Significance
Plants contain a unique family of membrane receptors, which are different from the ones found in bacteria and animals. These proteins are able to sense very different signals, such as steroid molecules, peptides, and proteins at the cell surface using a spiral-shaped ligand binding domain. Ligand binding allows the receptor to engage with a smaller coreceptor kinase, which is shared among different receptors. Here it is analyzed how one coreceptor protein can contribute to the sensing of two different ligands involved in plant growth and organ abscission and to activation of their cognate receptors.
Keywords: brassinosteroid signaling, floral abscission, membrane receptor kinase, leucine-rich repeat domain, receptor activation
Abstract
Plant-unique membrane receptor kinases with leucine-rich repeat ectodomains (LRR-RKs) can sense small molecule, peptide, and protein ligands. Many LRR-RKs require SERK-family coreceptor kinases for high-affinity ligand binding and receptor activation. How one coreceptor can contribute to the specific binding of distinct ligands and activation of different LRR-RKs is poorly understood. Here we quantitatively analyze the contribution of SERK3 to ligand binding and activation of the brassinosteroid receptor BRI1 and the peptide hormone receptor HAESA. We show that while the isolated receptors sense their respective ligands with drastically different binding affinities, the SERK3 ectodomain binds the ligand-associated receptors with very similar binding kinetics. We identify residues in the SERK3 N-terminal capping domain, which allow for selective steroid and peptide hormone recognition. In contrast, residues in the SERK3 LRR core form a second, constitutive receptor–coreceptor interface. Genetic analyses of protein chimera between BRI1 and SERK3 define that signaling-competent complexes are formed by receptor–coreceptor heteromerization in planta. A functional BRI1–HAESA chimera suggests that the receptor activation mechanism is conserved among different LRR-RKs, and that their signaling specificity is encoded in the kinase domain of the receptor. Our work pinpoints the relative contributions of receptor, ligand, and coreceptor to the formation and activation of SERK-dependent LRR-RK signaling complexes regulating plant growth and development.
Brassinosteroids are growth-promoting hormones with key roles in plant development (1). They are sensed by the receptor kinase with leucine-rich repeat ectodomain (LRR-RK) BRASSINOSTEROID INSENSITIVE 1 (BRI1) that is composed of a LRR ectodomain, a single membrane-spanning helix, and a cytoplasmic kinase module (2, 3). BRI1 directly binds brassinosteroids including the potent brassinolide (BL) (4). The steroid binding site is formed by the BRI1 LRR core and by the island domain inserted between LRRs 21 and 22 (5, 6). Steroid binding creates a docking platform for the LRR ectodomain of a member of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family of coreceptor kinases (SI Appendix, Fig. S1) (6–9). BRI1 forms a ligand-dependent heterodimeric complex with a SERK protein in vitro, with the SERK N-terminal capping domain completing the steroid binding pocket (10–12). Receptor–coreceptor complex formation at the cell surface brings the cytoplasmic domains of BRI1 and SERK in close proximity, allowing for transphosphorylation of the two kinases and activation of the brassinosteroid signaling pathway (11, 13, 14). The sequence-related LRR-RK HAESA shares the overall architecture with BRI1 but lacks the island domain (15). HAESA and HAESA-LIKE 2 (HSL2) bind the peptide hormone INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) to trigger abscission of floral organs in Arabidopsis and both receptors associate with SERKs in an IDA-dependent manner (16–18). HAESA/HSL2 and BRI1 bind drastically different ligands using different surface areas of their ectodomains. However, both receptors rely on the same SERK proteins for activation and the three-dimensional structures of the active signaling complexes are remarkably similar (11, 17, 19) (Fig. 1A). In addition, SERK proteins have been genetically and biochemically implicated in many other LRR-RK signaling pathways (20). How few SERK proteins allow for sensing of many different small molecule, peptide, and protein ligands and to the activation of their cognate receptors remains to be investigated. Here we analyze how SERKs can contribute to the specific binding of BL and IDA and to the activation of BRI1 and HAESA.
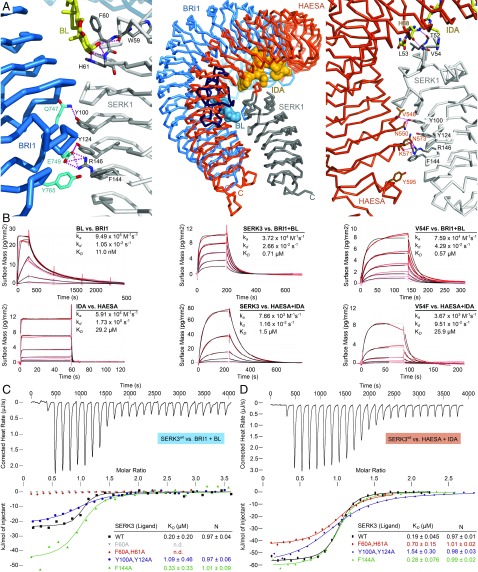
Fig. 1.
Two distinct SERK surface patches contribute to formation of different LRR-RK complexes. (A, Center) Structural superposition of BRI1–BL–SERK1 (PDB ID 4LSX) and HAESA–IDA–SERK1 (PDB ID 5IYX) complex structures, using the SERK1 ectodomain as reference (rmsd is ∼0.6 Å comparing 185 corresponding Cα atoms). Shown are Cα traces of the BRI1 LRR domain (blue, island domain in dark blue), the HAESA LRR domain (orange), the SERK1 ectodomain (gray) and the steroid (blue) and peptide (orange) ligands in surface representation, respectively. (A, Left) Close-up view of the BRI1–BL–SERK1 complex, highlighting the two distinct interaction surfaces and including selected interface interactions. BL is shown in bond representation (yellow). (A, Right) Details of the HAESA–IDA–SERK1 interface, with IDA shown in bonds representation (yellow). SERK residue numbering is according to Arabidopsis SERK3. (B) Grating-coupled interferometry (GCI)-derived binding kinetics for BRI1 and HAESA vs. their ligands and SERK3. Shown are sensorgrams with data in red and their respective fits in black (Materials and Methods). Table summaries of kinetic parameters are shown (ka, association rate constant; kd, dissociation rate constant; KD, dissociation constant). (C) Isothermal titration calorimetry (ITC) of the BRI1 LRR domain vs. wild-type and mutant SERK3 ectodomains and in the presence of BL. (D) ITC of the HAESA LRR domain vs. SERK3 proteins and in the presence of IDA. Table summaries for dissociation constants (KD) and binding stoichiometries (N) are shown (± fitting error; n.d., no detectable binding).
Results
A structural superposition of the previously reported BRI1–BL–SERK1 and HAESA–IDA–SERK1 complexes using the SERK1 ectodomain as reference [root mean square deviation (rmsd) is ∼0.6 Å comparing 185 corresponding Cα atoms] reveals that SERKs bind BRI1 and HAESA in similar positions, despite their drastically different ligand binding sites (Fig. 1A) (11, 17, 20). To characterize the interactions of BRI1 and HAESA with their ligands and coreceptors, we determined binding kinetics using a label-free surface biosensor based on grating-coupled interferometry (GCI) (Materials and Methods) (21). In addition, steady-state binding constants were derived from isothermal titration calorimetry (ITC) experiments. We found that BL binds BRI1 with a dissociation constant (KD) of ∼10 nM in GCI assays, similar to the previously reported binding affinity for radio-labeled BL interacting with BRI1-containing plant membrane fractions (Fig. 1B) (4). This tight interaction of the steroid with its receptor is characterized by a fast association rate (ka ∼ 1·106 M−1⋅s−1) and a relatively slow dissociation rate (kd ∼ 1·10−2⋅s−1). HAESA, in contrast, binds the IDA peptide with a KD of ∼30 μM (Fig. 1B), which is in good agreement with the previously reported KD determined by ITC (17). IDA rapidly associates with its receptor (ka ∼ 6·104 M−1⋅s−1), but in contrast to the steroid hormone the peptide has a fast dissociation rate (kd ∼ 2 s−1). Our kinetic analyses reveal that BRI1 and HAESA have evolved distinct modes of ligand binding, allowing them to sense steroid and peptide ligands with very different binding kinetics.
We assessed binding of the coreceptor SERK3 to the ligand-bound BRI1 and HAESA ectodomains. SERK3 is a bona fide BRI1 and HAESA coreceptor (8, 19, 22) and its ectodomain can be produced in sufficient quantities for biochemical studies (SI Appendix, Fig. S2). BRI1 shows no detectable binding to SERK3 in the absence of BL in vitro (11). In the presence of BL, SERK3 binds BRI1 with a KD of ∼0.7 μM (GCI) or ∼0.2 μM (ITC), and with 1:1 stoichiometry (Fig. 1 B and C). SERK3 binds HAESA–IDA with a KD of ∼0.2–1.5 μM as concluded from ITC (Fig. 1D) or GCI (Fig. 1B) experiments, respectively. The HAESA coreceptor SERK1 shows similar binding affinities (17). The kinetic parameters derived from the GCI experiments indicate that SERK3 binds BRI1 and HAESA in similar ways, with a relatively slow dissociation rate (kd ∼ 1–3 × 10−2⋅s−1) (Fig. 1B). The observed dissociation rates for SERK3 binding to BRI1/HAESA may be sufficiently long to allow for the receptor and coreceptor kinase domains to transphosporylate and activate each other in the cytosol (11, 13), as previously speculated (20).
We next analyzed which SERK residues interact with either the ligand or the cognate receptor in the previously reported BRI1–BL–SERK1 (11) and HAESA–IDA–SERK1 (17) complexes. Residues originating from the SERK N-terminal capping domain are in direct contact with the steroid and the peptide hormone, but different residues are used to recognize the respective ligands (Fig. 1A) (11, 17). In the case of BRI1, Phe60 (all amino acid numbering refers to SERK3) establishes a π-stacking interaction with the C ring of brassinolide, while side-chain and main-chain atoms from His61 and Trp59 are hydrogen bonding with the 2α–3α diol moiety of the ligand (6, 11, 12). In the case of HAESA, SERK residues Thr52 and Val54 form main-chain contacts with the conserved C terminus of the peptide hormone, with Phe60 and His61 not being part of the IDA binding site (Fig. 1A) (17). Mutation of Phe60 and/or His61 to Ala completely disrupts binding of SERK3 to BRI1–BL, suggesting that interactions between coreceptor and steroid hormone are critical for receptor–coreceptor complex formation (Fig. 1C and SI Appendix, Fig. S3). Importantly, SERK3F60A/H61A can still bind HAESA–IDA with moderately reduced affinity (Fig. 1D and SI Appendix, Fig. S3).
As IDA establishes only main-chain contacts with the SERK3 N-terminal cap loop, we replaced either Thr52 or Val54 with a bulky phenylalanine. While SERK3T52F still binds BRI1 and HAESA with wild-type affinity (SI Appendix, Fig. S3), mutation of Val54 to Phe selectively reduces binding of the mutant SERK3 protein to HAESA, but not to BRI1 (Fig. 1B). Thus, structure-based point mutations in SERK3 can selectively inhibit binding to either BRI1 or HAESA, highlighting that different surface patches in the SERK N-terminal cap can indeed allow for the sensing of specific ligands in different LRR-RK complexes (Fig. 1) (11, 17, 20).
In addition to SERKs forming part of the steroid and peptide hormone binding site, a direct receptor–coreceptor interface can be observed in the ternary complexes (11, 12, 17). In the case of BRI1, the interface involves SERKY100 that contacts BRI1Q747. SERKY124 and SERKR146 are making extensive polar interactions with BRI1E749. Phe144 in SERKs forms hydrophobic contacts with BRI1Y765 (Fig. 1A). These residues are invariant among Arabidopsis SERK1–4 and among SERKs from other species, establish similar contacts in the HAESA–IDA–SERK1 complex (Fig. 1A and SI Appendix, Fig. S1) (17), and are part of the receptor–coreceptor interface of related peptide hormone receptor complexes (23–25). Mutation of SERK3F144 to alanine has no drastic effect on BRI1–BL–SERK3 and HAESA–IDA–SERK3 interaction, but a SERK3Y100A/Y124A double mutant shows reduced binding to either BRI1–BL or HAESA–IDA (Fig. 1 and SI Appendix, Fig. S3). Thus, two different surface areas of the SERK ectodomain contribute to LRR-RK ligand sensing and complex formation.
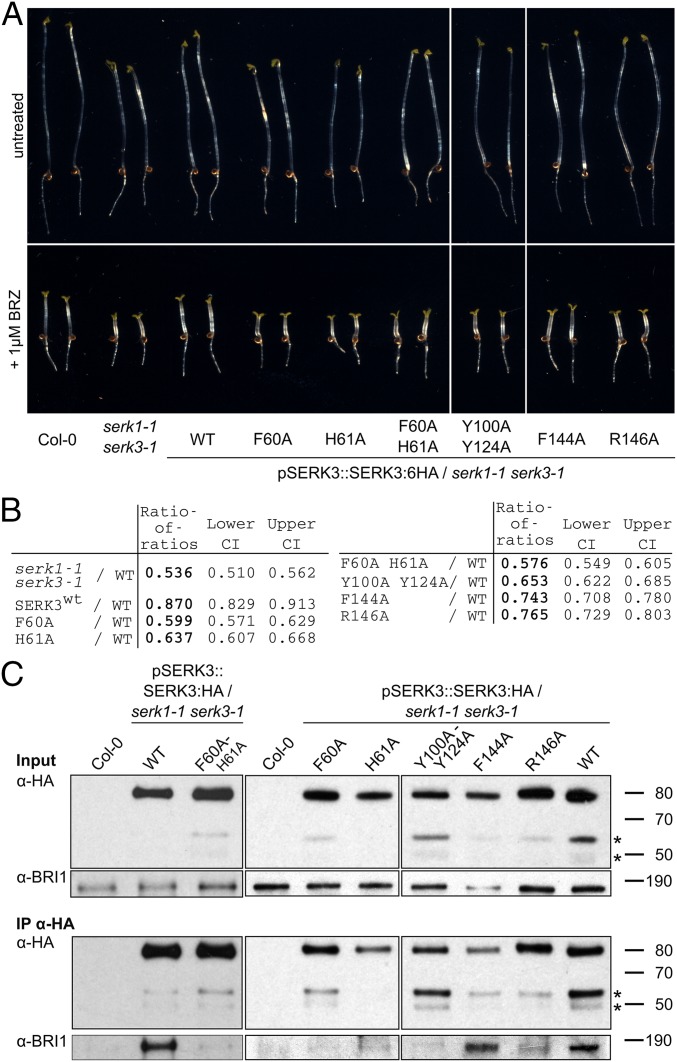
To assess the contribution of the SERK mutations analyzed in vitro to brassinosteroid signaling, we created stable transgenic lines expressing either wild-type or mutant SERK3 from a genomic construct under control of its native promoter and in a serk1-1 serk3-1 mutant background (SI Appendix, Table S1). Individual lines showing similar SERK3 protein levels were selected for subsequent analysis (compare SI Appendix, Fig. S4A). We did not quantify the effects of SERK3 point mutations on HAESA-mediated floral abscission, as detection of weak phenotypes is difficult in the established petal break-strength assay (17) and because higher-order SERK mutants display pleiotropic phenotypes at flowering stage (19, 26). The ability to complement the serk1 serk3 brassinosteroid signaling phenotype was evaluated in a quantitative hypocotyl growth assay (SI Appendix, SI Methods). In agreement with our biochemical experiments, mutation of Phe60 and/or His61 to alanine inactivates SERK3 brassinosteroid coreceptor function in planta (Fig. 2 A and B and SI Appendix, Fig. S5) and disrupts receptor–coreceptor complex formation in vivo, as judged from coimmunoprecipitation (co-IP) experiments (Fig. 2D). Mutation of SERK3 residues targeting the direct receptor–coreceptor interface results in moderate-to-severe brassinosteroid signaling defects, with SERK3Y100A/Y124A having the strongest effect (Fig. 2 A and B and SI Appendix, Fig. S5). This is in line with our biochemical findings, and our in vivo co-IPs confirm an important contribution of this direct receptor–coreceptor interface to BRI1 signaling complex integrity (Figs. 1C and 2D and SI Appendix, Fig. S3).
Fig. 2.
Mutations in the SERK3 ligand binding and receptor interfaces differentially impact SERK3 function and complex formation in vivo. (A) Hypocotyl growth assay in the presence and absence of the brassinosteroid biosynthesis inhibitor brassinazole (BRZ), from seedlings grown for 5 d in the dark. Serk1-1 serk3-1 mutants are BRZ hypersensitive and this phenotype can be complemented by expressing SERK3 in the mutant background (Col-0, untransformed wild type, n = 50). (B) Quantification of the data from A. The log-transformed endpoint hypocotyl length was analyzed by a mixed effects model for the ratio of the transgenic lines vs. wild type, allowing for heterogeneous variances. The ratio of the untreated and BRZ-treated hypocotyl length was calculated for wild type (rwt) and each mutant line (rm); the ratio of this ratio for wild type divided by the ratio for a given mutant line results in the ratio of ratios (RR = rwt/rm; CI, confidence interval). (C) Coimmunoprecipitation (Co-IP) of lines shown in A. SERK3:HA was immunoprecipitated from plant protein extracts using an anti-HA antibody (IP-HA) and BRI1 was detected with an anti-BRI1 antibody (14) in the IP elution.
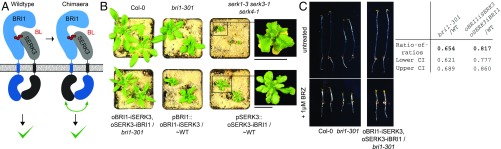
We next tested in a formal genetic way whether the receptor–coreceptor heteromers observed in vivo, in biochemical assays and in crystals are required for brassinosteroid signal activation (11, 14, 27). We created protein chimera between BRI1 and SERK3, in which the kinase domain of the receptor is swapped with that of the coreceptor and vice versa and expressed them under the control of either the BRI1 or SERK3 promoter in Arabidopsis (Fig. 3A and SI Appendix, Figs. S4 and S6 and Tables S1 and S4). Expression of both chimeric constructs together partially complements the bri1-301 (SI Appendix, Fig. S7) mutant in phenotypic and quantitative assays (28) (Fig. 3 B and C and SI Appendix, Figs. S4 and S6). Individual chimera cause dominant negative effects, corresponding to the mutant phenotype of their respective ectodomain: o(utside)BRI1–i(nside)SERK3 alone causes a bri1 mutant phenotype (Fig. 3B and SI Appendix, Fig. S6). oSERK3–iBRI1 plants are dwarf and show early senescence, reminiscent of serk1 serk3 serk4 triple mutants (Fig. 3B and SI Appendix, Figs. S4 and S6). Together, these experiments support an essential role for BRI1–SERK heteromers in brassinosteroid signaling.
Fig. 3.
BRI1 receptor activation requires heteromerization with a SERK coreceptor in planta. (A) Schematic view of signaling competent wild-type and chimeric BRI1–BL–SERK3 complexes. BRI1 domains are shown in blue, SERK3 domains in gray. (B) Stable transgenic lines expressing oBRI1–iSERK3:mCherry under the BRI promoter (pBRI1) and oSERK3–iBRI1:mCitrine under the SERK3 promoter (pSERK3) show a partially rescued bri1-301 growth phenotype when expressed together (Bottom Left). The expression of isolated chimera results in a dominant negative growth phenotype (Bottom Middle and Right). ∼WT corresponds to pBRI1::BRI1:mCitrine/bri1-null. (Scale bar, 1 cm.) (C) Hypocotyl growth assay in the presence and absence of BRZ (compare Fig. 2A). bri1-301 is hypersensitive to BRZ, a phenotype that is partially complemented by the expression of oBRI1–iSERK3 and oSERK3–iBRI1. Quantifications are shown, analyzed as in Fig. 2B.
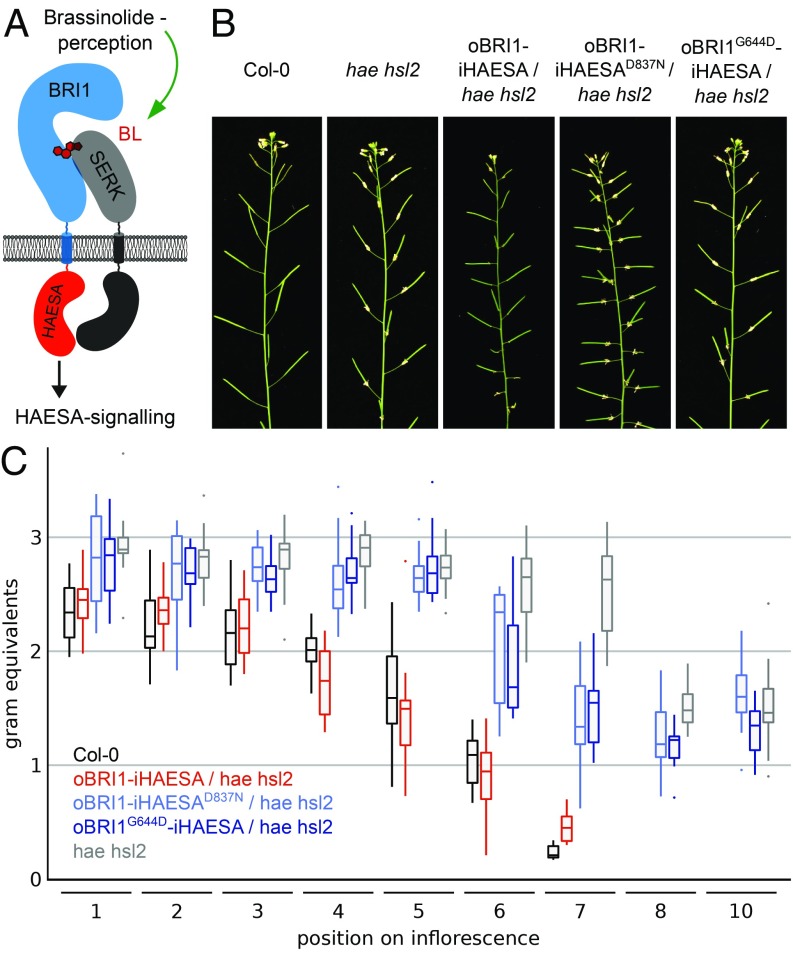
The structural and biochemical similarities between the BRI1–BL–SERK and HAESA–IDA–SERK signaling complexes prompted us to investigate whether the receptor activation mechanism is conserved among these pathways in planta (Fig. 1A). We complemented a hae hsl2 mutant, which shows strong floral abscission defects (18, 29), with a chimeric receptor, in which the BRI1 ectodomain is fused to the HAESA cytoplasmic domain and expressed under control of the HAESA promoter (Fig. 4A and SI Appendix, Fig. S6 and Tables S1 and S2). We found that in contrast to a kinase-inactive version (HAESAD837N), functional chimera can complement the hae hsl2 abscission phenotype (Fig. 4 B and C). Notably, expression of full-length BRI1 under the control of the HAESA promoter does not induce floral abscission in the hae hsl2 mutant (SI Appendix, Fig. S8). We hypothesized that endogenous brassinosteroids bind to the oBRI1 ectodomain in the Arabidopsis abscission zone and allow for SERK complex formation and HAESA signaling. To test this hypothesis, we generated a BRI1–HAESA protein chimera, in which the ectodomain carries the Gly644Asp missense mutation, which has been previously characterized in bri1-6 mutants (30). Brassinosteroid binding is disrupted in BRI1G644D in in vitro binding assays (SI Appendix, Fig. S9). A 2.6-Å crystal structure of the mutant protein revealed that the mutation does not affect the overall structure of BRI1 and leaves the conformation of the island domain intact, making the Gly644Asp mutation an ideal tool to specifically interfere with the ligand binding function of BRI1 in planta (SI Appendix, Fig. S9 and Table S3). Importantly, oBRI1G644D–iHAESA cannot complement the hae hsl2 mutant phenotype, suggesting that indeed brassinosteroid sensing and complex formation occurs in the abscission zone. Our oBRI1–iHAESA chimera indicates that the cytoplasmic signaling specificity of HAESA is encoded in its kinase domain and not in its extracellular domain.
Fig. 4.
The receptor’s kinase domain encodes for the signaling specificity of LRR-RK pathways. (A) Schematic overview of the oBRI1–iHAESA chimera and its envisioned signaling complex (colors as in Fig. 3A; the HAESA kinase domain is shown in orange). (B) Inflorescences of ∼9-wk-old Arabidopsis plants. Abscission of floral organs is impaired in hae hsl2 mutant plants compared with Col-0 wild type. (C) Box plots of BRI1–HAESA chimera vs. Col-0 and hae hsl2 lines. In a quantitative petal break-strength assay, the force required to remove a petal at a given position on the inflorescence is measured in gram equivalents (for each position, at least 15 independent measurements were taken). Statistical analysis is shown in SI Appendix, Table S2.
Discussion
The five SERKs in Arabidopsis have emerged as essential coreceptor kinases for different LRR-RK signaling pathways, controlling plant growth, development, and immunity (20). In the known LRR-RK–SERK complex structures, residues originating from the SERK N-terminal capping domain contact different small molecule and peptide ligands when bound to their cognate receptor, while the SERK LRR core forms variable interactions with the C-terminal half of the respective ectodomain of the receptor (11, 12, 17, 23–25). Here, we used the SERK-dependent LRR-RKs BRI1 and HAESA to assess the relative contributions of both coreceptor interaction surfaces to high-affinity ligand sensing, to complex formation, and to complex stability.
In the present study, we focused on analyzing SERK residues contributing to LRR-RK complex formation. We found that different residues from the SERK N-terminal capping domain interact with both the steroid hormone BL and the peptide hormone IDA and we could generate SERK3 mutants that selectively bind either BRI1–BL or HAESA–IDA. We speculate that these residues allow SERKs to discriminate between ligand-associated and apo forms of their cognate receptors. However, the sequence motifs identified in our study are not uniquely used for BRI1 or HAESA recognition: The SERK3 Phe60/His61-containing surface patch critical for interaction with BRI1 is also important for SERK recruitment to the plant phytosulfokine receptor, another island-domain containing LRR-RK (24). In addition, the SERK3 Thr52–Val54 loop is not only in direct contact with the C terminus of the peptide hormone IDA, but also mediates interaction with the LRR-RK peptide ligands flg22 (23), Pep1 (31), RGF (32), and CLE41 (25). This suggests that very different ligands are recognized by SERKs using only few invariant structural motifs (33). The requirement of several conserved surface patches in SERK proteins interacting with different ligands on one hand, and establishing direct complex interactions on the other hand, may rationalize why the SERK ectodomain sequences are highly conserved throughout plant evolution (SI Appendix, Fig. S1). Sequence variation alone thus cannot explain the fact that SERKs act only partially redundant in various developmental and immune signaling pathways, which may rather be caused by differences in expression and/or localization to specific membrane (nano) domains (34, 35).
The interaction of LRR-RK ectodomains has been quantified for selected ligands with dissociation constants spanning several orders of magnitude, ranging from the midmicromolar (17, 24) to the low-nanomolar range (32, 36, 37). In contrast, the reported KDs for the ligand-dependent SERK association with different receptors are very similar (∼0.2–1.5 μM) (17, 25). This indicates that differently shaped ligand binding pockets in LRR-RKs allow for specific sensing of chemically diverse ligands, but that the mode of coreceptor recruitment is conserved among the different signaling pathways. We characterized a second, direct receptor–coreceptor interface and found that it strongly contributes to BRI1–SERK3 complex stability (Fig. 1 and SI Appendix, Fig. S3). We speculate that SERKs target ligand-bound LRR-RKs using their N-terminal capping domain, and then form a larger, zipper-like interface with the receptor. The second, constitutive interface may stabilize the signaling-competent receptor–ligand–coreceptor complex. In this respect, it is of note that the dissociation rate kd for IDA binding to the isolated HAESA ectodomain is much faster than the dissociation of SERK3 from the HAESA–IDA complex (Fig. 1B). Together with the structural observation that SERKs bind on top of LRR-RK ligand binding sites, this suggests that a main function of the coreceptor may be to slow down dissociation of ligands from the receptor. Our work reveals that SERKs have evolved as adapter proteins that allow LRR-RKs to transduce information across membranes without the need for major conformation rearrangements.
The analysis of BRI1–SERK3 protein chimera in stable transgenic lines now provides formal genetic support for a heteromeric brassinosteroid signaling complex, extending previous studies on FLS2–SERK3 chimera analyzed in a transient expression system (38). The observation that expression of oBRI1–iSERK3 or oSERK3–iBRI1 chimera in isolation causes dominant negative growth phenotypes is likely due to their ability to form signaling incompetent complexes with endogenous LRR-RK pathway components. It is of note that our oSERK3–iBRI1 phenotypes are reminiscent of those caused by expression of the isolated SERK3 ectodomain (39). Our BRI1–HAESA chimera can rescue abscission defects in hae hsl2 mutant plants, suggesting that the activation mechanisms of BRI1 and HAESA are highly similar, and that the kinase domain of the receptor encodes the cytoplasmic signaling specificity of LRR-RK complexes. The recent finding that the rice immune receptor XA21 requires SERKs for activation (40) now rationalizes the earlier observation that a BRI1–XA21 chimera can trigger immune signaling upon BL sensing in rice cells (41). Based on our and previous findings (38, 41), LRR-RK receptor chimera could help in defining the functions of presently orphan receptor kinases and may represent useful tools to modulate plant growth and development in crops.
In conclusion, a ligand-induced and SERK-stabilized receptor–coreceptor heterodimer represents the minimal signaling unit of many LRR-RK complexes at the cell surface. It will now be interesting to define at the structural level, how the cytoplasmic kinase domains of receptor and coreceptor interact with each other in the cytosol. The detailed analysis of these interactions may enable us to understand the cytoplasmic signaling specificity of different LRR-RK pathways, and potentially, the signaling cross-talk between them (42).
Materials and Methods
See SI Appendix, SI Materials and Methods for complete details.
Grating Coupled Interferometry.
GCI experiments were performed with the Creoptix WAVE system (Creoptix AG), a label-free surface biosensor (21). The interaction between the BRI1 ectodomain and BL was measured using a 2PCH WAVEchip (long polycarboxylate matrix; Creoptix AG), while all other experiments were performed on a 2PCP WAVEchip (quasiplanar polycarboxylate surface; Creoptix AG). Chips were conditioned using borate buffer (100 mM sodium borate, pH 9.0, 1 M NaCl; Xantec), and the respective ligands were immobilized on the chip surface via a standard amine-coupling protocol, which consisted of a 7-min activation with a 1:1 mix of 400 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride and 100 mM N-hydroxysuccinimide (both Xantec), followed by injection of the ligands BRI1 or HAESA (1–50 μg/mL) in 10 mM sodium acetate, pH 5.0 (Sigma) until the desired density was reached, and quenched with 1 M ethanolamine, pH 8.0 for 7 min (Xantec). Since SERK3 showed nonspecific binding on the surface, BSA (0.5% in 10 mM sodium acetate, pH 5.0; BSA from Roche) was used to passivate the surface between BRI1/HAESA injection and ethanolamine quenching. Kinetic analysis of the BRI1–BL interaction was performed at 25 °C with a 1:4 dilution series (five dilutions in duplicates) from 0.4 μM BL in sodium citrate buffer, pH 5.0, using citrate buffer, pH 5.0, as running buffer, blank injections for double referencing, and DMSO calibration for bulk correction. A typical kinetic analysis of the SERK3 interaction was performed at 25 °C with a 1:2 dilution series from 2 μM in citrate buffer, pH 5.0, with 100 nM BL or 20 μM IDA in 20 mM sodium citrate, pH 5.0, 250 mM NaCl, blank injections for double referencing, and DMSO calibration for bulk correction. For BRI1 interaction experiments, the running buffer was supplemented with 100 nM BL while IDA was supplied only in the blanks and samples. Data correction and analysis was performed with the Creoptix WAVEcontrol software (corrections applied: X and Y offset, DMSO calibration and double referencing). Data were fitted to either one-to-one binding models or mass-transport limited models, using bulk correction.
ITC.
ITC experiments were performed at 25 °C using a Nano ITC (TA Instruments) with a 1.0-mL standard cell and a 250-μL titration syringe. Proteins were gel filtrated into ITC buffer (20 mM sodium citrate, pH 5.0, 150 mM NaCl) and Y-IDA (YVPIPPSA-Hyp-SKRHN) was dissolved in ITC buffer. A 10-mM BL stock solution in 100% (vol/vol) DMSO was diluted into ITC buffer. A typical experiment consisted of injecting 10-μL SERK3 wild-type or mutant protein aliquots (∼170 μM) into ∼17 μM BRI1 + 100 μM brassinolide or ∼20 μM HAESA + 200 μM IDA in the cell at 150-s intervals. ITC data were corrected for the heat of dilution by subtracting the mixing enthalpies for titrant solution injections into protein-free ITC buffer. Data were analyzed using the NanoAnalyze program (version 3.5) as provided by the manufacturer.
Plant Material and Generation of Transgenic Lines.
SERK3 and BRI1 were amplified from Arabidopsis thaliana (ecotype Col-0) genomic DNA and mutations were introduced by site-directed mutagenesis (SI Appendix, Table S4). Synthetic genes were constructed for the BRI1–SERK3 and BRI1–HAESA chimera (Invitrogen GeneArt) (SI Appendix, Fig. S4 and Table S1). Binary vectors were assembled using the multisite Gateway technology (Thermo Fisher Scientific). All constructs were introduced into Agrobacterium tumefaciens strain pGV2260, and Arabidopsis plants were transformed using the floral dip method (43). Plants were transformed with two vectors at the same time to create double transgenic lines containing pBRI1::oBRI1–iSERK3:mCherry and pSERK3::oSERK3–iBRI1:mCitrine. GABI_134E10 was used as a bri1-null allele (10), plants were grown in 50% humidity, 21 °C, in a 16-h light/8-h dark cycle, and analyzed in T3 generation.
Supplementary Material
Acknowledgments
We thank N. Geldner and Y. Jaillais for sharing Arabidopsis lines and plasmids; Y. Jaillais, R. Ulm, and C. S. Hardtke for commenting on the manuscript; and the staff at beam line PXIII of the Swiss Light Source (Villigen, Switzerland) for technical assistance during data collection. This work was supported by Swiss National Science Foundation Grant 31003A_156920, a Human Frontier Science Program Career Development Award (to M.H.), the European Molecular Biology Organization (EMBO) Young Investigator program (M.H.), and the Research Council of Norway Grant 230849/F20 (to M.A.B.). J.S. was supported by long-term fellowships from EMBO and the Federation of European Biochemical Societies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6FIF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714972115/-/DCSupplemental.
References
- 1.Singh AP, Savaldi-Goldstein S. Growth control: Brassinosteroid activity gets context. J Exp Bot. 2015;66:1123–1132. doi: 10.1093/jxb/erv026. [DOI] [PubMed] [Google Scholar]
- 2.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 6.Hothorn M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474:467–471. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt ED, Guzzo F, Toonen MA, de Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- 8.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 2008;148:611–619. doi: 10.1104/pp.108.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA. 2011;108:8503–8507. doi: 10.1073/pnas.1103556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santiago J, Henzler C, Hothorn M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 2013;341:889–892. doi: 10.1126/science.1242468. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, et al. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Bojar D, et al. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014;78:31–43. doi: 10.1111/tpj.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- 16.Butenko MA, et al. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15:2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santiago J, et al. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife. 2016;5:e15075. doi: 10.7554/eLife.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho SK, et al. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng X, et al. Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 2016;14:1330–1338. doi: 10.1016/j.celrep.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann U, Lau K, Hothorn M. The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol. 2017;68:109–137. doi: 10.1146/annurev-arplant-042916-040957. [DOI] [PubMed] [Google Scholar]
- 21.Kozma P, Hamori A, Cottier K, Kurunczi S, Horvath R. Grating coupled interferometry for optical sensing. Appl Phys B. 2009;97:5–8. [Google Scholar]
- 22.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015;525:265–268. doi: 10.1038/nature14858. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, et al. SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol Plant. 2016;9:1406–1414. doi: 10.1016/j.molp.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 26.Meng X, et al. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr Biol. 2015;25:2361–2372. doi: 10.1016/j.cub.2015.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bücherl CA, et al. Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 2013;162:1911–1925. doi: 10.1104/pp.113.220152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, Huang J, Li B, Li J, Wang Y. Is kinase activity essential for biological functions of BRI1? Cell Res. 2008;18:472–478. doi: 10.1038/cr.2008.36. [DOI] [PubMed] [Google Scholar]
- 29.Stenvik G-E, et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noguchi T, et al. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J, et al. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 2015;25:110–120. doi: 10.1038/cr.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song W, et al. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 2016;26:674–685. doi: 10.1038/cr.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song W, Han Z, Wang J, Lin G, Chai J. Structural insights into ligand recognition and activation of plant receptor kinases. Curr Opin Struct Biol. 2017;43:18–27. doi: 10.1016/j.sbi.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Bücherl CA, et al. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife. 2017;6:e25114. doi: 10.7554/eLife.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, et al. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol Plant. 2016;9:289–300. doi: 10.1016/j.molp.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Lin X, Han Z, Qu L-J, Chai J. Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 2016;26:543–555. doi: 10.1038/cr.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazak O, et al. Perception of root-active CLE peptides requires CORYNE function in the phloem vasculature. EMBO Rep. 2017;18:1367–1381. doi: 10.15252/embr.201643535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albert M, et al. A two-hybrid-receptor assay demonstrates heteromer formation as switch-on for plant immune receptors. Plant Physiol. 2013;163:1504–1509. doi: 10.1104/pp.113.227736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domínguez-Ferreras A, Kiss-Papp M, Jehle AK, Felix G, Chinchilla D. An overdose of the arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR of BIR1-1-dependent manner. Plant Physiol. 2015;168:1106–1121. doi: 10.1104/pp.15.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, et al. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol Plant. 2014;7:874–892. doi: 10.1093/mp/ssu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Z, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 42.Smakowska E, Kong J, Busch W, Belkhadir Y. Organ-specific regulation of growth-defense tradeoffs by plants. Curr Opin Plant Biol. 2016;29:129–137. doi: 10.1016/j.pbi.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.