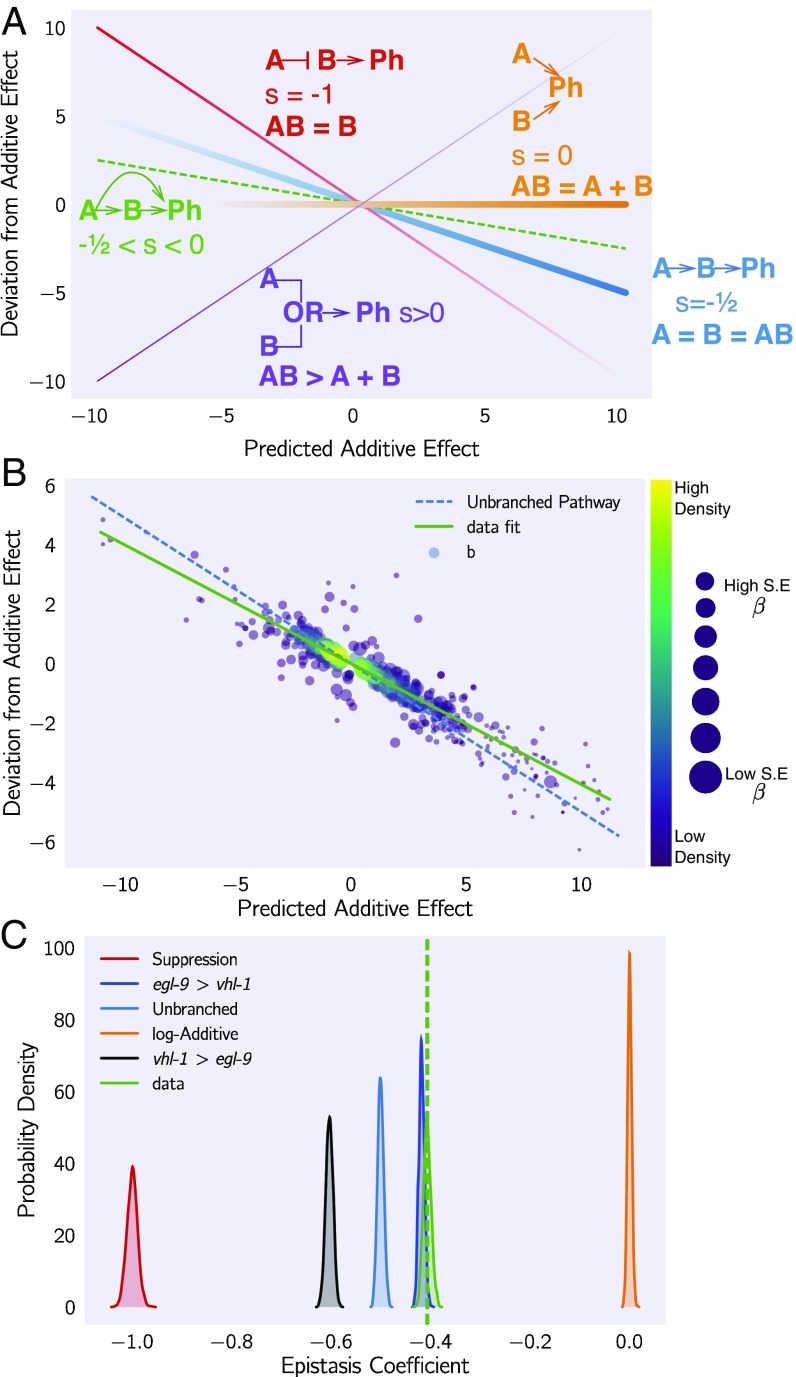
We found an approach that satisfies all of the above conditions and that can be graphed in an epistasis plot (see Fig. 5). In an epistasis plot, the x axis represents the expected coefficient for a given gene in a double mutant if and interact log-additively. In other words, each individual isoform’s x coordinate is the sum of the regression coefficients from the single mutants and . The y axis represents the deviations from the log-additive (null) model and can be calculated as the difference between the predicted and the observed coefficients. Only isoforms that are differentially expressed in all three genotypes are plotted. This attempts to ensure that the isoforms to be examined are regulated by both genes. These plots will generate specific patterns that can be described through linear regressions. The slope of these lines, to which we assign the mathematical notation , is the transcriptome-wide epistasis coefficient. Importantly, the transcriptome-wide epistasis coefficient is fundamentally distinct from Pearson or Spearman correlation coefficients and need not have a simple linear mapping. In other words, negative correlation coefficients do not imply a specific sign of the epistasis coefficient, and vice versa. For suppression to occur, for example, the only requirement is that the phenotype of the double mutant should match one, and only one, of the two single mutants. The value of the correlation coefficient is not relevant.
Fig. 5.
Quantification of epistasis transcriptome-wide. (A) Schematic diagram of an epistasis plot. The x axis on an epistasis plot is the expected coefficient for a double mutant under an log-additive model (null model). The y axis plots deviations from this model. Double mutants that deviate in a systematic manner from the null model exhibit transcriptome-wide epistasis (). To measure , we find the line of best fit and determine its slope. Genes that act log-additively on a phenotype (Ph) will have (null hypothesis, orange line), whereas genes that act along an unbranched pathway will have (blue line). Strong repression is reflected by (red line), whereas correspond to synthetic interactions (purple line). (B) Epistasis plot showing that the egl-9(lf); vhl-1(lf) transcriptome deviates significantly from a null additive. Points are colored qualitatively according to density (purple, low; yellow, high) and size is inversely proportional to the SE of the y axis. The green line is the line of best fit from an orthogonal distance regression. (C) Comparison of simulated epistatic coefficients against the observed coefficient. Green curve shows the bootstrapped observed transcriptome-wide epistasis coefficient for egl-9 and vhl-1. Dashed green line shows the mean value of the data. Simulations use only the single mutant data to idealize what expression of the double mutant should look like. means that the phenotype of is observed in a double mutant .
Transcriptome-wide epistasis coefficients can be understood intuitively for simple cases of genetic interactions if complete genetic nulls are used. If two genes act additively on the same set of differentially expressed isoforms, then all of the plotted points will fall along the line . If two genes act positively in an unbranched pathway, then all of the mutants should have the same phenotype. It follows that data from this pathway will form a line with a slope equal to . On the other hand, in the limit of complete genetic inhibition of by in an unbranched pathway (i.e., is in great excess over , such that under the conditions measured has no activity), the plots should show a line of best fit with a slope equal to . Genes that interact synthetically [i.e., through an odds ratio (OR)-gate] will fall along lines with slopes . When there is epistasis of one gene over another, the points will fall along one of two possible slopes that must be determined empirically from the single mutant data. We can use both single mutant data to predict the distribution of slopes that results for the cases stated above. Thus, the transcriptome-wide epistasis coefficient integrates information from many different isoforms into a single number (see Fig. 5).
In our experiment, we studied two double mutants, egl-9(lf) hif-1(lf) and egl-9(lf); vhl-1(lf). We wanted to understand how well an epistatic analysis based on transcriptome-wide coefficients agreed with the epistasis results reported in the literature, which were based on qPCR of single genes. Therefore, we determined the epistasis coefficient of the two gene combinations we studied (egl-9 and vhl-1, and egl-9 and hif-1). In addition to computing an epistasis coefficient from these factors, we would like to know which gene is suppressed in the double mutant. Suppression means that the double mutant should have exactly the phenotype of one and only one mutant; we can simulate the double mutant by replacing the double mutant data with either of the two single mutants and matching the simulated result to the observed result. The result that most closely matches the real data will reveal which gene is being suppressed, which in turn allows us to order the genes along a pathway.
We measured the epistasis coefficient between egl-9 and vhl-1, s(egl-9 vhl-1) (see SI Appendix, Quantifying Epistasis). Simulations using just the single mutant data showed that the double mutant exhibited the egl-9(lf) phenotype (see Fig. 5). We used Bayesian model selection to reject a linear pathway (OR ), which leads us to conclude egl-9 is upstream of vhl-1 acting on a phenotype in a branched manner. We also measured epistasis between egl-9 and hif-1, s(egl-9, hif-1) (see SI Appendix, Fig. S2), and we found that this behavior could be predicted by modeling hif-1 downstream of egl-9. We also rejected the null hypothesis that these two genes act in a positive linear pathway (OR). Taken together, this leads us to conclude that egl-9 strongly inhibits hif-1.
Epistasis Between Two Genes Can Be Predicted Using an Upstream Component.
Given our success in measuring epistasis coefficients, we wanted to know whether it would be possible to predict the epistasis coefficient between egl-9 and vhl-1 in the absence of the egl-9(lf) genotype. Since RHY-1 indirectly activates EGL-9, we reasoned that the rhy-1(lf) transcriptome should contain almost equivalent information to the egl-9(lf) transcriptome. Therefore, we generated predictions of the epistasis coefficient between egl-9 and vhl-1 by substituting in the rhy-1(lf) data, predicting s(rhy-1, vhl-1). Similarly, we used the egl-9(lf); vhl-1(lf) double mutant to measure the epistasis coefficient while replacing the egl-9(lf) dataset with the rhy-1(lf) dataset. We found that the epistasis coefficient using this substitution was . This coefficient was different from (OR ), reflecting the same qualitative conclusion that vhl-1 represents a branch in the hypoxia pathway. We were able to obtain a close prediction of the epistasis coefficient for two mutants using the transcriptome of a related, upstream mutant.
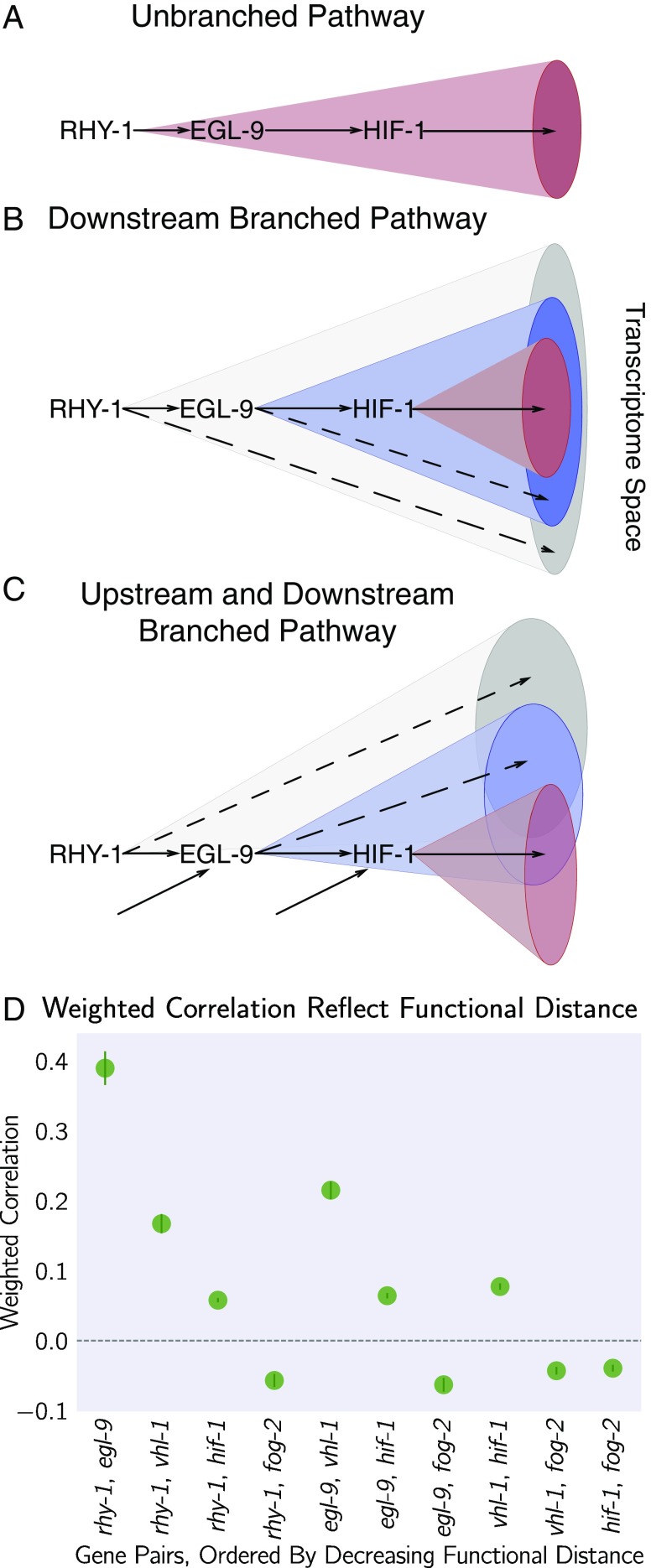
Transcriptomic Decorrelation Can Be Used to Infer Functional Distance.
So far, we have shown that RNA-seq can accurately measure genetic interactions. However, genetic interactions do not require two gene products to interact biochemically, nor even to be physically close to each other. RNA-seq cannot measure physical interactions between genes, but we wondered whether expression profiling contains sufficient information to order genes along a pathway.
Single genes are often regulated by multiple independent sources. The connection between two nodes can in theory be characterized by the strength of the edges connecting them (the thickness of the edge), the sources that regulate both nodes (the fraction of inputs common to both nodes), and the genes that are regulated by both nodes (the fraction of outputs that are common to both nodes). In other words, we expected that expression profiles associated with a pathway would respond quantitatively to quantitative changes in activity of the pathway. Targeting a pathway at multiple points would lead to expression profile divergence as we compare nodes that are separated by more degrees of freedom, reflecting the flux in information between them.
We investigated this possibility by weighting the robust Bayesian regression between each pair of genotypes by the size of the STP of each pair divided by the total number of isoforms differentially expressed in either mutant (). We plotted the weighted correlation of each gene pair, ordered by increasing functional distance (see Fig. 6). In every case, we see that the weighted correlation decreases monotonically due mainly, but not exclusively, to a smaller STP (see SI Appendix, Decorrelation Within Pathways).
Fig. 6.
Transcriptomes can be used to order genes in a pathway under certain assumptions. Arrows in the diagrams above are intended to show the direction of flow and do not indicate valence. (A) A linear pathway in which rhy-1 is the only gene controlling egl-9, which in turn controls hif-1, does not contain information to infer the order between genes. (B) If rhy-1 and egl-9 have transcriptomic effects that are separable from hif-1, then the rhy-1 transcriptome should contain contributions from egl-9, hif-1 and egl-9– and hif-1–independent pathways. This pathway contains enough information to infer order. (C) If a pathway is branched both upstream and downstream, transcriptomes will show even faster decorrelation. Nodes that are separated by many edges may begin to behave almost independently of each other with marginal transcriptomic overlap or correlation. (D) The hypoxia pathway can be ordered. We hypothesize the rapid decay in correlation is due to a mixture of upstream and downstream branching that happens along this pathway. Bars show the SE of the weighted coefficient from the Monte Carlo Markov Chain computations.
We believe that this result is not due to random noise or insufficiently deep sequencing. Instead, we propose a framework in which every gene is regulated by multiple different molecular species, which induces progressive decorrelation. This decorrelation in turn has two consequences. First, decorrelation within a pathway implies that two nodes may be almost independent of each other if the functional distance between them is large. Second, it may be possible to use decorrelation dynamics to infer gene order in a branching pathway, as we have done with the hypoxia pathway.
Classical Epistasis Identifies a Core Hypoxic Response.
We searched for genes whose expression obeyed the two epistatic equality relationships, hif-1(lf) = egl-9(lf) hif-1(lf) and egl-9(lf) = egl-9(lf); vhl-1(lf), since these equalities define the hypoxia pathway. We excluded genes whose expression deviated from this relationship by more than 2 standard deviations or that had opposite changes in direction. Using these criteria, we identified 1,258 genes in the hypoxia response. Tissue enrichment analysis showed that the intestine and epithelial system were enriched in this response ( for both terms), consistent with previous reports (27). Gene enrichment analysis (28) showed enrichment in the mitochondrion and in collagen trimers () (see SI Appendix, Enrichment Analysis of Hypoxia Pathway Data and SI Appendix, Figs. S3 and S4). This response included 15 transcription factors. Even though HIF-1 is an activator, not all of these genes were up-regulated. We reasoned that only genes that are up-regulated in HIF-1-inhibitor mutants are candidates for direct regulation by HIF-1. We found 264 such genes.
Feedback Can Be Inferred.
While some of the rank plots contained a clear positive correlation, others showed a discernible cross-pattern (see Fig. 4). In particular, this cross-pattern emerged between vhl-1(lf) and rhy-1(lf) or between vhl-1(lf) and egl-9(lf), even though vhl-1, rhy-1, and egl-9 are all inhibitors of hif-1(lf). Such cross-patterns could be indicative of feedback loops or other complex interaction patterns. If the above is correct, then it should be possible to identify genes that are regulated by rhy-1 in a logically consistent way: Since loss of egl-9 causes rhy-1 mRNA levels to increase, if this increase leads to a significant change in RHY-1 activity, then it follows that the egl-9(lf) and rhy-1(lf) should show anticorrelation in a subset of genes. Since we do not observe many genes that are anticorrelated, we conclude that is unlikely that the change in rhy-1 mRNA expression causes a significant change in RHY-1 activity under normoxic conditions. We also searched for genes with hif-1–independent, vhl-1–dependent gene expression and found 71 genes (Dataset S1).
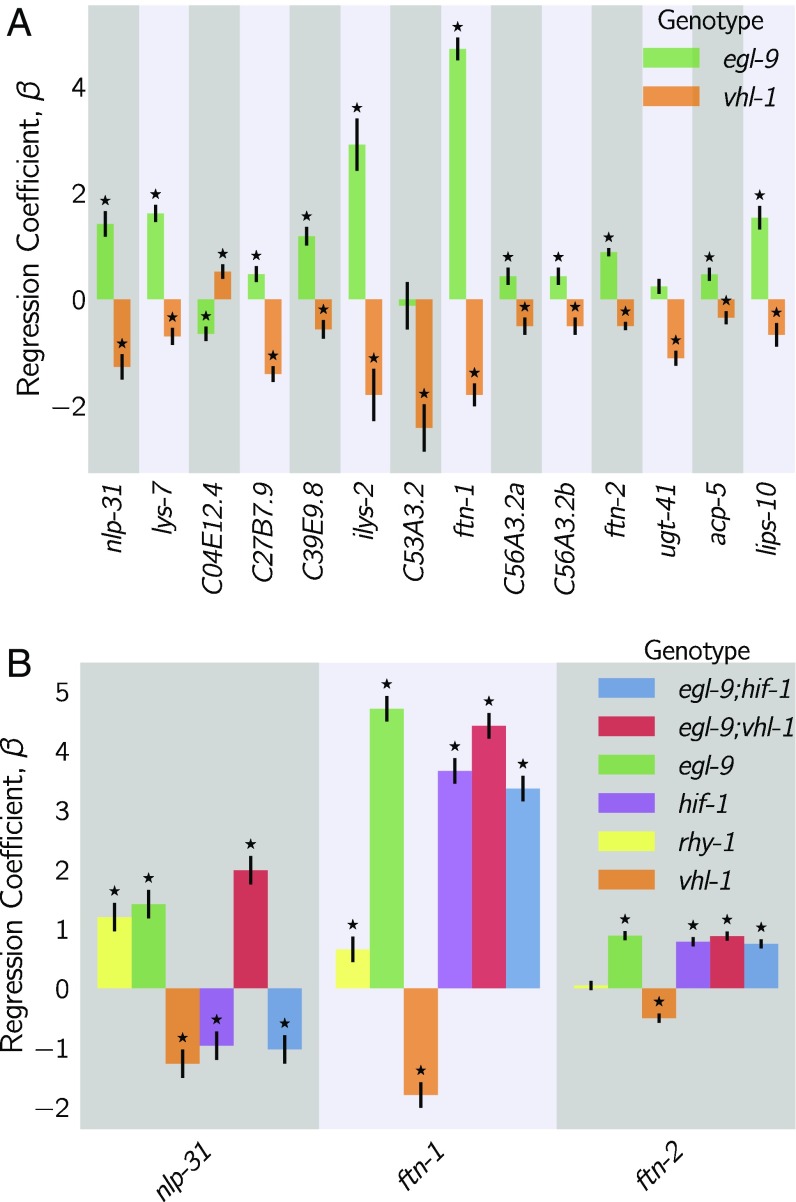
Identification of Nonclassical Epistatic Interactions.
hif-1(lf) has traditionally been viewed as existing in a genetic OFF state under normoxic conditions. However, our dataset indicates that 1,075 genes show altered expression when hif-1 function is removed in normoxic conditions. Moreover, we observed positive correlations between hif-1(lf) coefficients and egl-9(lf), vhl-1(lf), and rhy-1(lf) coefficients despite the negative regulatory relationships between these genes and hif-1. Such positive correlations could indicate a relationship between these genes that has not been reported previously.
We identified genes that exhibited violations of the canonical genetic model of the hypoxia pathway (see Fig. 7; see also SI Appendix). We searched for genes that changed in different directions between egl-9(lf) and vhl-1(lf) or, equivalently, between rhy-1(lf) and vhl-1(lf) [we assume that all results from the rhy-1(lf) transcriptome reflect a complete loss of egl-9 activity] without specifying any further conditions. We found 56 that satisfied this condition (see Fig. 7, Dataset S1). When we checked expression of these genes in the double mutant, we found that egl-9 remained epistatic over vhl-1 for this class of genes. This class of genes may in fact be larger because it overlooks genes that have wild-type expression in an egl-9(lf) background, altered expression in a vhl-1(lf) background, and suppressed (wild-type) expression in an egl-9(lf); vhl-1(lf) background. As a result, it could help explain why the hif-1(lf) mutant transcriptome is positively correlated with its inhibitors.
Fig. 7.
Fifty-six hif-1–dependent genes show nonclassical antagonistic effects of vhl-1 and egl-9. (A) A total of 56 genes in C. elegans exhibit nonclassical epistasis in the hypoxia pathway, characterized by opposite effects on gene expression, relative to the wild type, of the vhl-1(lf) compared with egl-9(lf) [or rhy-1(lf)] mutants. Shown are a random selection of 15 out of 56 genes for illustrative purposes. (B) Genes that behave noncanonically have a consistent pattern. vhl-1(lf) mutants have an opposite effect to egl-9(lf), but egl-9 remains epistatic to vhl-1 and loss-of-function mutations in hif-1 suppress the egl-9(lf) phenotype. Asterisks show values significantly different from 0 relative to wild type ().
Although this entire class had similar behavior, we focused on two genes, nlp-31 and ftn-1, which have representative expression patterns. ftn-1 is described to be responsive to mutations in the hypoxia pathway and has been reported to have aberrant behaviors; specifically, loss of function of egl-9 and vhl-1 have opposing effects on ftn-1 expression (29, 30). These studies showed the same ftn-1 expression phenotypes using RNAi and alleles, allaying concerns of strain-specific interference. We observed that hif-1 was epistatic to egl-9 and that egl-9 and hif-1 both promoted ftn-1 expression.
Analysis of ftn-1 expression reveals that egl-9 is epistatic to hif-1, that vhl-1 has opposite effects to egl-9, and that vhl-1 is epistatic to egl-9. Analysis of nlp-31 reveals similar relationships. nlp-31 expression is decreased in hif-1(lf) and increased in egl-9(lf). However, egl-9 is epistatic to hif-1. Like ftn-1, vhl-1 has the opposite effect to egl-9 yet is epistatic to egl-9. We propose in Discussion a model for how HIF-1 might regulate these targets.
Discussion
The C. elegans Hypoxia Pathway Can Be Reconstructed de Novo from RNA-Seq Data.
We have shown that whole-organism transcriptomic phenotypes can be used to reconstruct genetic pathways and to discern previously uncharacterized genetic interactions. We successfully reconstructed the hypoxia pathway including the order of action of the genetic components and its branching pattern. These results highlight the potential of whole-animal expression profiles for dissecting molecular pathways that are expressed in a large number of cells within an organism. While our results are promising, it remains to be seen whether our approach will also work for pathways that act in a few cells. We selected a previously characterized pathway because C. elegans is less amenable to high-throughput screens compared with cultured cells. That said, the striking nature of our results makes us optimistic that this technique could be successfully used to reconstruct unknown pathways.
Interpretation of the Nonclassical Epistasis in the Hypoxia Pathway.
The 56 genes that exhibit a striking pattern of nonclassical epistasis suggest the existence of previously undescribed aspects of the hypoxia pathway. Some of these nonclassical behaviors had been observed previously (29–31), but no satisfactory mechanism has been proposed to explain them. Previous studies (29, 30) suggested that HIF-1 integrates information on iron concentration in the cell to determine its binding affinity to the ftn-1 promoter but could not definitively establish a mechanism. It is unclear why deletion of hif-1 and deletion of egl-9 both cause induction of ftn-1 expression, but deletion of vhl-1 abolishes this induction. Moreover, Luhachack et al. (31) have previously reported that certain genes important for the C. elegans immune response against pathogens reflect similar noncanonical expression patterns. Their interpretation was that swan-1, which encodes a binding partner to EGL-9 (32), is important for modulating HIF-1 activity in some manner. The lack of a conclusive double mutant analysis in this work means the role of SWAN-1 in modulation of HIF-1 activity remains to be demonstrated. Other mechanisms, such as tissue-specific differences in the pathway (27), could also modulate expression, though it is worth pointing out that ftn-1 expression appears restricted to a single tissue, the intestine (33). Another possibility is that egl-9 controls hif-1 mRNA stability via other vhl-1–independent pathways, but we did not see a decrease in hif-1 level in egl-9(lf), rhy-1(lf), or vhl-1(lf) mutants. Another possibility, such as control of protein stability via egl-9 independently of vhl-1 (34), will not lead to splitting unless it happens in a tissue-specific manner.
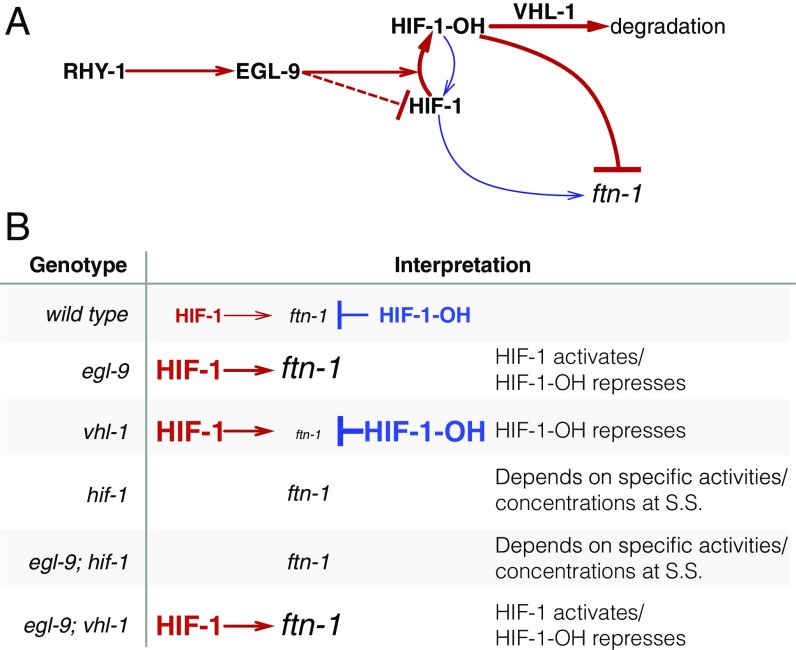
One parsimonious solution is to consider HIF-1 as a protein with both activating and inhibiting states. In fact, HIF-1 already exists in two states in C. elegans: unmodified HIF-1 and HIF-1-hydroxyl (HIF-1-OH). Under this model, the effects of HIF-1 for certain genes like ftn-1 or nlp-31 are antagonized by HIF-1-OH, which is present at only a low level in the cell in normoxia because it is degraded in a vhl-1–dependent fashion. This means that loss of vhl-1 stabilizes HIF-1-OH. If vhl-1 is inactivated, genes that are sensitive to HIF-1-OH will be inhibited as a result of the increase in HIF-1-OH, despite the increased levels of nonhydroxylated HIF-1. On the other hand, egl-9(lf) abrogates the generation of HIF-1-OH, stimulating accumulation of nonhydroxylated HIF-1 and promoting gene expression. Whether deletion of hif-1(lf) is overall activating or inhibiting will depend on the relative activity of each protein state under normoxia (see Fig. 8). HIF-1-OH is challenging to study genetically, and if it does have the activity suggested by our genetic evidence, this may have prevented such a role from being detected. No mimetic mutations are known with which to study the pure hydroxylated HIF-1 species, and mutations in the Von Hippel–Lindau gene that stabilize the hydroxyl species also increase the quantity of nonhydroxylated HIF-1 by mass action.
Fig. 8.
A hypothetical model showing a mechanism where HIF-1-OH antagonizes HIF-1 in normoxia. (A) Diagram showing that RHY-1 activates EGL-9. EGL-9 hydroxylates HIF-1 in an oxygen-dependent manner. HIF-1 is rapidly hydroxylated, and the product, HIF-1-OH, is rapidly degraded in a VHL-1–dependent fashion. EGL-9 can also inhibit HIF-1 in an oxygen-independent fashion. In our model, HIF-1 and HIF-1-OH have opposing effects on transcription. The width of the arrows represents rates in normoxic conditions. (B) Table showing the effects of loss-of-function mutations on HIF-1 and HIF-1-OH activity, showing how this can potentially explain the ftn-1 expression levels in each case. S.S., steady state.
Because HIF-1 is detected at low levels in cells under normoxic conditions (35), total HIF-1 protein levels are assumed to be so low as to be biologically inactive. However, our data show 1,075 genes change expression in response to loss of hif-1 under normoxic conditions, which establishes that there is sufficient total HIF-1 protein to be biologically active. Our analyses also revealed that hif-1(lf) shares positive correlations with egl-9(lf), rhy-1(lf), and vhl-1(lf) and that each of these genotypes also shows a secondary negative rank-ordered expression correlation with each other.
A homeostatic argument can be made in favor of the activity of HIF-1-OH. The cell must continuously monitor multiple metabolite levels. The hif-1–dependent hypoxia response integrates information from O2, -ketoglutarate, and iron concentrations in the cell. One way to integrate this information is by encoding it within the effective hydroxylation rate of HIF-1 by EGL-9. Then the dynamics in this system will evolve exclusively as a result of the total amount of HIF-1 in the cell. Such a system can be sensitive to fluctuations in the absolute concentration of HIF-1 (36). Since the absolute levels of HIF-1 are low in normoxic conditions, small fluctuations in protein copy number can represent a large fold-change in HIF-1 levels. These fluctuations might not be problematic for genes that must be turned on only under conditions of severe hypoxia—presumably, these genes would be activated only when HIF-1 levels increase far beyond random fluctuations.
For yet other sets of genes that must change expression in response to the hypoxia pathway, it may not be sufficient to integrate metabolite information exclusively via EGL-9–dependent hydroxylation of HIF-1. In particular, genes that may function to increase survival in mild hypoxia may benefit from regulatory mechanisms that can sense minor changes in environmental conditions and which therefore benefit from robustness to transient changes in protein copy number. Likewise, genes that are involved in iron or -ketoglutarate metabolism (such as ftn-1) may benefit from being able to sense, accurately, small and consistent deviations from basal concentrations of these metabolites. For these genes, the information may be better encoded by using HIF-1 and HIF-1-OH as an activator/repressor pair. Such circuits are known to possess distinct advantages for controlling output robustly to transient fluctuations in the levels of their components (37, 38).
Our RNA-seq data suggest that one of these atypical targets of HIF-1 may be RHY-1. Although rhy-1 does not exhibit nonclassical epistasis, all genotypes containing a hif-1(lf) mutation had increased expression levels of rhy-1. We speculate that if rhy-1 is controlled by both HIF-1 and HIF-1-OH, then this might imply that HIF-1 autoregulates both positively and negatively.
Strengths and Weaknesses of the Methodology.
We have described a set of methods that can in principle be applied to any multidimensional phenotype. Although we have not applied these methods to de novo pathway discovery, we believe that they will be broadly applicable to a wide variety of genetic problems. One aspect of our methodology is the use of whole-organism expression data. Data collection from whole organisms can be rapid with low technical barriers. On the other hand, a concern is that whole-organism data will average signals across tissues, which would limit the scope of this technology to the study of genetic pathways that are systemic or expressed in large tissues. In reality, our method may be applicable for pathways that are expressed even in a small number of cells in an organism. If a pathway is active in a single cell, this does not mean that it does not have cell-nonautonomous effects that could be detected on an organism-wide level. Thus, pathways that act in single cells could still be characterized via whole-organism transcriptome profiling. If the nonautonomous effects are long-lasting, then the profiling could take place after the time-of-action of this pathway. In fact, this is how the female-like state in C. elegans was recently identified (15): fog-2 is involved in translation repression of tra-2 in the somatic gonad, thereby promoting sperm formation in late larvae (39). Loss of this gene causes non–cell-autonomous effects that can be detected well after the time-of-action of fog-2 in the somatic gonad has ended. Therefore, we believe that our methodology will be applicable to many genetic cases, with the exception of pathways that act in complex, antagonistic manners depending on the cell type, or if the pathway minimally affects gene expression.
Genetic analysis of transcriptomic data has proved challenging as a result of its complexity. Although dimensionality reduction techniques such as PCA have emerged as powerful methods with which to understand these data, these methods generate reduced coordinates that are difficult or impossible to interpret. As an example, the first principal component in this paper (see Fig. 3) could be interpreted as HIF-1 pseudoabundance (40). However, another equally reasonable yet potentially completely different interpretation is as a pseudo–HIF-1/HIF-1-OH ratio. Another way to analyze genetic interactions is via GLMs that include interaction terms between two or more genes. GLMs can quantify the genetic interactions on single transcripts. We and others (14, 15) have used GLMs to perform epistasis analyses of pathways using transcriptomic phenotypes. GLMs are powerful, but they generate a different interaction coefficient for each gene measured. The large number of coefficients makes interpretation of the genetic interaction between two mutants difficult. Previous approaches (14) visualize these coefficients via clustered heatmaps. However, two clusters cannot be assumed to be evidence that two genes interact via entirely distinct pathways. Indeed, the nonclassical epistasis examples we described here might cluster separately even though a reasonable model can be invoked that does not require any new molecular players.
The epistasis plots shown here are a useful way to visualize epistasis in vectorial phenotypes. We have shown how an epistasis plot can be used to identify interactions between two genes by examining the transcriptional phenotypes of single and double mutants. Epistasis plots can accumulate an arbitrary number of points within them, possess a rich structure that can be visualized, and have straightforward interpretations for special slope values. Epistasis plots and GLMs are not mutually exclusive. A GLM could be used to quantify epistasis interactions at single-transcript resolution and the results then analyzed using an epistasis plot (for a nongenetic example, see ref. 15). A benefit of epistasis plots is that they enable the computation of a single, aggregate statistic that describes the ensemble behavior of a set of genes. This aggregate statistic is not enough to describe all possible behaviors in a system, but it can be used to establish whether the genes under study are part of a single pathway. In the case of the hypoxia pathway, phenotypes that are downstream of the hypoxia pathway should conform to the genetic equalities, egl-9(lf) hif-1(lf) = hif-1(lf) and egl-9(lf); vhl-1(lf) = egl-9(lf). Genes whose expression levels behave strangely yet satisfy these equalities are downstream of the hypoxia pathway. These anomalous genes cannot be identified via the epistasis coefficient, but the epistasis coefficient does provide a unifying framework with which to analyze them by constraining the space of plausible hypotheses.
Until relatively recently, the rapid generation and molecular characterization of null mutants was a major bottleneck for genetic analyses. Advances in genomic engineering mean that, for a number of organisms, production of mutants is now rapid and efficient. As mutants become easier to produce, biologists are realizing that phenotyping and characterizing the biological functions of individual genes is challenging. This is particularly true for whole organisms, where subtle phenotypes can go undetected for long periods of time. We have shown that whole-animal RNA-seq is a sensitive method that can be seamlessly incorporated with genetic analyses of epistasis.
Materials and Methods
Nematode Strains and Culture.
Strains used were N2 (Bristol), JT307 egl-9(sa307), CB5602 vhl-1(ok161), ZG31 hif-1(ia4), RB1297 rhy-1(ok1402), and CB6088 egl-9(sa307) hif-1(ia4) CB6116 egl-9(sa307); vhl-1(ok161). Lines were grown on standard nematode growth media Petri plates seeded with OP50 E. coli at 20 °C (41).
RNA Isolation.
Lines were synchronized by harvesting eggs via sodium hypochlorite treatment and subsequently plating eggs on food. Worms were staged and based on the time after plating, vulva morphology, and the absence of eggs. Between 30 and 50 nongravid young adults were picked and placed in 100 L of TE pH 8.0 (Ambion AM9849) in mL PCR tubes on ice. Worms were allowed to settle or spun down by centrifugation and L of supernatant removed before flash-freezing in liquid . These samples were digested with Recombinant Proteinase K PCR Grade (Roche Lot No. 03115 838001) for 15 min at 60° in the presence of 1% SDS and 1.25 L RNA Secure (Ambion AM7005). Five volumes of Trizol (Tri-Reagent Zymo Research) were added to the RNA samples and treated with DNase I using Zymo Research Quick-RNA MicroPrep R1050. Samples were analyzed run on an Agilent 2100 BioAnalyzer (Agilent Technologies). Replicates were selected that had RNA integrity numbers (RIN) equal to or greater than 9.0 and without bacterial ribosomal bands, except for the ZG31 mutant, where one of three replicates had a RIN of 8.3.
Library Preparation and Sequencing.
We reverse-transcribed 10 ng of total RNA from each sample into cDNA using the Clontech SMARTer Ultra Low Input RNA for Sequencing v3 kit (catalog no. 634848) in the SMARTSeq2 protocol (42). RNA was denatured at 70 °C for 3 min in the presence of dNTPs, oligo dT primer, and spiked-in quantitation standards (National Institute of Standards and Technology/External RNA Controls Consortium from Ambion, catalog no. 4456740). After chilling to 4 °C, the first-strand reaction was assembled using a LNA TSO primer (42) and run at 42 °C for 90 min, followed by denaturation at 70 °C for 10 min. The first-strand reaction was used as a template for 13 cycles of PCR using the Clontech v3 kit. Reactions were purified with Ampure XP SPRI beads (catalog no. A63880). After quantification using the Qubit High Sensitivity DNA assay, a 3 ng aliquot of the cDNA was run on the Agilent HS DNA chip to confirm the length distribution of the amplified fragments. The median value for the average cDNA lengths from all length distributions was 1,076 bp. Tagmentation of the full-length cDNA was performed using the Illumina/Nextera DNA library prep kit (catalog no. FC-121–1030). Following Qubit quantitation and Agilent BioAnalyzer profiling, the tagmented libraries were sequenced on an Illumina HiSeq2500 machine in single-read mode with a read length of 50 nt to a depth of 15 million reads per sample. Base calls were performed with RTA 1.13.48.0 followed by conversion to FASTQ with bcl2fastq 1.8.4.
Read Alignment and Differential Expression Analysis.
We used Kallisto (43) to perform read pseudoalignment and performed differential analysis using Sleuth (44). We fit a GLM for an isoform in sample :
| [1] |
where was the logarithm transformed counts of isoform in sample ; and were parameters of the model for the isoform , which could be interpreted as biased estimators of the log-fold change; were indicator variables describing the experimental conditions of the isoform in sample ; and was the noise associated with a particular measurement. After fitting the GLM, we tested isoforms for differential expression using the built-in Wald test in Sleuth (44), which outputs a value that has been corrected for multiple hypothesis testing.
Genetic Analysis, Overview.
The processed data were analyzed using Python 3.5. We used the Pandas, Matplotlib, Scipy, Seaborn, Sklearn, Networkx, PyMC3, and TEA libraries (45–52). Our analysis is available in Jupyter Notebooks (53). All code and processed data are available at https://github.com/WormLabCaltech/mprsq along with version-control information. Our Jupyter Notebook and interactive graphs for this project can be found at https://wormlabcaltech.github.io/mprsq/ in html format, or in SI Appendix. Raw reads were deposited in the Short Read Archive under the study accession no. SRP100886 and in the Gene Expression Omnibus (GEO) under accession no. GSE97355.
Weighted Correlations.
Correlations between mutants were calculated by identifying their STP. Transcripts were rank-ordered according to their regression coefficient, . Regressions were performed using a Student-T distribution with the PyMC3 library (48) (pm.glm.families.StudenT in Python). If the correlations had an average value , the average correlation coefficient was set to 1. Weights were calculated as the number of genes that were inliers divided by the number of DEGs present in either mutant.
Epistatic Analysis.
The epistasis coefficient between two null mutants a and b was calculated as:
| [2] |
Null models for various epistatic relationships were generated by sampling the single mutants in an appropriate fashion. For example, to generate the distribution for two mutants that obey the epistatic relationship , we substituted with and bootstrapped the result.
To select between theoretical models, we implemented an approximate Bayesian OR. We defined a free-fit model, , that found the line of best fit for the data:
| [3] |
where was the slope to be determined, are the coordinates of each point, and was the SE associated with the y value. We used Eq. 3 to obtain the most likely slope given the data, , via minimization (scipy.optimize.minimize in Python). Finally, we approximated the OR as:
| [4] |
where was the slope found after minimization, was the SD of the parameter at the point , and was the probability of the data given the parameter-free model, .
Enrichment Analysis.
Tissue, phenotype, and gene ontology enrichment analyses were carried out using the WormBase Enrichment Suite for Python (28, 51).
Supplementary Material
Acknowledgments
We thank Hillel Schwartz, Erich Schwarz, Jonathan Liu, Han Wang, and Porfirio Quintero for their advice throughout this project. This work was supported by the Howard Hughes Medical Institute, with whom P.W.S. is an investigator, and by the Millard and Muriel Jacobs Genetics and Genomics Laboratory at Caltech. Strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE97355). Raw reads were deposited in the Short Read Archive (accession no. SRP100886). All code and processed data are available in the GitHub database, https://github.com/WormLabCaltech/mprsq. Our Jupyter Notebook and interactive graphs for this project can be found at https://wormlabcaltech.github.io/mprsq/, or in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712387115/-/DCSupplemental.
References
- 1.Huang LS, Sternberg PW. Genetic dissection of developmental pathways. WormBook. 2006;14:1–19. doi: 10.1895/wormbook.1.88.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips PC. Epistasis—The essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz EM, Kato M, Sternberg PW. Functional transcriptomics of a migrating cell in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:16246–162451. doi: 10.1073/pnas.1203045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 8.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King EG, Sanderson BJ, McNeil CL, Long AD, Macdonald SJ. Genetic dissection of the Drosophila melanogaster female head transcriptome reveals widespread allelic heterogeneity. PLoS Genet. 2014;10:e1004322. doi: 10.1371/journal.pgen.1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 12.Capaldi AP, et al. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Driessche N, et al. Epistasis analysis with global transcriptional phenotypes. Nat Genet. 2005;37:471–477. doi: 10.1038/ng1545. [DOI] [PubMed] [Google Scholar]
- 14.Dixit A, et al. Perturb-seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angeles-Albores D, et al. The Caenorhabditis elegans female-like state: Decoupling the transcriptomic effects of aging and sperm status. G3 (Bethesda) 2017;7:2969–2977. doi: 10.1534/g3.117.300080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epstein ACR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 17.Shen C, Shao Z, Powell-Coffman JA. The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics. 2006;174:1205–1214. doi: 10.1534/genetics.106.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao Z, Zhang Y, Powell-Coffman JA. Two distinct roles for EGL-9 in the regulation of HIF-1-mediated gene expression in Caenorhabditis elegans. Genetics. 2009;183:821–829. doi: 10.1534/genetics.109.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci USA. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loenarz C, et al. The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 2011;12:63–70. doi: 10.1038/embor.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 22.Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci USA. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang LE, Arany Z, Livingston DM, Franklin Bunn H. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem. 1996;271:32253–32259. doi: 10.1074/jbc.271.50.32253. [DOI] [PubMed] [Google Scholar]
- 24.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Ma DK, Vozdek R, Bhatla N, Horvitz HR. CYSL-1 interacts with the O 2-sensing hydroxylase EGL-9 to promote H 2S-modulated hypoxia-induced behavioral plasticity in C. elegans. Neuron. 2012;73:925–940. doi: 10.1016/j.neuron.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- 27.Budde MW, Roth MB. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol Biol Cel. 2010;21:212–217. doi: 10.1091/mbc.E09-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angeles-Albores D, Lee RY, Chan J, Sternberg PW. 2017. Phenotype and gene ontology enrichment as guides for disease modeling in C. elegans. bioRxiv:10.1101/106369.
- 29.Ackerman D, Gems D. Insulin/IGF-1 and hypoxia signaling act in concert to regulate iron homeostasis in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002498. doi: 10.1371/journal.pgen.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romney SJ, Newman BS, Thacker C, Leibold EA. HIF-1 regulates iron homeostasis in Caenorhabditis elegans by activation and inhibition of genes involved in iron uptake and storage. PLoS Genet. 2011;7:e1002394. doi: 10.1371/journal.pgen.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luhachack LG, et al. EGL-9 controls C. elegans host defense specificity through prolyl hydroxylation-dependent and -independent HIF-1 pathways. PLoS Pathog. 2012;8:e1002798. doi: 10.1371/journal.ppat.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao Z, Zhang Y, Ye Q, Saldanha JN, Powell-Coffman JA. C. elegans swan-1 binds to egl-9 and regulates hif-1- mediated resistance to the bacterial pathogen Pseudomonas aeruginosa PAO1. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YI, Cho JH, Yoo OJ, Ahnn J. Transcriptional regulation and life-span modulation of cytosolic aconitase and ferritin genes in C. elegans. J Mol Biol. 2004;342:421–433. doi: 10.1016/j.jmb.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 34.Chintala S, et al. Prolyl hydroxylase 2 dependent and Von-Hippel-Lindau independent degradation of Hypoxia-inducible factor 1 and 2 alpha by selenium in clear cell renal cell carcinoma leads to tumor growth inhibition. BMC Cancer. 2012;12:293. doi: 10.1186/1471-2407-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–20518. [PubMed] [Google Scholar]
- 36.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart Y, Antebi YE, Mayo AE, Friedman N, Alon U. Design principles of cell circuits with paradoxical components. Proc Natl Acad Sci USA. 2012;109:8346–8351. doi: 10.1073/pnas.1117475109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hart Y, Alon U. The utility of paradoxical components in biological circuits. Mol Cell. 2013;49:213–221. doi: 10.1016/j.molcel.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Clifford R, et al. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- 40.Lönnberg T, et al. Single-cell RNA-seq and computational analysis using temporal mixture modeling resolves TH1/TFH fate bifurcation in malaria. Sci Immunol. 2017;2:eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulston JE, Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974;77:95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 43.Bray NL, Pimentel HJ, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 44.Pimentel HJ, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-Seq incorporating quantification uncertainty. Nat Methods. 2016;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 45.McKinney W. 2011 pandas: A foundational Python library for data analysis and statistics. Available at http://www.dlr.de/sc/Portaldata/15/Resources/dokumente/pyhpc2011/submissions/pyhpc2011_submission_9.pdf. Accessed March 1, 2018.
- 46.Oliphant TE. SciPy: Open source scientific tools for Python. Comput Sci Eng. 2007;9:10–20. [Google Scholar]
- 47.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Machine Learn Res. 2012;12:2825–2830. [Google Scholar]
- 48.Salvatier J, Wiecki T, Fonnesbeck C. Probabilistic programming in Python using PyMC. PeerJ Computer Sci. 2015;2:e55. doi: 10.7717/peerj-cs.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Der Walt S, Colbert SC, Varoquaux G. The NumPy array: A structure for efficient numerical computation. Comput Sci Eng. 2011;13:22–30. [Google Scholar]
- 50.Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9:99–104. [Google Scholar]
- 51.Angeles-Albores D, N.Lee RY, Chan J, Sternberg PW. Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics. 2016;17:366. doi: 10.1186/s12859-016-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waskom M, et al. 2016. seaborn: v0.7.0. Zenodo, 10.5281/zenodo.883859.
- 53.Pérez F, Granger B. IPython: A system for interactive scientific computing Python: An open and general- purpose environment. Comput Sci Eng. 2007;9:21–29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.