Significance
Actin filaments are dynamic cytoskeletal elements that assemble upon ATP binding. Actin homologs are present in all domains of life, and all share a similar 3D structure of the assembling subunit, but evolutionary changes to the subunit have generated many different actin filament structures. The filament structure of the bacterial actin AlfA, which positions plasmids—small, circular DNA molecules that encode important genes—ensures that each daughter cell receives at least one copy at cell division. AlfA is different from all other actins in two critical ways: it binds to ATP in a unique way and it is missing a quarter of the conserved structural core. These differences explain unusual AlfA assembly dynamics that underlie its ability to move plasmids.
Keywords: bacterial actin, cytoskeleton, plasmid segregation, cryo-EM
Abstract
Bacterial actins are an evolutionarily diverse family of ATP-dependent filaments built from protomers with a conserved structural fold. Actin-based segregation systems are encoded on many bacterial plasmids and function to partition plasmids into daughter cells. The bacterial actin AlfA segregates plasmids by a mechanism distinct from other partition systems, dependent on its unique dynamic properties. Here, we report the near-atomic resolution electron cryo-microscopy structure of the AlfA filament, which reveals a strikingly divergent filament architecture resulting from the loss of a subdomain conserved in all other actins and a mode of ATP binding. Its unusual assembly interfaces and nucleotide interactions provide insight into AlfA dynamics, and expand the range of evolutionary variation accessible to actin quaternary structure.
Actin is one of the most highly conserved eukaryotic proteins, with critical roles in processes as diverse as motility (1), cell shape (2, 3), organelle positioning (4), and cell division (5). Bacterial actins are involved in many of the same processes (6–8), and share evolutionarily conserved functional properties with eukaryotic actin: they form filaments whose assembly and disassembly is controlled by ATP binding and hydrolysis (9–14), their assembly dynamics are modulated by regulatory proteins (11, 15), and the filaments can serve as the basis for larger cellular structures (9, 16, 17). Actins all share a conserved structural core that has a complex topology of two domains (I and II), each arranged as two subdomains (Ia, Ib, IIa, and IIb), with an ATP binding site between domains I and II (18). Five conserved sequence motifs (phosphate 1, connect 1, phosphate 2, adenosine, connect 2) in domains Ia and IIa surround the ATP binding site and have served to define members of the family (19). The fold is also shared with Hsp70 and sugar kinases, which bind and hydrolyze ATP but do not form filaments. All members of this broader family undergo functionally important conformational changes upon ATP binding and hydrolysis that, in the actins, underlie assembly dynamics.
Despite these conserved features, bacterial actins exhibit far lower levels of sequence conservation than their eukaryotic counterpart. Unlike eukaryotic actin, where a single filament form has been adapted to multiple functions through a host of regulatory binding proteins, bacteria have evolved specialized actins for specific purposes that require fewer interaction partners. This has relaxed evolutionary constraints and allowed bacterial actins to explore a greater range of sequence diversity. The result is extensive divergence of bacterial actins at the sequence level, with corresponding variation in filament architecture, function, and dynamics.
A diverse subset of bacterial actins is involved in separation of plasmid DNA. Large, low-copy number plasmids often encode active segregation systems to ensure against stochastic loss when the host cell divides. Most segregation systems are composed of three elements encoded on the plasmid itself: a cytomotive filament to provide the force for plasmid movement, an adaptor protein that couples filament movement to the plasmid, and a centromere-like DNA region bound by the adaptor (20). Several different types of ATP-dependent cytomotive filaments have been adapted for plasmid segregation (21–23), with bacterial actins among the most widely distributed (12).
The most well studied actin-based segregation system is the par operon of the R1 multidrug resistance plasmid in Escherichia coli. ParM filaments are dynamically unstable: they assemble upon ATP binding and hydrolyze ATP with kinetics that lag behind assembly, so that when the hydrolysis front reaches the end of a growing filament it catastrophically disassembles due to reduced stability of the ADP-bound state (10). ParM makes use of dynamic instability in a search and capture mechanism to segregate plasmids. Filaments nucleate spontaneously, and those that fail to encounter an adaptor–DNA complex eventually disassemble. When the ends of ParM filaments do encounter adaptor–DNA complexes, their dynamic instability is suppressed, allowing processive growth that separates plasmids by pushing them toward opposite poles (15, 24).
The plasmid segregating actin AlfA, encoded by the Bacillus subtilis plasmid pLS32, was initially identified as an actin on the basis of the five conserved actin motifs, and like other actins AlfA forms ATP-dependent filaments both in vivo and in vitro (9, 13). Unlike ParM, however, AlfA is not dynamically unstable, forming stable filaments that remain assembled indefinitely in the ADP-bound state (9). Moreover, unlike ParM, which is structurally polar but grows at equal rates from both ends (10), AlfA filaments grow unidirectionally (25). AlfA filaments associate laterally into mixed polarity bundles, and extension of plasmid-bound filaments along bundles provides the mechanism of plasmid segregation. The adaptor protein AlfB regulates AlfA dynamics: free AlfB suppresses AlfA growth and promotes disassembly of ADP-bound filaments, while AlfB–DNA complexes nucleate AlfA filaments (25). These combined activities suppress spontaneous nucleation and ensure that filaments grow primarily from plasmids. Consistent with its unusual dynamics, our initial low-resolution structural studies of AlfA filaments revealed an unusual filament architecture, more ribbon-like and twisted than other actins (9). However, the relationship between this architecture and the dynamic properties of AlfA has remained unclear, as no high-resolution structures of the AlfA filament nor crystal structure of monomeric AlfA has been determined.
Here, we report the structure of AlfA filaments determined by electron cryo-microscopy (cryo-EM) at near-atomic resolution, which reveals the basis for its unique structural and functional characteristics. We show that AlfA lacks the canonical actin subdomain IIb, which plays important structural and functional roles in all other actins. AlfA polymerization interfaces have diverged extensively from other actins, and AlfA binds ATP through completely novel interactions with the adenosine base. These unique features of AlfA explain how it assembles stable filaments despite the loss of subdomain IIb, and why the filaments remain stable after ATP hydrolysis.
Results and Discussion
AlfA Lacks Subdomain IIb.
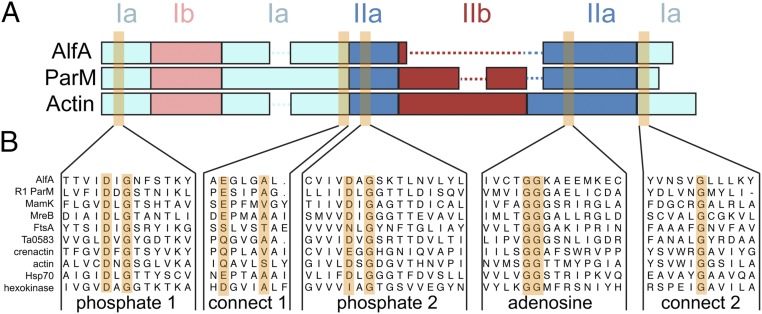
In seeking clues to the unusual architecture from the AlfA sequence, we carried out extensive sequence searches and multiple sequence alignment with bacterial actins. While AlfA was clearly identified as an actin on the basis of the five conserved actin motifs (13), sequence alignment of regions outside these motifs can be challenging due to the very low level of sequence identity. Beginning with alignments of only the closest relatives to AlfA and expanding the size of the sequence set, we were able to generate robust alignments showing that AlfA is missing the canonical subdomain IIb (Fig. 1 and Fig. S1). The closest homologs, with an average identity of ∼20% to AlfA, constitute a family defined by the lack of IIb, consisting of actins primarily from Gram-negative bacteria, and from several bacillus phages. The relatively limited number of actins in this family suggests that they have experienced a deletion of IIb during their evolutionary divergence from other actins.
Fig. 1.
AlfA sequence conservation. (A) Diagram of domain arrangements for AlfA, the bacterial actin ParM from the E. coli R1 plasmid, and vertebrate actin. The five conserved actin sequence motifs surrounding the actin binding cleft are highlighted in orange. (B) Sequence alignments of AlfA with other actins, Hsp70, and hexokinase in the regions surrounding the five conserved motifs.
Subdomain IIb is critical for function in all other actins described to date. While IIb does not include any of the conserved ATP-binding motifs, it forms half of the binding pocket for the ATP adenosine base and contributes a significant fraction (17–35%) of the total surface area buried in filament assembly interfaces. In other actins, ATP binding allosterically regulates polymerization by stabilizing a filament-bound ATP conformation whose major difference with the unbound conformation is the relative positioning of subdomains Ib and IIb (Fig. S2). The importance of IIb assembly interactions is highlighted by their role in ParM dynamic instability, where structural changes associated with ATP hydrolysis break IIb interactions with neighboring protomers, destabilizing the filament (26). The integral role of IIb in actin filament structure and function raises several questions: How can AlfA bind ATP with half of the canonical binding site missing? How is it able to form stable filaments without the IIb assembly interfaces? And how does ATP binding trigger polymerization? To answer these questions, we determined the structure of the AlfA filament using cryo-EM.
AlfA Filament Structure.
At physiological salt concentrations AlfA filaments spontaneously assemble into bundles of variable thickness that are not well suited to high-resolution structure determination. However, bundles can be dissociated into single two-stranded filaments at high salt concentrations (9). We initially attempted to determine the cryo-EM structure of single AlfA filaments in 1 M KCl, but background from high salt concentration limited the resolution of these reconstructions to about 12 Å. We then turned to a mutant we had previously designed, (two pairs of lysines—K21,K22 and K101,K102—mutated to alanines) that forms single filaments that do not form bundles in vitro, except in the presence of high concentrations of crowding agents (25). These mutants can be imaged as single filaments at lower salt concentrations, making the sample better suited to high-resolution structure determination (Fig. S3).
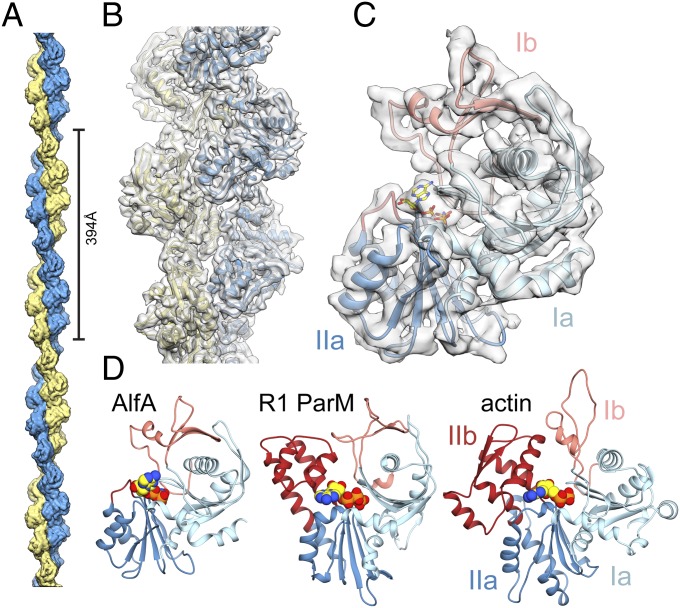
We assembled the nonbundling AlfA mutant with the nonhydrolyzable ATP analog AMPPNP, and determined the structure of the filaments at 4.2-Å resolution by cryo-EM (Fig. 2 and Fig. S4). The refined helical symmetry of the two-stranded filament was 157.7° rotation and 24.4 Å rise per subunit, giving a repeat distance of 394 Å (Fig. 2A). The two strands are parallel and offset by a half-subunit stagger. The repeat distance is considerably shorter than found in other actins, which range from 512 to 834 Å (27–32), yielding a highly twisted AlfA filament with only eight subunits per turn of the two-start helix. The filament has a left-handed two-start twist, confirming our previous determination of handedness by tomographic reconstruction of negatively stained AlfA filaments (9).
Fig. 2.
Cryo-EM structure of the AlfA filament. (A) The two-stranded AlfA filament with a pitch of 394 Å. The pointed end is at top and barbed end at bottom throughout all of the figures. (B) The atomic model fit into a segment of cryo-EM density. (C) A single AlfA protomer from the reconstruction colored by subdomain. (D) Protomers of different actin filaments, with bound nucleotides in yellow, share a conserved fold, but AlfA is missing subdomain IIb.
The cryo-EM map has a clearly defined secondary structure throughout, with bulky side-chains visible in some regions. We generated an atomic model using comparative modeling (33) followed by automated refinement (34), which covers the entire AlfA sequence and includes bound AMPPNP (Fig. 2 B and C and Fig. S4 D and E). Comparison of our atomic model with other actins confirmed that subdomain IIb is missing, replaced by a short five-residue loop (Fig. 2D). It is likely that the lack of IIb has reduced constraints on the helical symmetry of AlfA, making the more highly twisted architecture possible. The rest of the AlfA protomer has the typical actin fold, with subdomains Ia and IIa, each built by a pair of α-helices packed against a five-stranded mixed-polarity β-sheet, and subdomain Ib consisting of a small three-stranded antiparallel β-sheet and a short helix. Like ParM from the R1 (35) and pSK41 (36) plasmids, Bacillus thurigensis (37), and the archael actin Ta0853 (38), the subdomain Ib three-stranded β-sheet of AlfA wraps around helix1 (residues 82–99), making contacts with a pair of antiparallel β-strands inserted after helix2 (residues 128–143) and burying most of helix1. This similarity suggests that this group of bacterial actins may share a more recent common evolutionary ancestor with each other than actins like MreB, MamK, crenactin, and eukaryotic actin that lack these features.
A Nucleotide Binding Mode in the AlfA Filament.
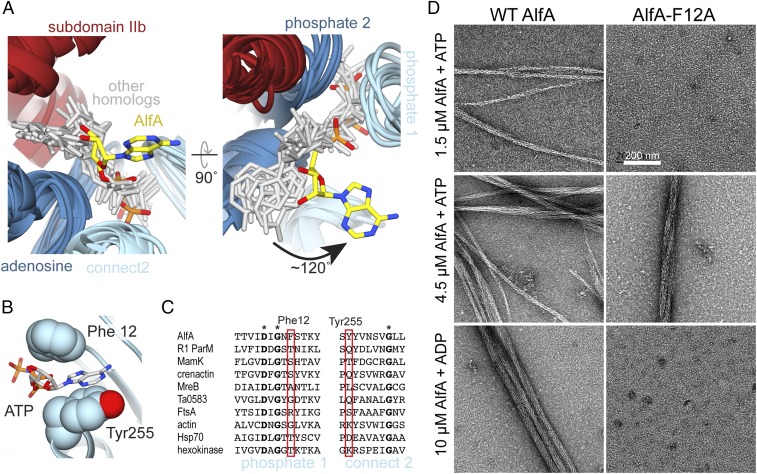
The density for AMPPNP is clearly defined in the cryo-EM map (Fig. S4E). The overall backbone configuration of AlfA in the ATP binding region is conserved with existing actin structures, with average RMSD of 1.8 Å for backbone atoms between AlfA and protomers of other actin filaments. The three phosphates of AMPPNP are bound as in other actin structures, interacting with residues of the phosphate 1, connect 1, and phosphate 2 motifs. Strikingly, however, the adenosine base is rotated ∼120° from the position it occupies in all other actin structures (Fig. 3A, Fig. S5, and Movie S1). Rather than packing against the adenosine motif in subdomain IIa, in AlfA the adenosine base is sandwiched between the phosphate 1 and connect 2 motifs in subdomain Ia. The ATP base stacks against the side-chain of Tyr255, while Phe12 packs against both the base and the ribose sugar (Fig. 3B). Both Phe12 and Tyr255 appear to be unique to AlfA (Fig. 3C). This ATP binding mode, with the base sandwiched between two parts of subdomain Ia, explains how AlfA binds ATP despite the lack of subdomain IIb.
Fig. 3.
AlfA binds ATP through novel interactions. (A) Structural alignment of ATP binding sites of AlfA and other homologs of the actin/Hsp70/sugar kinase family bound to nucleotide, with the proteins rendered as ribbons (colored by subdomain as in Fig. 1), and nucleotides rendered as sticks (gray and yellow). The aligned structures are from cryo-EM filament reconstructions (PDB ID codes: actin, 5JLF; MamK, 5JLV; R1 ParM, 5AEY; crenactin, 5MW1), and crystal structures (PDB ID codes: MreB, 4CZJ; Ta0583, 2FSN; FtsA, 1E4G; Hsp70, 3KVG; hexokinase, 2E2Q). Structural alignment was performed using just the regions around the conserved actin sequence motifs. (B) In the AlfA filament structure the adenosine base is sandwiched between Phe12 and Tyr255 in subdomain Ia. (C) Sequence alignment of the phosphate 1 and connect 2 actin motifs, with positions of Phe12 and Tyr255 highlighted in red. Invariant positions are marked with an asterisk. (D) Negative-stain electron micrographs of AlfA wild-type and F12A mutant in the presence of ATP and ADP. The F12A mutation is capable of assembling filaments but cannot maintain stable filaments in ADP.
AlfA has only a fourfold difference in critical concentration between ATP and ADP (9), indicating that it does not distinguish strongly between di- and triphosphate nucleotides, and explaining why filaments remain assembled in the ADP-bound state. This is in stark contrast to ParM, which experiences dynamic instability and rapidly disassembles after ATP hydrolysis. We reasoned that the unique contacts AlfA makes with the ATP adenosine base provide the basis for its reduced discrimination between ATP and ADP. To test this, we generated a point mutant of Phe12 to alanine (AlfA-F12A) and tested its ability to assemble under different nucleotide conditions (Fig. 3D, Fig. S6, and Table S2). The mutation increased the apparent critical concentration for assembly to about 4 μM from the previously reported value of 2.5 μM (9). However, while wild-type AlfA assembles in ADP with a critical concentration of ∼10 μM, no assembly was observed with ADP for AlfA-F12A up to 30 μM protein (Fig. S6). This highlights the importance of Phe12 in nucleotide binding and suggests that the unique ATP binding mode in AlfA is linked to the increased stability of AlfA filaments in ADP.
AlfA Assembly Interactions.
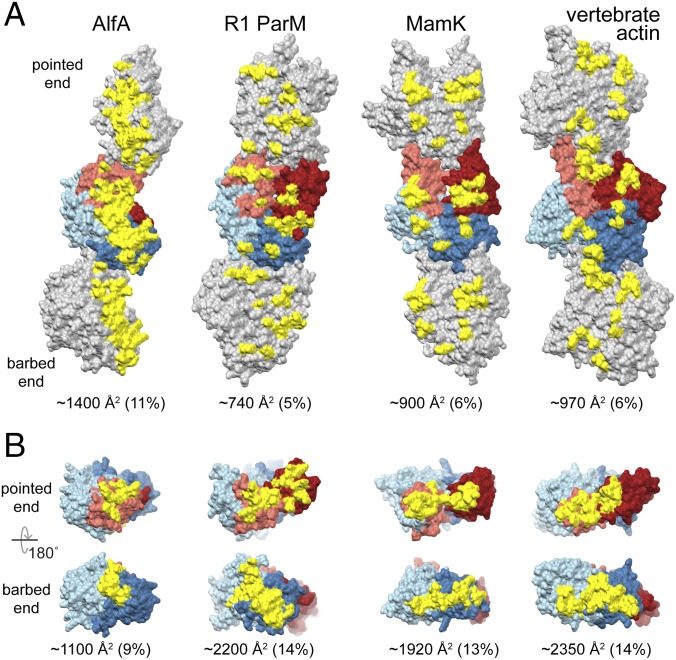
Actin filaments polymerize through two types of interface: head-to-tail longitudinal interactions that run along a single strand, and cross-strand lateral interactions. Longitudinal interactions define the structural polarity of actin filaments, with the two ends generally referred to as the “barbed end” (subdomains Ia and IIa) and the “pointed end” (subdomains Ib and IIb). In the AlfA filament, longitudinal interactions bury about 1,100 Å2 of surface area per protomer, while cross-strand contacts bury about 1,400 Å2 (Fig. 4). Nearly all of the interaction surfaces are within subdomains Ib and IIa, with only very minor contributions from subdomain Ia, which plays a larger role in assembly of other actins (Fig. S7). While the total interface area per protomer is lower for AlfA than other actins, the fraction of its total surface involved in interfaces (20%) is comparable. However, the distribution of interfaces across the surface of AlfA is strikingly different from other actins.
Fig. 4.
Increased AlfA interstrand contacts compensate for missing subdomain IIb interactions. (A) Three protomers from single strands of AlfA and three other actins are shown, with the central protomer colored as in Fig. 1. Residues involved in cross-strand interaction surfaces are colored yellow. (B) Single protomers with residues involved in longitudinal interacting surfaces colored yellow. The size of each interface is given as both absolute area per protomer and as a fraction of the total surface area of the protomer.
The lack of subdomain IIb means that AlfA longitudinal interfaces are less than half the size of the equivalent interfaces in other actins. Longitudinal interactions consist primarily of contacts between a loop (residues 37–45, the equivalent of the D-loop in vertebrate actin) and short α-helix in subdomain Ib (residues 59–65) in one protomer, and loops 162–164 and 179–188 in subdomain IIa in an adjacent protomer. These include primarily hydrogen-bonding interactions, and a small hydrophobic cluster formed by Leu39 and Phe64 on one protomer and Ile180 and Met187 on the adjacent protomer (Movie S2).
To compensate for reduced longitudinal contacts due to the missing subdomain IIb, AlfA has greatly extended cross-strand interfaces, which are 50–100% larger than those of other actins. The cross-strand interface is also mostly a single large patch running across subdomains Ib and IIa, compared with the distributed contacts scattered across all four subdomains in other actin filaments (Fig. 4A). The cross-strand interface is formed primarily between a loop in subdomain Ib (residues 69–82) of one protomer with a helix in subdomain IIa (residues 201–211). The aliphatic chains of lysines 206 and 210 on one side of the interface pack against Phe74 and Met82 on the other side of the interface, with the remainder of the contacts being primarily hydrogen-bonding interactions (Movie S3). The equivalent surfaces in other actin filaments are well separated, but the strong left-handed twist of the AlfA strands positions these regions closer to each other in the filament. Overall, both the lateral and longitudinal interfaces consist of primarily of hydrophobic and hydrogen bonding interactions; the lack of ionic interactions likely explains why AlfA filaments are stable under very high salt concentrations (9).
We verified the AlfA assembly interfaces by generating mutations predicted to disrupt polymerization (Fig. S8). The mutations were designed at an early stage of cryo-EM structure determination, with a preliminary structure at ∼12-Å resolution and a simple homology model of AlfA built by mapping the AlfA sequence onto the structure of R1 ParM. The mutations had varying effects on filament assembly, ranging from no effect to nearly complete inhibition of assembly, as assessed by filament pelleting assays and negative-stain EM (Fig. S8 C and D). Inspection of the mutation sites in the final high-resolution atomic model reveals that the severity of the effect on polymerization correlates with the position of the mutated residue relative to the interface (Fig. S8B).
Consequences of AlfA Filament Architecture for Dynamics and Function.
In other actins the protomers are flattened in the filament relative to their free conformation, with the major change being a rotation between domains I and II around the connect 1 and connect 2 motifs. This conformational change results in large changes to the juxtaposition of subdomains Ib and IIb at the pointed end that would not be relevant in AlfA due to the loss of IIb (Fig. S2). However, more subtle changes occur upon polymerization between Ia and IIa at the barbed end of the protomer. Assuming that AlfA undergoes a structural conversion similar to other actins, these smaller barbed-end changes may be relevant to promoting filament assembly. The detailed nature of polymerization-associated conformational changes in AlfA awaits a high-resolution structure of the unpolymerized AlfA protomer.
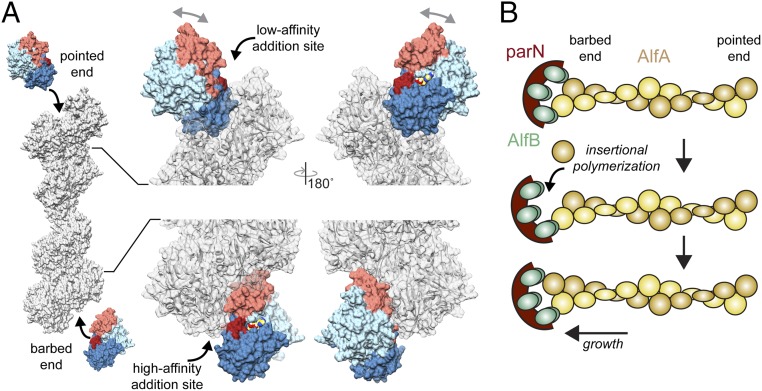
AlfA is unusual in that it exhibits extreme kinetic polarity, elongating almost entirely from one end, although which is the growing end has not been established (25). The structure of the filament provides a likely explanation for the asymmetry of subunit addition (Fig. 5A). In other actin filaments, assembly interactions involve both domains I and II, whether adding at the barbed or pointed end. Interaction of both domains with the end of the filament stabilizes the flattened filament conformation of newly added protomers at either end. Similarly, at the AlfA barbed end the terminal protomer is bound through both subdomains Ib and IIa, which would create a conformationally stable new helical addition site. However, the terminal protomer at the pointed end is bound only via subdomain IIa. Being bound by only a single subdomain would potentially allow the terminal protomer to sample multiple conformational states and create a poorly defined, flexible helical addition site. This suggests that unidirectional growth of AlfA occurs at the barbed end, which would make it similar to actin, which grows more rapidly from the barbed than the pointed end, and distinct from ParM, which grows at indistinguishable rates from both ends (10).
Fig. 5.
Structural differences between protomer addition at barbed and pointed ends. (A) New protomers would add to the pointed end only through interaction with subdomain IIa, which leaves domain I free to rotate relative to domain II (gray arrows). In contrast, addition at the barbed end involves interactions with both subdomains Ib and IIa, stabilizing a filament-bound conformation. Flexibility at the terminal pointed end protomer would create a very low-affinity addition site, while the more rigid conformation of the terminal protomer at the barbed end would create a more defined high-affinity site, potentially explaining the observed unidirectional elongation of AlfA. (B) Model for growth by insertional polymerization at the barbed end of AlfA when bound to the AlfB-parN adaptor–DNA complex.
The adaptor protein AlfB regulates AlfA dynamics, nucleating AlfA filaments from plasmid DNA while simultaneously suppressing spontaneous nucleation. Moreover, after nucleation AlfB processively tracks the growing end of AlfA filaments, providing the basis for plasmid segregation (25). From the structural analysis of AlfA polarity above, which suggests filaments grow from the barbed end, we would also predict that AlfB binds at the barbed end. This would be similar to interaction between ParM and its adaptor ParR, which binds in a cavity between subdomains Ia and IIa that is only fully exposed at the barbed end. A comparable situation may exist for AlfB binding to AlfA, which has a cavity in the region corresponding to the ParR binding site that partially overlaps with a longitudinal assembly interface. A similar mechanism of insertional polymerization has also been proposed for ParM, in which transient dissociation of one of the multivalent adaptor–filamen interactions allows for insertion of a new protomer on one filament strand while the adaptor remains bound to the second strand (Fig. 5B). Interactions between AlfB and the barbed end of AlfA protomers may be the key to the dual role of AlfB in both suppressing (through sequestration of AlfA monomers as free AlfB) and promoting (through AlfA end-binding when attached to the plasmid) AlfA polymerization; understanding the molecular mechanism of this activity will require further biophysical characterization of AlfA–AlfB interactions.
Conclusions
The combination of altered domain architecture and a mechanism of ATP binding give rise to the uniquely stable, highly twisted filament structure and unusual polymer dynamics of AlfA. The lack of a canonical actin subdomain is not unprecedented among bacterial actins, as FtsA, part of the cell division machinery, lacks subdomain Ib (39). However, in FtsA another domain is inserted at the barbed end of domain Ia, which makes contacts that compensate for the lost interaction surfaces (40). On the other hand, AlfA has compensated through altered assembly interfaces, including a more extensive and continuous cross-strand interface. The divergent structure of AlfA highlights the extreme evolutionary plasticity of actin filament quaternary structure. This property has been exploited by bacteria to generate a broad range of actin filaments with unique dynamic and functional properties tuned to a wide variety of specific cellular functions. Given that only a small number of bacterial actins have been structurally and functionally characterized, it is likely that further functionally important variation in filament morphology and dynamics remains to be discovered.
Methods
Sequence Comparison.
AlfA homologs were identified by a BLAST search (41) and multiple sequence alignments were calculated with MAFFT (42).
AlfA Expression Constructs.
Previously described untagged expression constructs using a codon-optimized alfa gene were used to express wild-type (pJKP100) and nonbundling (pJKP102) AlfA (9, 25). AlfA-F12A was generated by site-directed mutagenesis of pJKP100. For nonassembling mutants, the AlfA coding region was cloned into pSMT3-Kan (43), which inserted a His-SMT3/SUMO tag at the N terminus of AlfA. The tag can be cleaved by ULP1 protease, leaving only two residual nonnative residues at the N terminus.
Protein Expression and Purification.
Recombinant AlfA was expressed using previously described codon-optimized expression constructs (9, 25). Assembly mutants were expressed as His-SMT3/SUMO tagged constructs and purified by Ni-NTA and size-exclusion chromatography.
Electron Microscopy and Structure Determination.
For negative-stain imaging, wild-type and mutant AlfA samples were polymerized for 15 min at room temperature with 1 mM nucleotide and 1 mM MgCl2. For cryo-EM, 5 μM AlfA was polymerized in AMPPNP. Cryo-EM data were collected on a Titan Krios microscope (FEI) and recorded on a K2 Summit detector in super resolution mode (Gatan). Motion correction and dose weighting were performed with MotionCor2 (44). Filaments were automatically identified and extracted then subjected to 2D classification and refinement using RELION (45).
An initial structure of the AlfA protomer was generated asymmetrically using RosettaCM (33) and the structure was refined using helical symmetry constraints using the protocol described by Wang et al. (34). The sizes of interacting surfaces between domains in AlfA and other actin filaments were calculated using the PDBePISA server (46).
Supplementary Material
Acknowledgments
We thank Daniel Southworth and the Life Sciences Institute at the University of Michigan for access to their microscopy facility; and Julien Bergergon for helpful discussions. This work was supported by Canadian Institutes for Health Research Grant 298465 (to J.M.K.), Human Frontier Science Program RGY0076 (to J.M.K.), and The Scientific and Technological Research Council of Turkey 2213 Graduate Scholarship Program for Studies Abroad (G.D.U.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6BQW) and the map deposition in the EMDataBank (EMDB ID code: EMD-7134).
See Commentary on page 3205.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715836115/-/DCSupplemental.
References
- 1.Rottner K, Stradal TE. Actin dynamics and turnover in cell motility. Curr Opin Cell Biol. 2011;23:569–578. doi: 10.1016/j.ceb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Heuser JE, Kirschner MW. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J Cell Biol. 1980;86:212–234. doi: 10.1083/jcb.86.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussey PJ, Ketelaar T, Deeks MJ. Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol. 2006;57:109–125. doi: 10.1146/annurev.arplant.57.032905.105206. [DOI] [PubMed] [Google Scholar]
- 4.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 5.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 8.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 11.Draper O, et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82:342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker E, et al. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 2006;25:5919–5931. doi: 10.1038/sj.emboj.7601443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salje J, Zuber B, Löwe J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509–512. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- 17.Popp D, et al. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397:1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 19.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter JC, Funnell BE. Plasmid partition mechanisms. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.PLAS-0023-2014. [DOI] [PubMed] [Google Scholar]
- 21.Sonkaria S, et al. Insight into the assembly properties and functional organisation of the magnetotactic bacterial actin-like homolog, MamK. PLoS One. 2012;7:e34189. doi: 10.1371/journal.pone.0034189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan W, Xie C, Lv J. Screening for the interacting partners of the proteins MamK & MamJ by two-hybrid genomic DNA library of Magnetospirillum magneticum AMB-1. Curr Microbiol. 2012;64:515–523. doi: 10.1007/s00284-012-0099-2. [DOI] [PubMed] [Google Scholar]
- 23.Taoka A, Asada R, Wu LF, Fukumori Y. Polymerization of the actin-like protein MamK, which is associated with magnetosomes. J Bacteriol. 2007;189:8737–8740. doi: 10.1128/JB.00899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell CS, Mullins RD. In vivo visualization of type II plasmid segregation: Bacterial actin filaments pushing plasmids. J Cell Biol. 2007;179:1059–1066. doi: 10.1083/jcb.200708206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polka JK, Kollman JM, Mullins RD. Accessory factors promote AlfA-dependent plasmid segregation by regulating filament nucleation, disassembly, and bundling. Proc Natl Acad Sci USA. 2014;111:2176–2181. doi: 10.1073/pnas.1304127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galkin VE, Orlova A, Rivera C, Mullins RD, Egelman EH. Structural polymorphism of the ParM filament and dynamic instability. Structure. 2009;17:1253–1264. doi: 10.1016/j.str.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bharat TA, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izoré T, Kureisaite-Ciziene D, McLaughlin SH, Löwe J. Crenactin forms actin-like double helical filaments regulated by arcadin-2. eLife. 2016;5:e21600. doi: 10.7554/eLife.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Löwe J, He S, Scheres SH, Savva CG. X-ray and cryo-EM structures of monomeric and filamentous actin-like protein MamK reveal changes associated with polymerization. Proc Natl Acad Sci USA. 2016;113:13396–13401. doi: 10.1073/pnas.1612034113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergeron JR, et al. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. 2017;26:93–102. doi: 10.1002/pro.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozyamak E, Kollman J, Agard DA, Komeili A. The bacterial actin MamK: In vitro assembly behavior and filament architecture. J Biol Chem. 2013;288:4265–4277. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 33.Song Y, et al. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang RY, et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife. 2016;5:e17219. doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Ent F, Amos LA, Löwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 36.Popp D, et al. Structure and filament dynamics of the pSK41 actin-like ParM protein: Implications for plasmid DNA segregation. J Biol Chem. 2010;285:10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S, et al. Novel actin filaments from Bacillus thuringiensis form nanotubules for plasmid DNA segregation. Proc Natl Acad Sci USA. 2016;113:E1200–E1205. doi: 10.1073/pnas.1600129113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roeben A, et al. Crystal structure of an archaeal actin homolog. J Mol Biol. 2006;358:145–156. doi: 10.1016/j.jmb.2006.01.096. [DOI] [PubMed] [Google Scholar]
- 39.van den Ent F, Löwe J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 2000;19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boratyn GM, et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 44.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He S, Scheres SHW. Helical reconstruction in RELION. J Struct Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.