Significance
In this study, we identified the direct interaction region between Drosophila Piwi and Papi. We further determined the crystal structures of Papi-eTud in the apo form, in complex with unmethylated Piwi peptide, and in complex with symmetrically dimethylated Piwi peptide at arginine-10, which demonstrated that Papi interacts with an RGRRR motif on the N terminus of Piwi in a sequence-specific manner both in vitro and in vivo. This recognition sequence, which determines the specificity of Papi–Piwi interactions, is different from all previously reported (G/A)R repeats. Our studies provide mechanistic insights into the important role of Papi–Piwi interactions in the 3′ end-trimming process of PIWI-interacting RNA biogenesis and facilitate the identification of new PIWI-interacting partners of Tudor domain-containing proteins.
Keywords: Papi, Piwi, eTud domain, piRNA biogenesis, 3′-end trimming
Abstract
The Tudor domain-containing (Tdrd) family proteins play a critical role in transposon silencing in animal gonads by recognizing the symmetrically dimethylated arginine (sDMA) on the (G/A)R motif of the N-terminal of PIWI family proteins via the eTud domains. Papi, also known as “Tdrd2,” is involved in Zucchini-mediated PIWI-interacting RNA (piRNA) 3′-end maturation. Intriguingly, a recent study showed that, in papi mutant flies, only Piwi-bound piRNAs increased in length, and not Ago3-bound or Aub-bound piRNAs. However, the molecular and structural basis of the Papi–Piwi complex is still not fully understood, which limits mechanistic understanding of the function of Papi in piRNA biogenesis. In the present study, we determined the crystal structures of Papi-eTud in the apo form and in complex with a peptide containing unmethylated or dimethylated R10 residues. Structural and biochemical analysis showed that the Papi interaction region on the Drosophila Piwi contains an RGRRR motif (R7–R11) distinct from the consensus (G/A)R motif recognized by canonical eTud. Mass spectrometry results indicated that Piwi is the major binding partner of Papi in vivo. The papi mutant flies suffered from both fertility and transposon-silencing defects, supporting the important role conferred to Papi in piRNA 3′ processing through direct interaction with Piwi proteins.
The Tudor domain was first identified in the Drosophila Tudor (Tud) protein, which plays an important role in germ cell formation during oogenesis (1). The core of the Tudor domain adopts an oligonucleotide/oligosaccharide-binding (OB) fold, which is a β-barrel formed by four anti-parallel β-strands (2). A conserved aromatic cage is usually located on the surface to accommodate the methylated ligands. According to the bound ligands, the Tudor domains are classified into two major groups: a methyllysine-binding group and an arginine-binding group (2, 3). Tudor domains that recognize methyllysine usually act as histone readers in chromatin biology (4). However, Tudor domains that bind methylated arginine are usually involved in RNA processing such as splicing regulation and piRNA (PIWI-interacting RNA)-mediated biogenesis (5). For example, the Tudor domain of spinal muscular atrophy disease protein SMN could bind symmetrically dimethylated arginine (sDMA) residues of spliceosome component Sm proteins (6). Moreover, in the piRNA pathway, Tudor domains hybridized with the staphylococcal nuclease (SN) domain, designated as “extended-Tudor” (eTud) or “Tudor-sn” domains, were reported to regulate transposon silencing in the germ cells through recognizing the N terminus of PIWI (P-element–induced wimpy testis) proteins with sDMA modification (7, 8).
PIWI proteins are a clade of evolutionarily conserved Argonaute family proteins usually found in animal gonads (9, 10). PIWI proteins can interact with piRNAs to form a piRNA-induced silencing complex (piRISC complex) to silence transposons (11–13). There are three PIWI proteins in Drosophila melanogaster, namely Piwi, Aubergine (Aub), and Ago3 (13–15). In Drosophila ovaries, the precursor piRNAs (prepiRNAs) that are transcribed from piRNA clusters are exported to the nuage in the cytoplasm, where the prepiRNAs are cleaved into piRNA intermediates by a mitochondrial outer membrane protein Zucchini (Zuc), followed by 3′-end trimming and methylation to yield mature piRNAs (12, 16–20). Subsequently, the mature piRNAs are loaded into PIWI proteins to form various piRISCs. Piwi-piRISCs enter the nucleus to silence the transposons. Alternatively, mature piRNAs can also be loaded into Aub to trigger the ping-pong cycle that cleaves both the transposon mRNAs and the piRNA transcripts (21, 22).
The Tudor domain-containing (Tdrd) proteins, which are conserved among flies, worms, and mammals, play important roles in PIWI localization and function through recognizing the sDMA-modified PIWI proteins by eTud domains (2, 15, 23). Silkworm Qin and Spn-E, also named “Tdrd4” and “Tdrd9,” take part in this ping-pong pattern, facilitating the loading of piRNAs to Siwi (the silkworm Aub) (24, 25). Mutations in Drosophila Qin cause homotypic Aub:Aub interactions instead of the normal Aub:Ago heterotypic ping-pong interactions (26). Tejas (Tej), also called “Tdrd5,” interacts with the RNA helicase Vasa to regulate the localization of some piRNA effectors, such as Spn-E, Aub, Ago, Krimper, and Maelstrom, to nuage and engages in the formation of nuage for piRNA production (27). Recent studies have shown that silkworm Papi (partner of PIWIs/Tdrd2, also known as “BmPapi”) recruits PNLDC1, a PARN family 3′–5′ exoribonuclease, to trim the 3′ end of the piRNA intermediates produced by Zuc cleavage for piRNA maturation (28, 29). Moreover, depletion of TDRKH (the mouse Papi homolog) results in an obvious 3′-end extension of piRNAs and male sterility (28, 30). Similarly, in Drosophila, Papi may be involved in primary piRNA production. Zuc-mediated cleavage of prepiRNA directly generates mature piRNA, while prepiRNAs released from the ping-pong cycle require further resection by Nibbler, which belongs to another 3′–5′ exoribonuclease family (31, 32). The depletion of Papi increases the length of Piwi-bound piRNA somewhat, leaving both Aub-bound and Ago3-bound piRNAs unaffected (31). However, the molecular mechanism underlying the function of the Papi–Piwi complex remains unclear. Here, we identified the region of Piwi that interacts with the eTud of Papi. We then determined the crystal structures of Papi-eTud both in the apo form and in complex with unmethylated Piwi (Piwi-unme) or symmetrically dimethylated Piwi at arginine-10 (Piwi-R10me2s). Our structural and biochemical data showed that, unlike the consensus eTud recognition (G/A)R motif, Papi recognizes the RGRRR motif of Piwi in a sequence-specific manner both in vitro and in vivo. Moreover, deletion of papi results in fly fertility defects. In vivo fly rescue experiments further establish that the binding pocket of Papi is required for its function in fertility and transposon silencing, in accordance with the structural and biochemical analysis. Our findings provide important insight into the role of the interactions between Papi and Piwi in piRNA maturation, supporting the recent discovery that after Zuc cleavage Papi-assisted piRNA trimming is consistent with its role in silkworms and mice (28, 30).
Results
The eTud Domain of Papi Preferentially Binds to the N Terminus of Piwi.
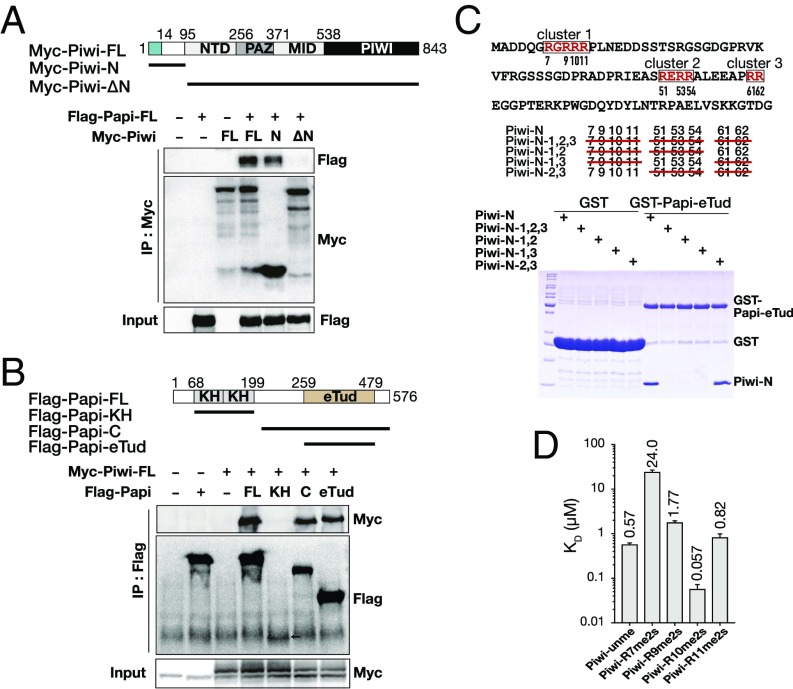
The interaction between the Piwi N terminus (residues 1–491) and Papi was revealed by yeast two-hybrid assay in a previous report (33). However, the molecular basis of the interactions between Papi and Piwi has not yet been characterized in Drosophila. Therefore, we sought to characterize the binding affinity and specificity of Piwi by Papi. First, we applied coimmunoprecipitation (co-IP) assays to determine the key regions in each protein for interaction. According to previously reported Siwi structure and secondary structure prediction of Piwi (34), we generated three N-terminal Myc-tagged Piwi constructs and four C-terminal Flag-tagged Papi constructs, i.e., Myc-Piwi-FL (full length; residues 1–843), Myc-Piwi-N (N terminus; residues 1–94), Myc-Piwi-ΔN (N-terminal deletion; residues 95–843), Papi-FL-Flag (residues 1–576), Papi-KH-Flag (the tandem K homology domain of Papi; residues 64–210), Papi-C-Flag (C terminal; residues 211–576), and Papi-eTud-Flag (residues 259–479) (Fig. 1 A and B). Co-IP results showed the eTud domain of Papi could form complexes with Piwi-FL and Piwi-N, but not with Piwi-ΔN, indicating that the 94 N-terminal amino acids of Piwi are essential and sufficient for binding to Papi.
Fig. 1.
Papi binds to the 14 N-terminal amino acids of Piwi via its eTud domain. (A and B) Co-IP assays to map the interaction regions between Papi and Piwi in S2 cells. Domain architectures of Papi and Piwi are schematically presented. (A) Co-IP of Myc-tagged wild-type Piwi or truncations with Flag-Papi. (B) Co-IP of Flag-tagged wild-type Papi or truncations with Myc-Piwi. (C, Upper) The sequence of 94 N-terminal amino acids of Piwi. R-rich clusters are shown in red boxes. (Middle) Red lines strike out Rs replaced by As. (Bottom) The wild type and Piwi-N mutants were used in GST pull-down assays to check the interactions with GST-Papi-eTud or mock control GST. (D) Bar graph of the ITC results determining the Kd of the interaction between Papi-eTud and indicated Piwi peptides.
Previous studies all showed that the eTud of Tdrd proteins prefers to bind sDMA in (G/A)R repeats (2, 5). Three arginine-rich clusters were found when we searched the GR motif in the N terminus of Piwi (Fig. 1C). To determine which cluster binds directly to Papi-eTud, we performed a GST pull-down assay using the purified Papi-eTud domain. Piwi-N (residues 1–94) with an N-terminal His-tag and a C-terminal Strep tag was expressed and purified from Escherichia coli. To identify important arginine residues in Piwi-N protein, mutants were generated to replace arginine with lysine in each cluster (Fig. 1C). GST-Papi-eTud could interact directly with wild-type Piwi-N. A mutation at cluster 2 or 3 did not affect GST-Papi-eTud interactions. In contrast, the mutation in cluster 1 abolished the binding.
There are four arginine residues in cluster 1 (RGRRR, residues 7–11), namely R7, R9, R10, and R11. We performed isothermal titration calorimetry (ITC) assays to measure the dissociation constant (Kd) of Papi-eTud against the Piwi peptides (residues 4–14), which are symmetrically dimethylated on different arginine residues (Fig. 1D). Papi-eTud binds Piwi-unme peptides at a high affinity, with a Kd of 0.57 μM. However, methylation at R7, R9, or R11 reduces the binding affinity to 42-fold, threefold, and 1.4-fold, respectively. Nevertheless, methylation at R10 in Piwi-R10me2s increased the binding affinity∼10-fold compared with the unmethylated peptide.
Thus, co-IP, GST pull-down, and mutagenesis assays suggest that the eTud domain of Papi specifically recognizes the N-terminal region (residues 4–14) of Piwi. Moreover, methylation on R10 significantly enhances the binding, whereas methylation at other positions impairs the binding.
Overall Structure of the Papi-eTud–Piwi-R10me2s Complex.
To gain further insights into the recognition of Piwi by Papi, we first solved the crystal structure of Papi-eTud in the apo form (Fig. S2A and Table S1). The crystal that belongs to the space group P6522 was solved by molecular replacement using the Tudor core structure of human Tdrd2 [the human Papi homolog, Protein Data Bank (PDB) ID code 3FDR] as a search model. The remaining amino acid sequence of Papi-eTud was traced after two cycles of initial refinement (Table S1). However, our initial attempts at crystallizing Papi-eTud and Piwi complex failed. By analyzing the structure, we mutated D287 to alanine to reduce the potential surface entropy for crystallization. The Kd of D287A to Piwi-unme and Piwi-R10me2s was determined to be 0.62 μM and 0.054 μM, respectively, indicating that the mutation at D287 had no impact on the binding (Fig. S2B). The crystal structure of the Papi-eTud-D287A mutant, which belongs to the space group P41, was also determined (Fig. S2C and Table S1). The rmsd between these two structures was 0.66 Å, indicating that the D287A mutant induces only slight conformational changes (Fig. S2D). Papi-eTud-D287A was successfully cocrystallized with Piwi N-terminal peptides. Thus, for convenience, we also called the D287A mutant “Papi-eTud.”
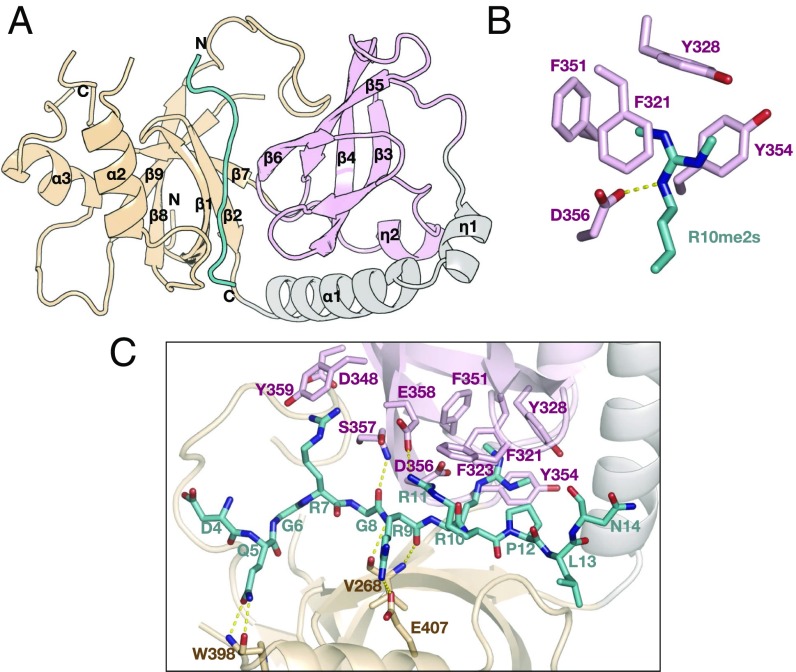
The structure of the Papi-eTud and Piwi-R10me2s complex was determined at 1.55 Å. Papi-eTud is a hybrid domain with a Tudor core (β3–β6) and an SN domain bridged by an α-helix (α1) (Fig. 2A). The Papi-eTud apo and complex forms were superimposed quite well, with an rmsd of 0.36 Å (Fig. S2E). One Piwi-R10me2s peptide bound to the concave surface of Papi-eTud with a buried surface area of 820.0 Å2 (Figs. S1A and S2F). Both the Tudor and the SN domains contributed to the binding (Fig. 2B and Fig. S1A). The interactions between Papi and Piwi were mostly governed by hydrogen bonds and cation-π interactions (Fig. 2B). Symmetrically dimethylated R10 stretches into an aromatic cage surrounded by F321, Y328, F351, and Y354 (Fig. 2C). F321 and Y354 are located in parallel on each side of the guanidino group of R10me2s and are stabilized by π-cation-π sandwich interactions. Y328 and F351 lie perpendicularly on the front and left sides, respectively, of the symmetrical dimethyl group as two walls of the aromatic cage. The two methyl groups on the R10 face toward Y328 and F351 (Fig. 2C). D356 further neutralizes the positive charge of R10me2s. In addition to R10, three more arginine residues, R7, R9, and R11, form salt bridges with D348, E407, and E358, respectively. Y359 and F323 further stabilize R9 and R11 via cation-π interactions. Moreover, the main chain of G8 and R9 contacts S357 and V268 via hydrogen bonds, and Q5 forms hydrogen bonds with W398. Electrostatic potential analysis showed that the Piwi-bound surface of Papi-eTud was negatively charged to facilitate the accommodation of the positively charged Piwi peptide (Fig. S1B).
Fig. 2.
Structural basis of Piwi recognition by Papi-eTud. (A) Overall structure (1.55 Å) of Papi-eTud complexed with Piwi-R10me2s peptide (residues 4–14). Color codes: Tudor core, pink; SN domain, wheat; and Piwi-R10me2s peptide, teal. (B) Details of intermolecular contacts between Papi-eTud and Piwi-R10me2s. (C) Details of the R10me2s recognized by the aromatic cage of Papi-eTud.
Papi-eTud Specifically Recognizes the RGRRR Motif of Piwi.
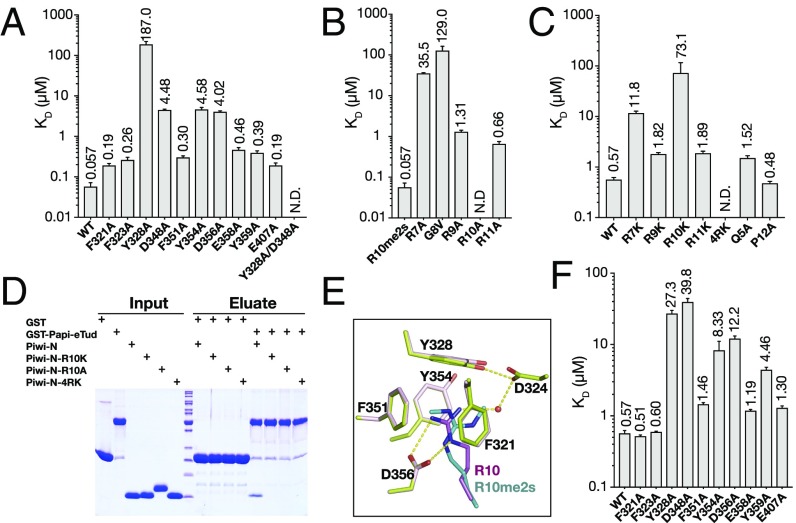
Next, we tried to validate the intermolecular contacts in the Papi–Piwi complex using site-specific mutagenesis. The replacements of key amino acids by alanine in Papi-eTud largely abolished or decreased binding as determined by ITC assays (Fig. 3A). The mutations F321A, Y328A, F351A, and Y354A at the aromatic cage, which bound the symmetrically dimethyl group of R10me2s, reduced the binding about threefold, 3,280-fold, fivefold, and 80-fold, respectively. D356A, which interacts with R10me2s via electrostatic interactions, lowered the binding capacity about 70-fold. Alanine substitution of D348 and Y359 that interact with R7 decreased the binding about 80-fold and sevenfold, respectively. The combined Y328A and D348A mutant showed no binding at all. However, the mutations in residues that interact with R9 and R11 (E407A, F323A, and E358A) reduced the binding about threefold, fivefold, and eightfold, respectively. This indicates that R9 and R11 may be less important than R7 and R10 in the sequence-specific recognition.
Fig. 3.
Structure-guided mutations decrease Papi binding to Piwi in vitro. (A–C and F) Quantification of the Kds between structure-guided mutations in Papi-eTud and Piwi peptides by ITC. (A) Mutations of key residues in Papi-eTud reduce the binding to Piwi-R10me2s dramatically. (B) Mutations of R to A in Piwi-R10me2s peptides reduce the binding to Papi-eTud. (C) Mutations of R to K in Piwi-unme peptides reduce the binding to Papi-eTud. (D) Pull-down assay of wild-type and mutant Piwi-N by GST-Papi-eTud. (F) Mutations of key residues in Papi-eTud reduce the binding to Piwi-unme. (E) Superimposition of the aromatic cages from Papi–Piwi-unme and Papi–Piwi-R10me2s.
Mutations of the arginine residues on Piwi were also studied (Fig. 3B). R7A, R9A, and R11A significantly weakened the binding about 623-fold, 23-fold, and 12-fold, respectively. However, no binding could be detected for R10A. Notably, the mutation of G8 to valine decreased the binding ability about 2,300-fold, indicating that the bulk side chain at this position may cause steric hindrance for the adaptation of the Piwi peptide by Papi-eTud. To further explore whether the interactions between Papi-eTud and Piwi are solely electrostatic forces, replacements of arginine residues with lysine were also tested (Fig. 3C). In agreement with the alanine substitutions, R7K reduced the binding ability more dramatically than R9K and R11K. Moreover, R10K decreased the binding about 128-fold, consistent with the binding data of Papi-eTud mutants, showing that R10, in addition to R7, is a key residue for Papi recognition. The quadruple mutant (Piwi-4RK) that replaces all four arginine residues with lysine disrupts the binding completely. Moreover, Q5A reduces the binding by approximately threefold, and P12A has a negligible effect on the binding. These results show that the interactions between Papi and Piwi are sequence specific. The critical role of R10 in the recognition by Papi was verified by GST pull-down assays (Fig. 3D). Only wild-type Piwi-N could interact with GST-Papi-eTud. Mutations in either R10K or R10A disrupt the binding, indicating the indispensable role of R10 in the absence or presence of sDMA modification.
We determined the crystal structure of Papi-eTud in complex with the unmethylated Piwi N-terminal peptide as well (Fig. S3A and Table S1). The unmethylated Piwi peptide bound on the same concave surface as the Piwi-R10me2s peptide. The rmsd between these two structures was 0.15 Å (Fig. S3B). The binding details for Piwi-unme were similar to Piwi-R10me2s, except that the side chain of R11, instead of R9, formed salt bridges with E407 (Fig. S3C). Unmethylated R10 still inserts into the aromatic cage (Fig. 3E). However, no hydrophobic interactions between the two methyl groups in R10me2s and residues Y328 and F351 were observed. Instead, the guanidinium moiety of R10 was found to interact with D356 and D324 via electrostatic interactions and water-mediated hydrogen bonds, respectively.
Papi-eTud aromatic cage mutations also show significant impacts on the binding to Piwi-unme peptide, although the reduced fold change of binding affinities was not as dramatic as the corresponding fold change against Piwi-R10me2s (Fig. 3F). F321, which lies over the guanidinium moiety of R10, changes only slightly upon alanine substitution, indicating that F321 may not be involved in the recognition of unmethylated R10. The mutation in D356A also reduces the binding about 20-fold, showing its important role in the recognition of R10. Other mutations, including D348, E358A, Y359A, and E407A, all resulted in a decline in binding to Piwi-unme, showing effects similar to those of the binding to methylated Piwi peptide.
Thus, the above results suggest that Papi-eTud recognizes the N-terminal RGRRR motif (R7–R11) of Piwi in a sequence-dependent manner. Every arginine residue contributes to the binding, especially R7 and R10. sDMA modification at R10 enhances the binding, whereas methylation at the consensus (G/A)R motif, such as R7me2s and R9me2s, decreases the binding, suggesting a distinct mode of recognition of Papi-eTud.
A Protruding Loop May Prevent the Binding of Papi to the (G/A)R Motif.
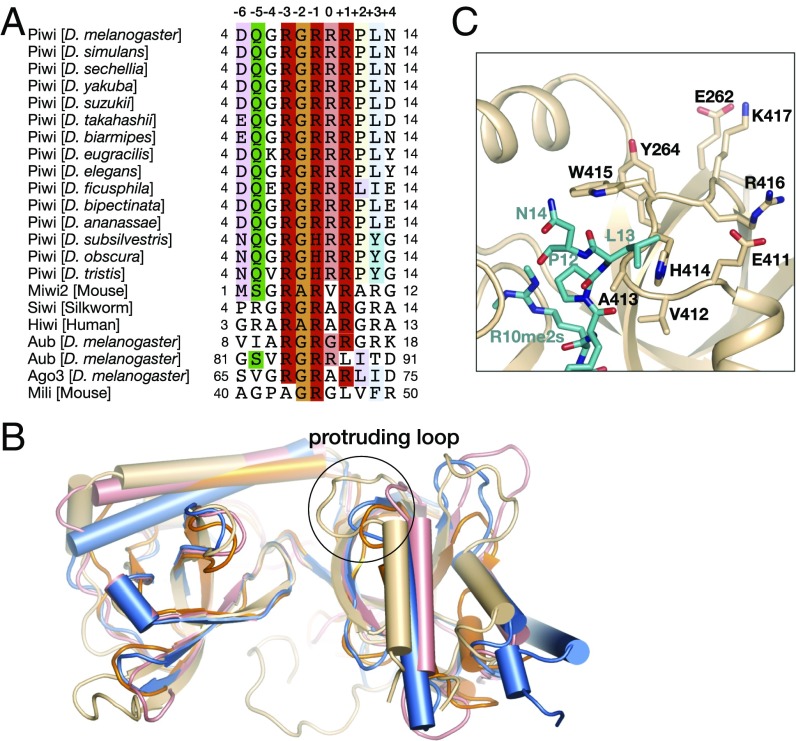
To date, several structures of PIWI proteins recognized by the eTud domains of Tdrd family proteins have been reported, all of which bind to the consensus (G/A)R repeats in the N terminus of PIWI proteins (35–37). Although the overall structures of eTud domains are quite similar, the orientation of the bound PIWI peptides may vary. Interestingly, in either structure the methylated arginine or unmodified arginine sidechain is stretched into the aromatic cage from a different direction (Fig. S4). Residues involved in the aromatic cage formation are highly conserved among Tdrd family proteins (Fig. S5). However, the residues that recognize the neighboring sequence are unique in Drosophila Papi proteins. For example, D348 and Y359, which specifically recognize R7, are not conserved in the other eTud domains. Moreover, D356, which plays important roles in interacting with R10 or R10me2s, exists only in the Drosophila Papi and its homologs but not in other eTud domains.
Conversely, the RGRRR motif of Piwi recognized by Papi-eTud is quite different from the previously reported (G/A)R motif (Fig. 4A) (2, 5). In the consensus sequence, if we designate the dimethylated arginine as position 0, alanine or glycine is usually found at positions −1 and +1, and arginine residues occupy positions −2 and +2. However, our structural and biochemical data showed that the corresponding residues at positions −1 and +1 are R9 and R11, respectively, and glycine is at position −2.
Fig. 4.
Interactions between Papi and the RGRRR motif of Piwi are conserved among Drosophila species. (A) Sequence alignment of Piwi among Drosophila species. (B) Superimposition of the eTud domain structures of Papi, Tud, Tdrd1, and SND1. The protruding loop is circled. Color coding: Papi-eTud, wheat; Tud-eTud, orange; Tdrd1-11th eTud, pink; SND1-eTud, blue. The PDB ID code of each structure is given in Supporting Information. (C) Close-up view of the protruding loop circled in B.
To determine why Papi prefers not to bind to the (G/A)R motif, we compared our structure with previously reported eTud-PIWI complex structures (35–37). Superimposition of these structures revealed that a loop connecting α2 and β8 (residues 411–417) protruded toward the Piwi peptide (Fig. 4B). This protruding loop is stabilized by E411 and R416, which form two salt bridges with R416 and E262, respectively. Moreover, V412, A413, and H414 interacted with P12 and L13 via hydrophobic interactions. Therefore, W415 stretched out and lay over the aromatic cage to which the Rme2s bind, thus blocking the entrance of the aromatic cage from the top (Fig. 4C).
Papi Interacts Specifically with Piwi in Vivo.
Next, we explored the binding of Piwi to Papi-eTud in S2 cells (Fig. 5A). We generated three Flag-tag Papi mutants, Y328A, D348A, and a double mutant Y328A/D348A. Y328 and D348 are involved in the interaction with R10 and R7, respectively. These mutants were cotransfected with Myc-tagged Piwi in S2 cells, and the abilities of these Papi mutants to bind to Piwi were assessed by co-IP. The results showed a reduced level of Piwi introduced by either Y328A or D348A mutation. As expected, a significantly lower level of Piwi was observed in the co-IP assay with the double mutant.
Fig. 5.
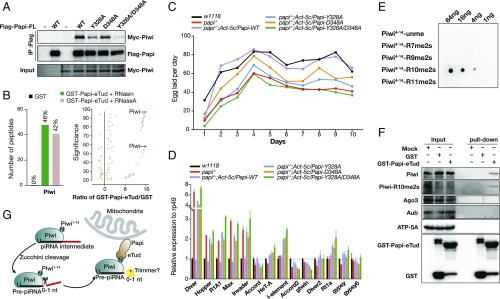
Papi specifically interacts with Piwi in vivo. (A) Co-IP of Flag-tagged wild-type or mutant Papi with Myc-Piwi in S2 cells. (B) MS analysis for Papi-eTud–associated proteins in the fly ovary. (C) Fertility test (number of eggs laid) in w1118, papi−/− and papi mutant rescue flies. (D) Transposon expression levels of ovaries from w1118, papi−/−, and papi mutant rescue flies were quantified by RT-PCR and normalized to rp49. (E) Dot blot assay showing the selectivity of anti–Piwi-R10me2s antibody against various Piwi peptides. (F) Papi-eTud is specific associated with Piwi in Drosophila ovaries, and Piwi-R10me2s exists in fly ovaries. (G) Model of the role of Papi in the piRNA 3′-end trimming downstream of Zuc via direct interaction with the Piwi N terminus by the Papi-eTud domain.
We also used GST-tagged Papi-eTud to pull down the interacting proteins from w1118 fly ovary lysate with or without RNaseA treatment and assessed the result by MS (Fig. 5B). Piwi is identified as the most significant protein in the presence or absence of RNaseA treatment compared with the GST control. Identified Piwi peptides cover 46% and 42% of the Piwi sequence. However, no Aub or Ago3 peptides were detected in the top 200 hits. Therefore, in the Drosophila ovary, Papi-eTud interacts specifically with Piwi independent of RNA.
Effect of piRNA 3′-End Trimming and Fertility Defect upon Loss of papi.
To determine Papi’s function in vivo, we generated papi-null mutants using CRISPR/Cas9 with two different single-guide RNAs (sgRNAs) (Fig. S7 A–E). Loss of Papi leads to an ∼40% decline in female fertility (eggs laid by 15 females in 10 d: 10,319 by wild-type controls versus 6,220 by papi−/− mutants) (Fig. 5C). We then examined the transposon expression in papi-deletion ovaries using quantitative RT-PCR (Fig. 5D). Here, 13 transposons were selected, and five of these 13 transposons were found to be mildly up-regulated more than twofold over the wild-type level. These were Diver, Hopper, R1A1, Max, and Invader. Transgenic expression of wild-type Papi successfully rescued both the fertility and transposon activation in papi−/− flies, but that of Papi mutants, including Y328A, D348A and Y328A/D348A, did not (Fig. 5 C and D and Fig. S7F). This indicates that Piwi-R10 binding is required for Papi’s function in vivo.
To further investigate the role of Papi in piRNA 3′-end processing, we reanalyzed the data using the method previously reported (31). The average length of Piwi-bound piRNA in papi mutant flies is extended by about 0.45 nt (Fig. S8C), consistent with the previously reported value (31). No obvious change was observed for the piRNA production in papi−/− flies (Fig. S8 A and B). However, piRNAs derived from the 13 selected transposons all showed an increase in length (Fig. S8D). Thus, although the loss of papi did not affect the piRNA production and changed the length of the Piwi-bound piRNAs only slightly, transposon expressions were mildly up-regulated, and fertility was partially compromised.
Presence of Piwi-R10me2s in Fly Ovary.
R10 is located in the middle of three continuous arginine residues (R9–R11). Symmetrical dimethylation on the second arginine of three continuous arginine residues has not yet been reported in PIWI proteins. Therefore, we expressed a Piwi-N (residues 1–94) with an N-terminal His-tag and a C-terminal Strep-tag as well as full-length Piwi in Sf9 cells (Fig. S6). sDMA modification on R10 was detected on the both Piwi-N and full-length Piwi, indicating that R10 could be dimethylated in insect cells.
To identify the sDMA modification status on R10, we use GST-Papi-eTud to enrich Piwi protein from w1118 fly ovary lysate. However, MS results failed to identify sDMA modification of R10, which may be due to the low abundance of modified Piwi among endogenous Piwi. Next, we generated an antibody against Piwi-R10me2s (residues 4–15) and assessed its specificity by dot blot assay (Fig. 5E). The anti–Piwi-R10me2s antibody distinguished Piwi-R10me2s from unmethylated Piwi peptide as well as Piwi-R7me2s, Piwi-R9me2s, and Piwi-R11me2s. We subsequently detected the R10me2s signal by Western blot using anti–Piwi-R10me2s antibody. Both Piwi and Piwi-R10me2s were detected in the eluate pulled down by GST-Papi-eTud, but Ago3 or Aub were not (Fig. 5F). Therefore, there is dimethylation on R10 of Piwi in vivo, albeit in low abundance. In addition, pull-down results support the binding specificity of Piwi rather than Aub or Ago3 by Papi.
Discussion
In the present study, we identified the interaction region between Papi and Piwi and determined the crystal structure of the Papi-eTud domain both in the apo form and in complex with the symmetrically dimethylated Piwi or unmethylated Piwi. Structural analysis revealed that the Papi-eTud domain recognizes the RGRRR motif of Piwi in a sequence-dependent manner, which is significantly different from the consensus eTud domain-binding motif. We further showed that Papi specifically interacts with Piwi both in vitro and in vivo. We found that deletion of papi results in a subset of transposon activation and fly fertility defects. Our findings reveal an unexpected binding mode for Papi-eTud to the N terminus of Piwi, which broadens the understanding of the binding specificity of eTud domains. The structural information is very likely extendable to other species and should be of general interest in identifying new partners of Tdrd family proteins and PIWI proteins in species other than flies.
sDMA-dependent protein interactions in the biogenesis and function of piRNAs have been studied for a long time (2). However, the structural details of the sDMA site on Piwi have not yet been elucidated. We identified an sDMA modification on R10 of Piwi and provided a mechanistic insight for the molecular interaction between Papi and Piwi-R10me2s. Papi binds Piwi tightly with or without dimethylation on R10, although there is a 10-fold difference between the binding affinities (0.057 μM for Piwi-R10me2s and 0.57 μM for Piwi-unme). Both Piwi and Piwi-R10me2s could be enriched by Papi from fly ovary lysate. The exact function of the R10 sDMA modification in Piwi remains unclear and awaits further investigation. Moreover, we deliberate on the presence of other nuclear Tudor domain proteins that may recognize the Piwi-R10me2s marker.
Previous studies have reported sDMA sites on other PIWI proteins (2). In Drosophila, Aub has been reported to be dimethylated on R11, R13, and R15 within the sequence ARGRGRGR (residues A10–R17) (5, 35, 38). Three sDMA sites, R4, R68, and R70, were identified for Ago3 (5, 38). Moreover, the sDMA sites have also been reported in the mouse PIWI proteins Mili and Miwi (5, 39, 40). The common feature of these modified arginine residues is that they are within the (G/A)R repeats (41). Recently, the structure of Tdrd2 in complex with Hiwi was reported. Tdrd2 is the homolog of Papi in human with an incomplete aromatic cage that binds the unmethylated (G/A)R motif of Hiwi (42). Superimposing the Papi–Piwi-R10me2s, Papi–Piwi-R10unme, and Tdrd2–Hiwi structures shows that the binding modes of the RGRRR motif in Piwi and the (G/A)R motif in Hiwi are different (Fig. S9 A and B). Four (G/A)R repeats of Hiwi (spanning residues 3–10) bind to the concave surface of Tdrd2 in an “arch” shape (Fig. S9 B and C), while Piwi binds to the corresponding surface of Papi with one residue less than Hiwi (Fig. S9 B and D). Moreover, we tested the binding affinities of Papi to Aub and Ago3, including Ago366–76-unme (VGRGRARLIDT, residues 66–76), Aub9–18-unme (IARGRGRGRK, residues 9–18), and Aub81–90-unme (GSVRGRRLIT, residues 81–90) (Fig. S9E). No binding was detected for Ago366–76 series peptides, whether they were methylated on arginine or not (Fig. S9F). Papi binds to Aub peptides with significantly reduced binding affinity compared with Piwi-unme (Fig. S9F). Therefore Papi in different species may have different sequence specificity to its binding partner and undergo different binding modes.
Although the recognition motifs are apparently different in mouse and silkworm PIWI proteins (28, 30), the interactions between Papi and Piwi are conserved. We expressed and purified the eTud domains of mTDRKH/Tdrd2 and BmPapi, the mouse and silkworm homologs of Papi, respectively (Fig. S10A), and examined their binding affinities to Miwi22–11-unme (residues 2–11) and Siwi5–17-unme (residues 5–17) peptides. mTDRKH and BmPapi were shown to recognize the (G/A)R motif in Miwi2 and Siwi peptides, respectively (Fig. S10 B and C). These results concur with the previous report that the role of Papi in assisting the 3′-end trimming of piRNAs is conserved in fly, mouse, and silkworm (28–30, 39, 40).
In silkworms, PNLDC1 couples with BmPapi to trim the 3′ end of prepiRNAs (29). However, PNLDC1 does not exist in Drosophila (29, 31, 43). Previously, Hayashi et al. (31) reported that there are two pathways in Drosophila for the 3′-end processing of prepiRNAs (29). In the Zuc-mediated phasing pathway, most prepiRNAs are directly cleaved to produce mature piRNAs (18, 19, 31). In the other pathway, the piRNA intermediates generated by the Aub:Ago3 ping-pong require Nibbler to trim the 3′ end (31, 32). Hayashi et al. (31) showed that Papi and Zuc are colocalized on mitochondrial outer membrane (28). The depletion of papi only affects the length of Piwi-bound piRNAs, not that of Ago3-bound or Aub-bound piRNAs (31). Our data have shown that Papi can interact specifically with Piwi in Drosophila ovaries, consistent with the fact that Papi is involved in the Zuc-mediated phasing pathway (Fig. 5 B and F). Overall, our study provides one possible molecular mechanism by which Papi recruits Piwi to the piRNA biogenesis machinery (Fig. 5G).
Materials and Methods
Crystals were obtained by the sitting-drop vapor-diffusion method at 16 °C. Protein expression, purification, crystallization, structure determination, biochemical assays, and fly experiments are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the fly facility of the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, for generating the Papi-KO fly; Prof. Lei Zhang (Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) for providing the pUAST-Flag and pUAST-Myc plasmids; Prof. Laixin Xia (School of Basic Medical Science, Southern Medical University) for providing the pUASp-attb plasmid; the Shanghai Scientific Research Center for instrumental support and technical assistance; and the staff of the BL19U1 and BL17U1 beamlines at the Shanghai Synchrotron Radiation Facility for assistance with data collection. This work was supported by National Natural Science Foundation of China Grants 91640102 (to Y.H.) and 31230041 (to J.M.), Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08010202 (to Y.H.), Chinese Academy of Sciences Facility-based Open Research Program, and the State Key Laboratory of Molecular Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Structural coordinates have been deposited in the Protein Data Bank (PDB) database [PDB ID codes 5YGC (Papi-eTud apo), 5YGB (Papi-eTud-D287A), 5YGD (Papi-eTud-D287A–Piwi-R10me2s complex), and 5YGF (Papi-eTud-D287A–Piwi-unme complex)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717116115/-/DCSupplemental.
References
- 1.Boswell RE, Mahowald AP. Tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 3.Maurer-Stroh S, et al. The tudor domain ‘royal family’: Tudor, plant agenet, chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 4.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siomi MC, Mannen T, Siomi H. How does the royal family of tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One. 2012;7:e30375. doi: 10.1371/journal.pone.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripsianes K, et al. Structural basis for dimethylarginine recognition by the tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18:1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 9.Weick EM, Miska EA. piRNAs: From biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 10.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki YW, Siomi MC, Siomi H. PIWI-interacting RNA: Its biogenesis and functions. Annu Rev Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- 14.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 15.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 16.Andersen PR, Tirian L, Vunjak M, Brennecke J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature. 2017;549:54–59. doi: 10.1038/nature23482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohn F, Handler D, Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimasu H, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 21.Czech B, Hannon GJ. One loop to rule them all: The ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster A, et al. Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by Krimper. Mol Cell. 2015;59:564–575. doi: 10.1016/j.molcel.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development. 2012;139:2255–2266. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- 24.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Ke A. PIWI takes a giant step. Cell. 2016;167:310–312. doi: 10.1016/j.cell.2016.09.043. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. Heterotypic piRNA ping-pong requires qin, a protein with both E3 ligase and tudor domains. Mol Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the tudor domain protein Tejas. Curr Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 28.Honda S, et al. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA. 2013;19:1405–1418. doi: 10.1261/rna.040428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi N, et al. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell. 2016;164:962–973. doi: 10.1016/j.cell.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi R, et al. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature. 2016;539:588–592. doi: 10.1038/nature20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3′ ends in Drosophila. Development. 2016;143:530–539. doi: 10.1242/dev.128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Qi H, Wang J, Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto N, et al. Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell. 2016;167:484–497.e9. doi: 10.1016/j.cell.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, et al. Structural basis for methylarginine-dependent recognition of Aubergine by tudor. Genes Dev. 2010;24:1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K, et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended tudor domain. Proc Natl Acad Sci USA. 2010;107:18398–18403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathioudakis N, et al. The multiple tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA. 2012;18:2056–2072. doi: 10.1261/rna.034181.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishida KM, et al. Functional involvement of tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirino Y, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, et al. Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2. Proc Natl Acad Sci USA. 2017;114:12483–12488. doi: 10.1073/pnas.1711486114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang W, Tu S, Lee HC, Weng Z, Mello CC. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell. 2016;164:974–984. doi: 10.1016/j.cell.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.