Significance
Imaging provides an insight into biological patho-mechanisms of diseases. However, the link between the imaging phenotype and the underlying molecular processes is often not well understood. Methods such as metabolomics and proteomics reveal detailed information about these processes. Unfortunately, they provide no spatial information and thus cannot be easily correlated with functional imaging. We have developed an image-guided milling machine and unique workflows to precisely isolate tissue samples based on imaging data. The tissue samples remain cooled during the entire procedure, preventing sample degradation. This enables us to correlate, at an unprecedented spatial precision, comprehensive imaging information with metabolomics and proteomics data, leading to a better understanding of diseases.
Keywords: imaging, omics, tissue extraction
Abstract
Phenotypic heterogeneity is commonly observed in diseased tissue, specifically in tumors. Multimodal imaging technologies can reveal tissue heterogeneity noninvasively in vivo, enabling imaging-based profiling of receptors, metabolism, morphology, or function on a macroscopic scale. In contrast, in vitro multiomics, immunohistochemistry, or histology techniques accurately characterize these heterogeneities in the cellular and subcellular scales in a more comprehensive but ex vivo manner. The complementary in vivo and ex vivo information would provide an enormous potential to better characterize a disease. However, this requires spatially accurate coregistration of these data by image-driven sampling as well as fast sample-preparation methods. Here, a unique image-guided milling machine and workflow for precise extraction of tissue samples from small laboratory animals or excised organs has been developed and evaluated. The samples can be delineated on tomographic images as volumes of interest and can be extracted with a spatial accuracy better than 0.25 mm. The samples remain cooled throughout the procedure to ensure metabolic stability, a precondition for accurate in vitro analysis.
Many efforts have been made over recent years to accurately merge in vivo imaging, tissue biopsy samples, and ex vivo histology in both biomedical research and clinical diagnosis. The aims of these efforts have been cross-validation of findings or increased precision of imaging information (1, 2). Without a doubt, the comprehensive assessment of in vivo imaging parameters, such as metabolic profile, receptor status, perfusion, and morphology, with ex vivo tissue characterization at cellular and molecular levels is extremely beneficial. However, these methodologies require different processing workflows and reveal data at different temporal and spatial scales, making an accurate match nearly impossible to date. Thus, no reliable technology or workflow exists to allow a spatially accurate match of in vivo imaging and ex vivo data. Specifically, in the field of oncology it is recognized that tumors are not homogeneous masses but exhibit large spatial and temporal pheno- and genotypic variations and a complex expression profile of receptors and biomarkers within a single lesion or between metastases (3–6). Variability in oncogenic mutations can be the source of such differences, but epigenetic variations and differences in the tumor microenvironment also play a role and may have a strong impact on the proteomic and metabolomic phenotype of distinct tumor areas (7). Tumor heterogeneity has a clear impact on cancer diagnosis and therapeutic response (8); accurately characterizing and quantifying heterogeneity is therefore important. Tumor heterogeneity can be observed with various imaging technologies in vivo (9), and different imaging biomarkers can depict heterogeneous patterns within the tumor (10, 11). Thus, molecular and functional imaging can guide the extraction of biopsies. However, obtaining a tissue sample from an accurately defined area, guided by imaging, is not a trivial task. Image-guided biopsies are already commonplace in clinical practice, notably in the prostate with, in some cases, interventions using robotics (12). Such technologies are not readily available to study animal models of disease at the desired precision. The most common approach is excision of an entire tumor and subsequent processing for evaluation. It is difficult to keep the orientation of the tumor and limit tissue deformations (13) or to obtain a sample from an exact region of interest. Several approaches have been undertaken, including 3-D printing of tissue molds (14, 15), immobilization devices with subsequent registration (16, 17), and registration of histology to imaging without additional aids. Good accuracy can be achieved in tissues with clearly defined structures such as the brain (18), but results may be inadequate in other types of tissue. When tissues that are not readily accessible are studied, such as orthotopic tumor models or metastatic disease, registration approaches are particularly susceptible to orientation changes and deformations.
With the increasing importance of multiomics approaches for accurate characterization of the metabolic, proteomic, or genetic tissue profile, tissue excision not only requires high spatial accuracy from regions of interest defined on in vivo imaging but also needs to be very fast because the composition of the tissue changes rapidly, immediately after the onset of death, and is extremely susceptible to temperature variations. As the tissue enters an ischemic state and enzymes are still functional, large changes in the transcriptomic, proteomic, and metabolomic profile occur in a relatively short time (19). One approach to quench metabolism is to freeze the tissue very quickly. For s.c. tumors, this can be achieved by rapidly excising the tumor and snap-freezing it in liquid nitrogen. For orthotopic tumors (e.g., lesions in the liver or prostate) or multiple lesions within one animal, the extraction can be more cumbersome and time-consuming. When the samples are taken manually, the time between death and the sample being frozen may vary, increasing the variability of the results. Thus, in this case, only quick freezing of the entire animal is an option.
We present a comprehensive solution that addresses the prompt cooling of the tissue post mortem, the navigational aspect of image-guided tissue sampling, and accurate tissue excision. We developed an image-guided milling machine (IGMM) that lets us obtain samples of any dimension and shape within wide limits from any location within a small animal, guided by in vivo imaging. The rodent is killed after imaging and is instantly frozen, following a standardized workflow to halt metabolite and protein changes post mortem. Here, we describe the technical setup and workflow and report its accuracy and the quality of the prepared samples for NMR-based metabolomics and histology. Since our methodology could be combined with fluorescence and brightfield microscopy, similar to Roy et al. (20), and thus could be used for 3D cryo-imaging, we also performed planar imaging in one animal as an example of a further application.
Materials and Methods
IGMM.
Several significant adaptations were made to a three-axis computerized numerical control (CNC) milling machine (PFE 500-PX; BZT Maschinenbau) to extract tissue from a mouse. The machine has a 720 × 525 × 135 mm3 working area and a manufacturer’s specified resolution of 2.5 μm and repeatability of ±25 μm.
An overview of the IGMM is shown in Fig. 1 A and B. The most conspicuous modification is a stainless steel bath for liquid nitrogen that is thermally insulated with expanded foam. The evaporating liquid nitrogen keeps the mouse cold during the milling process. To ensure sufficient chilling, gaseous nitrogen is blown in the liquid nitrogen during milling to release more cold nitrogen vapor. Moreover, the IGMM features a cover around the bath and the spindle, keeping the vapor contained but leaving sufficient mobility for milling.
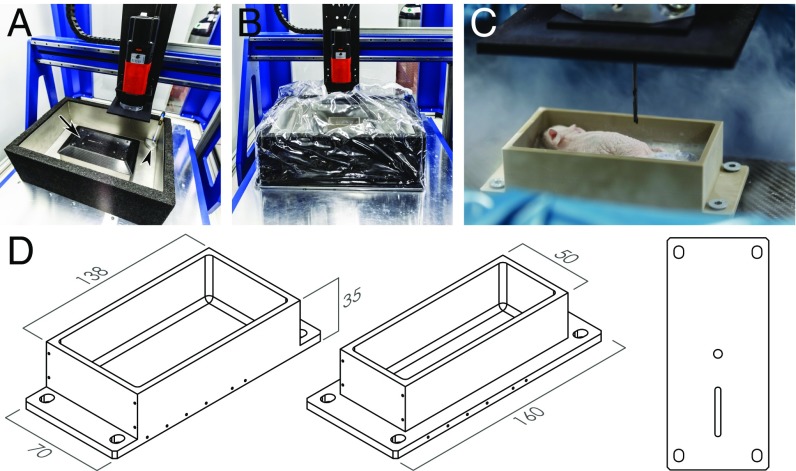
Fig. 1.
Components of the IGMM. (A) Overview of the IGMM showing the milling machine, the liquid nitrogen bath, the inflow valve for the gaseous nitrogen (arrowhead), and the attachment plate for the holders (arrow). (B) Cover around the milling area to keep the cold nitrogen vapor contained. (C) Close-up image of a large holder with an embedded, frozen mouse. Notice the nitrogen vapor surrounding the milling area and the milling bit shown at the top. (D) Drawing of a large (Left) and small (Center) holder and the underside (Right) with two notches ensuring accurate fixation.
Custom-built animal holders were used in conjunction with the IGMM to enable accurate extraction of the tissue. The holder has the following features to orient and fix the animal at a defined position. A frozen animal can be fixed with ice to the holder. The holder can be placed in only one position and orientation and is secured with screws. Finally, the holder allows the placement of 14 glass capillaries with a 1.0-mm outer diameter; these capillaries can be filled with a radioactive solution and used as fiducial markers for image coregistration. The holders were made from polyether ether ketone (PEEK), a material with good mechanical properties, chemical resistance, and a relatively low thermal expansion coefficient. Two variations were made with either a 0.15- or 0.22-L functional volume (Fig. 1 C and D).
Animals.
All animal experiments were performed in accordance with the German animal welfare act, and the animal use and care protocol was approved by the Regierungspraesidium Tuebingen (no. 35/9185.81-2). All animals were housed in individually ventilated cages under standard conditions with access to food and water ad libitum and a 12/12-h light/dark cycle.
Animals used for spatial accuracy measurements.
Twenty-five Swiss nude mice were obtained from Oncodesign. These mice had s.c. CR-IC-002P tumors implanted in the right flank as described by Julien et al. (21). Experiments were performed when the tumors reached a volume of at least 200 mm3.
Animals used to implement, test, and optimize different tissue-processing workflows.
Twenty-one 12-wk-old female CD-1 nude mice (Charles River) were used to implement, compare, and optimize different workflows involving manual and IGMM-automated tissue extraction. One of these mice was used for temperature measurements of the tissue during processing to confirm that the tissue is maintained well below the freezing point during all steps. One animal was used to obtain samples for histology, one was used for testing planar optical imaging (OI), two were used for RNA isolation and sample quality assessment by NMR, and one was used for the comparison of tracer uptake determined in vivo with PET and ex vivo gamma-counting.
Comprehensive Imaging, Animal Handling, and IGMM Workflow.
To quantify the accuracy of our methodology and to investigate several use cases, we implemented and tested a workflow as depicted in Fig. 2A. Each of the individual steps involved in the process is described below. For specific experiments, some of these steps were modified or were not applied, as mentioned in the respective sections.
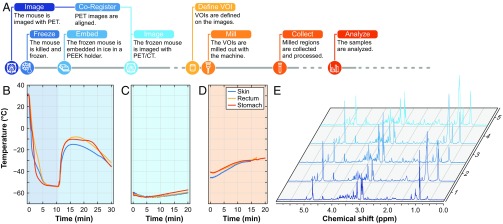
Fig. 2.
Overview of the milling procedure. (A) Flowchart depicting the procedure for image-guided tissue extraction. (B–D) The temperature measured during the entire procedure with sensors in the stomach, in the rectum, and embedded under the skin is depicted below for freezing and embedding (B), at the time when the mouse is placed at room temperature covered with dry ice (C), and during milling (D). (E) An example of NMR spectra from samples obtained with the IGMM.
Alive PET imaging.
The animals were anesthetized with 2% isoflurane evaporated in O2 at a flow rate of 0.5 L/min, and ∼12 MBq of 18F-fluorodeoxyglucose (18F-FDG) or 18F-fluorothymidine (18F-FLT) was administered through a tail-vein catheter, followed by a saline flush. Afterward, 18F-FDG–injected mice remained anesthetized and were warmed for 55 min. Mice injected with 18F-FLT were allowed to wake up after the injection and were anesthetized again after 90 min. Then, each animal was placed on a warmed bed, and PET imaging was performed on a dedicated small-animal PET-scanner (Inveon; Siemens Healthineers). PET images were acquired for 10 min and were reconstructed using two iterations of the ordered subset expectation maximization 3D algorithm, followed by 18 iterations of the fast maximum a posteriori algorithm. Attenuation and scatter correction were not performed. The reconstructed voxel size was 0.39 × 0.39 × 0.80 mm3.
Freezing and embedding.
After in vivo imaging, animals were killed through cervical dislocation and rapidly submersed for 10 min in isopropyl alcohol (SAV Lipid Production GmbH) that was cooled to −60 to −50 °C by the addition of adequate amounts of dry ice (Westfalen AG). The temperature of the isopropyl alcohol was monitored by an electronic temperature sensor (PCE-T317; PCE Instruments). Subsequently, the frozen mouse was placed in a prechilled holder that had previously been fitted with radioactive fiducial markers. A small amount of ice-cold water was poured around the mouse. The holder was then placed in dry ice to let the water freeze and fix the mouse to the holder. Finally, the holders were stored at −80 °C.
Frozen PET/computed tomography imaging.
After the freezing procedure, the holders were covered with dry ice pellets, wrapped in aluminum foil, and imaged with PET/computed tomography (CT). The CT scans were performed with the following settings: 80 kV peak tube voltage, 0.15 mAs per projection, and 180 projections in 2° steps; reconstruction was performed with the Feldkamp algorithm (Cobra version 6.3.39; Exxim Computing Corporation). The reconstructed voxel size was 0.10 × 0.10 × 0.10 mm3. The PET scans were performed as described above for alive PET imaging.
Image registration.
To align the first in vivo PET scan from each animal to the post mortem PET scan of the frozen animal, the tumor was segmented in both images with a standard uptake value (SUV) threshold ≥0.4 in Inveon Research Workplace (IRW; Siemens). The threshold segmentations were manually corrected to include low-uptake tumor areas (e.g., necrosis) and to exclude nontumor tissues (e.g., lymph nodes and muscle). The final segmented images were exported and loaded in MATLAB (version R2013a; MathWorks) and converted to the Neuroimaging Informatics Technology Initiative (NIfTI) file format. Finally, the two images from each mouse were nonrigidly aligned with Elastix (22). The alignment quality was assessed visually, based on the agreement of the alive and post mortem PET tracer uptake and on the rate of voxel volume changes. The same workflow can be applied if more than one region is of interest.
Volume of interest definition.
The PET/CT images of the frozen mice were manually aligned to a computer model (0.1 mm isotropic voxel size) of the holder in IRW. The volumes of interest (VOIs) that were defined in the tumors, livers, hearts, or kidneys of the mice were cylinders with a volume of 10 μL (2.0 mm diameter, 3.2 mm height).
The VOIs were exported as Digital Imaging and Communications in Medicine (DICOM) radiotherapy structures and converted to the Initial Graphics Exchange Specification (IGES) file format using in-house–developed software in MATLAB through the Inventor Component Object Model (COM) interface (version 2015; Autodesk). These files were loaded in a computer-aided manufacturing program (SprutCAM 8; SPRUT Technology) to define the milling path in G-code.
Milling and sample collection.
The bath on the IGMM was filled with ∼8 L of liquid nitrogen. Individual holders containing one or two embedded mice were placed and secured with screws (Fig. 1C), and the plastic enclosure was positioned around the mill (Fig. 1B). Afterward, gaseous nitrogen was blown into the liquid nitrogen for 3 min to create sufficient amounts of vapor to keep the animal frozen; then the milling process was started. The duration of the procedure depended on the dimensions of the working area and the depth of the sample, i.e., the total volume that needed to be milled away, and was usually between 5 and 10 min. After milling, the shavings left behind around the sample (consisting of ice and tissue) were carefully removed with a brush. To determine the accuracy of the milling process, a postmilling CT was acquired from the mice, again with dry ice pellets covering the mouse to maintain the low temperature and avoid degradation of the metabolites (more details are given below).
Afterward, the samples were collected by placing a disposable NMR insert (BL4; Bruker BioSpin) over the exposed tissue cylinders and breaking off the sample. All samples were processed as described in the NMR section below.
NMR data acquisition.
Immediately after collection, samples were weighed and then centrifuged at 4 °C for 10 s at 17,000 × g. Ten microliters of cold D2O (Euriso-top) were added to the NMR sample tube; then it was closed and submerged in liquid nitrogen. Afterwards, the samples were stored at −80 °C until further processing. Shortly before acquisition of the NMR spectra, each sample tube was placed in a ZrO2 rotor that was closed with a polychlorotrifluoroethylene drive cap (Bruker BioSpin).
The NMR experiments were performed on an Avance III 600 MHz (Bruker BioSpin). To assess the effects of different sample-handling conditions, intact biopsies were directly measured with a 4-mm double-resonance (1H and 13C) high-resolution magic angle-spinning (HR-MAS) probe (Bruker BioSpin), providing characteristic profiles of metabolites, lipids, and macromolecule background. Samples were spun at 5 kHz and measured at 4 °C. A total of five 1H spectra with presaturation were acquired from each sample in sequence, similar to Beckonert et al. (23). A one-pulse sequence (zgpr) was used to optimize the water suppression, followed by a 1D NOESY (noesygppr1d), a T2 filtered Carr–Purcell–Meiboom–Gill (CPMG) sequence (cpmgpr1d), and a diffusion-edited sequence (ledbpgppr2s1d). The last spectra were obtained with a 2D J-resolved sequence (JRES; jresgpprqf). Acquisition, preprocessing and Fourier transformation of the free induction decay (FID), and subsequent phase correction and baseline correction were all performed in TopSpin (v3.2; Bruker BioSpin).
For sample quality assessment (quenching efficiency and biochemical stability), extracts from a representative liver and brain biopsy were measured with NOESY, CPMG, and JRES experiments on a 5-mm triple-resonance (1H, 13C, 15N) TXI probe (Bruker BioSpin) at 4 °C as described in the Histology, sample quality assessment, and comparison of PET and gamma-counter uptake section below.
NMR data analysis.
The NMR data were analyzed using the MVAPACK (24) package in GNU Octave (25). The FID files were apodized, phase corrected, and Fourier transformed under the default settings. For comparison, the real parts of the Fourier transformed spectra were normalized with the multiple scatter correction (MSC) (24), histogram matching (HM) (26), standard normal variate normalization (SNV) (24), and probabilistic quotient normalization (PQN) (27) algorithms. The normalized spectra were binned using an adaptive binning algorithm (28) after suppressing signals for water and isopropanol. Then, principal component analyses (PCAs) were performed on the normalized and binned spectra.
Quality Control.
To validate the IGMM and the freezing procedure of the mice, temperature measurements were performed, and the spatial accuracy of the milled regions was determined. Additionally, metabolomic comparisons between manual and automated methods, OI experiments, and histological staining of IGMM-generated tissue samples were performed.
Accuracy measurements.
Additional CT images were acquired immediately after the milling process and were aligned to the computer model of the holder in IRW. Ten-microliter cylindrical VOIs were placed over the freestanding cylinders, and their centroids were compared with those obtained from the VOIs defined before the milling. The difference between the original and postmilling VOIs was measured in three directions, and the overall accuracy was determined by the mean of these differences as well as the mean of the absolute differences.
Temperature measurements.
Temperature measurements were performed with an analog circuit based on 10-kΩ negative temperature coefficient surface-mount device (SMD) thermistors (B573; EPCOS) connected to an Arduino computer (UNO rev. 3; Arduino). The data were acquired at 2 Hz at 10-bit precision. The sensors were calibrated to a PCE-T317 thermometer in isopropyl alcohol that was warmed from approximately −50 °C to 20 °C. A Steinhart–Hart equation (29) with four coefficients was used to convert the measured resistance to temperature.
One animal was killed by cervical dislocation, and sensors were immediately inserted in the rectum, in the stomach, and underneath the skin of the mouse. Afterward, the mouse was processed as described in the freezing and milling section. To simulate a frozen PET/CT scan between freezing and milling, the frozen animal embedded in a holder was covered with dry ice, wrapped in aluminum foil, and left at room temperature for ∼45 min. The temperature development was recorded for all three sensors during the whole procedure.
Comparison of manual sample preparation and preparation with IGMM.
Fifteen animals were killed through cervical dislocation after 10 min of isoflurane anesthesia. Five of these animals were processed as described in the workflow but without tracer injection and PET imaging. To compare results from the IGMM with manual tissue sampling, 10 mice were killed and processed manually. The livers of the mice were dissected and snap-frozen in liquid nitrogen at different time points after cervical dislocation. In five mice, the liver was extracted as quickly as possible (<90 s); in five other mice, one half of the liver was frozen 10 min postmortem, and one half was frozen 20 min postmortem. Liver samples of ∼10 mg were later taken manually from the frozen livers at −20 °C using a 2-mm-diameter biopsy punch (Miltex) and were transferred into disposable NMR inserts.
Planar imaging.
One animal was injected with FDG as described in the alive PET imaging section and 55 min later was injected with 100 µL of Cy5 (Vybrant DiD; Invitrogen) via a tail-vein catheter. Another ∼5 min later, the mouse was killed by cervical dislocation and frozen and embedded in an animal holder as described in the freezing section. A frozen PET/CT scan was performed, and a flat plane was positioned and milled on the coronal midsection of the animal. The mouse was placed in an optical imaging system (IVIS Spectrum; PerkinElmer), and a fluorescence image was acquired. In addition, a normal light photograph of the plane was acquired.
Histology, sample quality assessment, and comparison of PET and gamma-counter uptake.
Four mice were processed as described in the general workflow. Standard 10-μL VOIs were placed in the heart and kidney (histology experiment), in the heart, kidney, and brain (RNA and NMR analysis experiment), or in several organs (gamma-counter experiment). Importantly, for the gamma-counter experiment, attenuation correction was performed after the frozen PET scan using the built-in 57Co transmission source. After the milling procedure, the tissue samples were carefully broken off with cooled forceps.
For histology, the samples were immediately placed in 4.5% neutral buffered formalin (SAV Liquid Production GmbH) for 24 h at room temperature. Following dehydration in a tissue processor (TP1020; Leica Biosystems), the samples were embedded in paraffin. Subsequently, 5-µm sections were prepared and stained with H&E according to standard procedures.
For RNA isolation, samples were immediately placed in a precooled cryotube, submersed in liquid nitrogen, and stored at −80 °C. RNA isolation and quality control were performed by a commercial company (c.ATG). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. Briefly, 5–10 mg of frozen tissue was dissociated using 350 µL of RLT buffer (Qiagen) in a 2-mL extraction tube containing 5-mm-diameter beads and was agitated twice for 2 min at 30 Hz in a TissueLyser II (Qiagen). Optional DNase digestion and centrifugation steps were performed to prevent DNA and ethanol contamination. Elution was performed using 30 µL of RNase-free water. The RNA concentration was measured using the Qubit RNA BR Assay Kit (Thermo Fisher Scientific), and RNA purity was assessed using A260/A280 and A260/A230 ratios using a spectrophotometer (NanoDrop ND-1000; PEQLAB). The RNA integrity number (RIN) was estimated using a RNA 6000 Nano Kit (Agilent) and a Bioanalyzer 2100 (Agilent). Overall RNA integrity was performed using electrophoretic separation on microchips and using the ratio of 18S to 28S ribosomal subunits using the Bioanalyzer software.
For sample quality assessment by NMR, samples were immediately placed in a precooled cryotube, submersed in liquid nitrogen, and stored at −80 °C. To extract metabolites, each sample was placed in a cryogenic impactor (CP02 Cryoprep Pulverizer; Covaris) to cryofracture it mechanically. Afterwards the pulverized tissue was transferred into a 1.5-mL Eppendorf tube to which 500 µL of a 100-µM TSP solution (Sigma-Aldrich) in D2O was added. The samples were vortexed for 1 min and then centrifuged for 5 min at 16,600 × g. The supernatant was then transferred into a 5-mm NMR tube and directly measured at 4 °C.
For comparison of PET and gamma-counter uptake, the samples were weighed and transferred to an automatic gamma-counter (WIZARD2 2480; PerkinElmer) for determination of radioactivity and calculation of the activity concentration. All values were then decay-corrected to the time of the gamma-counter measurement.
Results
Quality Control.
Temperature.
The temperature profile measured in a mouse during all steps of the procedure is shown in Fig. 2 B–D. The sensors placed s.c., in the stomach, and in the rectum reached subzero temperatures within 30 s, 50 s, and 69 s, respectively. When the mouse was taken out of the cooling bath after ∼10 min, the temperature had stabilized at around −54 °C. During the subsequent embedding process, the sensors briefly registered a maximum temperature of −7 °C before decreasing again (Fig. 2B). When the frozen, embedded mouse was placed at room temperature for ∼45 min, the temperature inside the mouse increased to −43 °C. In a typical scenario, a PET/CT scan of the mouse can be acquired within 20 min, during which the temperature increases only marginally (Fig. 2C and Fig. S1). The milling procedure was started at a much higher temperature than normally would be encountered because of the long period at room temperature. Nonetheless, the maximum temperature measured during milling was −27 °C, as shown in Fig. 2D. To mill tissue as close to the sensors as possible, two of the three sensors were destroyed in the milling process.
Accuracy.
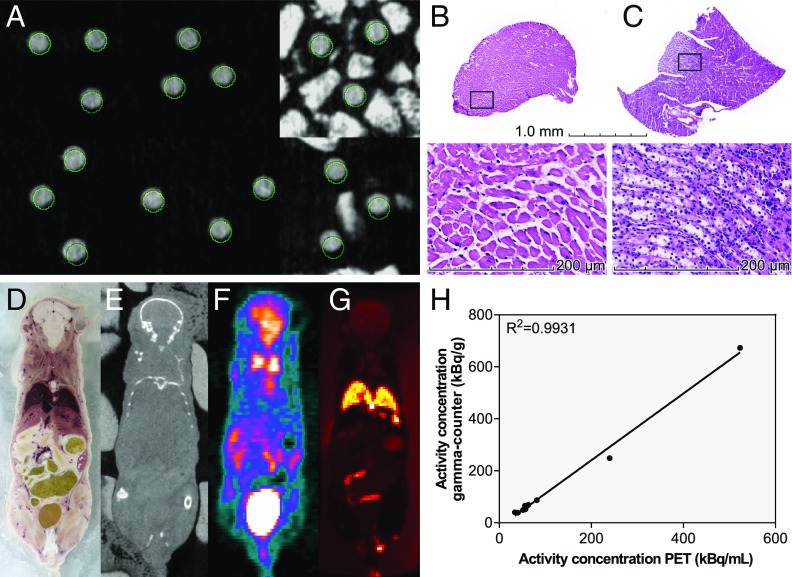
In total, the accuracy of 70 milled cylinders in 25 mice was established by comparing the postmilling CT with the intended VOIs. Of these, 13 cylinders partially broke off, but their accuracy could still be measured. An additional three cylinders broke off completely and have not been taken into account. The absolute deviations in the x and y directions were 0.24 ± 0.15 mm and 0.23 ± 0.16 mm, respectively (Fig. 3A); the absolute difference in the z direction was 0.18 ± 0.14 mm. These numbers are influenced to a large extent by the current average bias of 0.22, 0.16, and 0.16 mm for x, y, and z, respectively. To further prove the accuracy of the IGMM, we milled a total of 13 cylinders in one mouse; two of these cylinders broke off. The remaining cylinders were collected for ex vivo analysis in a gamma-counter. The activity concentration (kBq/g) of the samples as determined by the gamma-counter measurement was correlated with the activity concentration (kBq/mL) of the VOIs determined in the frozen PET scan. The two parameters show a linear correlation with a high coefficient of determination of 0.9931, further proving the accuracy of the IGMM (Fig. 3H). The slightly higher activity concentration measured in the gamma-counter compared with the PET scan can be explained mainly by partial volume effects occurring in the PET scan.
Fig. 3.
Spatial accuracy and planar imaging. (A) CT images acquired postmilling from six mice with three regions each. The VOIs, as defined before milling, are circled in green. (B and C) H&E stains of IGMM-derived tissue samples from a VOI placed in the heart (B) and from a VOI placed in the kidney (C). (D–G) A regular light photograph (adjusted for white-balance and slight geometric distortions) (D), CT (E), PET (F), and OI (G) in a plane through a mouse. D and G were obtained after milling. (H) Activity concentrations obtained with PET and a gamma-counter in 11 cylinders.
RNA isolation.
To test RNA integrity in the frozen material, we extracted total RNA from kidney. Both processed samples presented a yield over 5 µg of RNA and a RIN over 8 (8.8 and 8.1), therefore allowing whole-genome gene-expression profiling without possible degradation-related biasing in the measurement (30). We also extracted high-quality RNA from the more difficult tissues, brain (tissue rich in lipids; RIN 9.2 and 8.9) and liver (tissue with high glycogen content; RIN 8.0 and 7.7), without adapting the standard RNA-extraction protocol. However, yield and quality could have been improved by respectively using TRIzol extraction and decreasing the ethanol concentration during the binding.
Planar imaging and histology.
The histological sections obtained using the IGMM are shown in Fig. 3 B and C. The tissue clearly shows the morphology of myocardium and renal medullary tissue. A sagittal slice through a mouse, photographed after milling, is shown in Fig. 3 D–G together with OI and its matching slice in CT and PET.
Image registration.
One example of the image alignment is shown in Fig. 4.
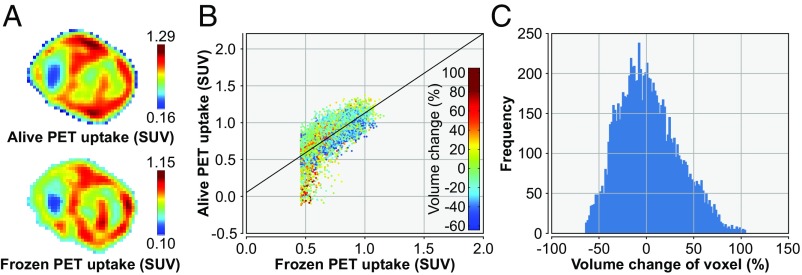
Fig. 4.
An example of the alignment of the PET scans acquired pre- and postfreezing. (A) Frozen PET (Lower) and aligned prefreezing PET (Upper). (B) Scatter plot of the voxel values from the frozen PET (horizontal axis) and the aligned prefreezing PET (vertical axis), with color indicating the amount of deformation required. (C) Histogram of the required change in voxel size for proper alignment.
NMR.
Several examples of HR-MAS spectra from the samples obtained with the IGMM are shown in Fig. 2E. Two tissue-extract spectra from the sample quality assessment are shown in Fig. 5A.
Fig. 5.
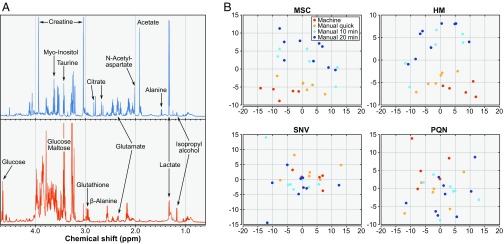
Results from NMR data acquisition. (A) Sample quality assessment: NMR extract spectra (CPMG) from a murine brain (blue) and liver sample (orange) obtained with the IGMM with important metabolites and isopropyl alcohol highlighted. (B) Effect of handling conditions: PCA of the NOESY spectra from samples obtained with the IGMM and through manual sampling. The spectra were binned, and four different normalization techniques were used: HM, MSC, PQN, and SNV.
Contaminants.
Because of the freezing procedure, isopropyl alcohol is the most likely candidate for contamination. Fig. 5A shows the contribution of isopropyl alcohol to the acquired 1H NMR spectra. While a prominent doublet at 1.17 ppm can be clearly identified and excluded from further analysis, the multiplet at 4.01 ppm does not need to be considered, since it falls into the spectral noise. Otherwise, the acquired NMR spectra after the IGMM procedure and established workflow appear without abnormalities.
Comparison with manual sampling.
To compare the quality of the samples collected by the IGMM with samples collected by hand, four groups of NMR spectra were defined. Group 1 comprised samples collected by the IGMM, group 2 consisted of samples collected immediately by hand, and groups 3 and 4 included samples collected by hand after 10 and 20 min at room temperature, respectively. As shown in Fig. 5B, the PCA of the NOESY spectra normalized using SNV and PQN algorithms could not separate the four groups. Nevertheless, the PCA of the NOESY spectra normalized using HM and MSC algorithms yielded separation of groups 1 and 2 from groups 3 and 4. There was an overlap between groups 3 and 4, while groups 1 and 2 were clustered together. This indicated that the samples collected by the IGMM have not undergone metabolite degradation to a larger extent than those collected very quickly by hand. However, the clear separation of groups 3 and 4 indicates that time is a critical component of the tissue extraction workflow.
Discussion
In this study, we present an imaging-guided approach to obtain tissue samples that can be used for further analysis using a wide range of methods, including NMR, MS, or deep sequencing, to address metabolomic, proteomic, transcriptomic, or genomic questions. We have proven that the samples can be rapidly frozen and remain well below the freezing point at all times of the procedure to maintain metabolic stability. The samples can be extracted and defined by morphological, functional, and molecular imaging information yielded from CT, MRI, or PET with a spatial accuracy in the submillimeter range. This enables unprecedented possibilities to link imaging to omics data at a time when the different fields of omics are attracting increasing interest. The plethora of information produced by these methods has proven to be valuable in the understanding of diseases such as cancer, neurodegenerative diseases, and many others (31–35). The ability to link these procedures to imaging with high spatial accuracy will lead to a better understanding of the imaging data.
In the fields of omics, measured samples should represent normal or pathological physiology as best possible, and changes occurring post mortem should be reduced to a minimum. The rapid freezing of the animals and the subsequent uninterrupted cold chain minimizes these alterations. Furthermore, instead of certain organs or tissues being excised manually post mortem, the animals are frozen immediately after death. Since this can be performed rapidly and in a standardized manner, the time between death and freezing of the organ of interest will be shorter than the time required for manual organ excision. In addition, variations induced by the individual experimenter can be excluded almost entirely, possibly leading to higher reproducibility of the samples. Because of metabolomic changes occurring postmortem, the liver tissue samples obtained at different time points through manual sampling could be distinguished. This is in contrast with the work of Haukaas et al. (36), who concluded that the metabolic profile of tumor samples is robust to changes for up to 30 min at room temperature. This discrepancy can be explained, at least in part, by differences in tissue type, with liver tissue undergoing rapid changes under ischemic conditions (37). The samples obtained with the IGMM were shown to be similar to the samples that were obtained as quickly as possible manually. Furthermore, we were able to isolate RNA from samples excised with the IGMM and could show its high quality. Since RNA is very prone to degradation, this can be seen as an indicator of the high preservation of the samples due to the fast freezing procedure. This is also supported by the NMR data, showing that many organ-specific metabolites with fast turnover rates are still present at physiological ratios in our sample quality-assessment spectra (Fig. 5A). In the brain sample, we found high levels of citrate and lactate, both metabolites known to be intermediates of the energy metabolism. Furthermore, high concentrations of myo-inositol were found, a key metabolite in intracellular osmoregulation, which is decreased in many pathologies. Finally, we determined relatively low levels of N-acetyl aspartate (NAA), a specific marker for neuronal integrity. In the liver sample we could detect high remaining levels of carbohydrates (e.g., glucose, maltose) and only low levels of lactate, indicating that the tissue was effectively quenched and not subjected to a long period of hypoxia and ongoing enzymatic degradation. Furthermore, Fig. 5B indicates that samples obtained with the IGMM are of at least equal quality to those obtained with quick manual preparation. However, other methods specifically developed for the immediate quenching of the metabolism, including freeze-clamping (38), will preserve more of the metabolites that degrade quickly, such as ATP.
The potential of the developed IGMM goes beyond only linking imaging and multiomics. It also enables the correlation of imaging results with histology or histochemistry at a spatial accuracy that could never be achieved before. As an alternative technology, coregistration of imaging with histology using 3D printed tissue molds has proven to be a promising approach and has significantly improved the alignment of the different modalities. For this technique, the tissue is excised, placed in an individually printed mold based on MRI or CT images to avoid deformations, and then is cut and finally is frozen or fixed. However, a prerequisite for this technology is tissue excision, during which the material is not cooled. Therefore, subsequent metabolomic or transcriptomic analysis is unfeasible. However, although small deformations due to the cutting or the fixation process can still occur, it is especially suitable for the alignment of histological sections with imaging. With this technique the successful preparation of 4.5- to 6-mm-thick sections has been reported (14, 15).
The achieved accuracy in the submillimeter range is therefore outstanding compared with current manual methods and leaves little room for further optimization because of technical limitations. It is not feasible to have the frozen mice in the holders at exactly the same temperature during the milling and frozen PET/CT scan. The resulting temperature difference between the two processes will inevitably lead to minimal thermal deformations. In addition, the limited resolution and the small deformations that may occur in the reconstructed images limit the attainable accuracy. However, the main inaccuracies are introduced by the experimenter when manually aligning the PET/CT scan with the computer model of the holder and when manually adjusting the zero point of the milling machine. The latter is most likely responsible for the current bias in the milling process. Therefore, choosing the zero point more accurately could significantly improve the accuracy. The ±25-μm inaccuracy of the milling machine, as reported by the manufacturer, has a negligible effect on the final outcome.
Overall, the accuracy is more than satisfying, especially with the limited spatial resolution of the PET scanner with an FWHM of ∼1.5 mm (39). In addition, with the help of the postmilling CT, the location from which the sample was taken can be determined precisely. With the high spatial accuracy presented here, we will be able to determine variations in the concentration of metabolites, lipids, proteins, RNA molecules, and genomic mutations on a very small spatial scale, and differences that exist between tissue classes can be identified. A recent paper describes the collection of MRI-guided biopsies in living animals (40). However, this has been shown only for brain tissue and has not been tested for other organs. In addition, such procedures exhibit severe deformations, which do not occur in our approach.
During the milling process, several different tissues are milled away, increasing the risk of contaminating the sample. The amount of contamination might be decreased by changing or cleaning the drill immediately before the VOI is reached, but this appeared to be impractical, considering that the amount of contamination in the samples is expected to be negligible. The isopropyl alcohol contamination introduced during the freezing process could be avoided only by changing the freezing procedure, e.g., to liquid nitrogen. However, this method frequently induces cracks and destroys the physical integrity of the animals. Therefore, we considered the isopropyl contamination to be acceptable, since this compound generates defined peaks in NMR or MS spectra and can be easily excluded from analysis.
With this method, tissue extraction is possible from any location in the animals, and it can be easily applied to different species or disease models. Of special interest here are models of neurological disorders to excise specific brain regions because changes in the tissue composition are particularly fast after ischemia onset (41). However, inflammation or diabetes models could also benefit from this approach. Furthermore, in the field of oncology, in particular, studies in endogenous tumor models for which a high heterogeneity was previously reported (42, 43) could benefit from our methodology. Significant effort is being put forth in quantifying tumor heterogeneity and clustering the tumor into biologically distinct regions based on imaging (44–46). Validation of such approaches is not trivial and often relies heavily on the proper alignment of the histological sections to the imaging (45). With our approach, samples can be taken selectively for validation. Finally, in metastasizing tumor models, it is now possible to obtain tissue samples of several distant metastases that were frozen quickly at very similar times after death; such samples would be impossible to generate with manual preparation.
We show the correspondence of regular light photographs and OI with both CT and FDG-PET images acquired in a single coronal flat plane through a mouse (Fig. 3 D–F). This bears some resemblance to the digimouse atlas as described by Dogdas et al. (47). By milling multiple planes, each a small distance apart, our approach could be extended to construct a 3D dataset as well, even for individual animals. Furthermore, such a procedure could be performed with a specific fluorescent dye or a fluorescent-labeled molecule.
Specimens larger than mice could, in principle, also be used in our IGMM, although several adaptations and additional tests are required. A larger holder would be necessary, as is an imaging device that is able to fit it. Because of the decreased surface-to-volume ratio in larger animals such as rats, rabbits, and guinea pigs, it will take longer for these animals to freeze completely when submersed in cooled isopropyl alcohol. Therefore, it needs to be verified whether the freezing time is still within an acceptable range, especially in deeper tissues. Finally, because of the larger volume, the aforementioned deformations resulting from thermal expansion will have a greater effect, possibly leading to decreased accuracy.
In the current study, we used PET/CT imaging, but single-photon emission CT (SPECT)/CT could be used instead without any changes in the experimental setup. Although longer scan times are usually required for SPECT because of the lower sensitivity, we showed that the holders can be placed at room temperature for at least 45 min when covered with dry ice, without the risk of thawing. In contrast, using MRI in this setup is not feasible, since no MRI can be acquired after the animal has been frozen. Although net magnetization increases linearly with lower temperatures, the signal intensity decreases exponentially below the freezing point because of free water crystallization and T2* shortening (48). However, especially in light of the rapid proliferation of PET/MRI technology in recent years (49), there is a clear potential for alignment of simultaneously acquired datasets, including MRI.
Some of the cylinders broke off during the milling process, and the respective samples were lost. This may be a result of the often limited structural integrity of tumor tissues, but we will also address the issue by adjusting parts of the milling process. In future experiments, the breakage could be reduced by using different drill heads, rotation speeds, cutting depth, and feed velocities of the drill head.
To further improve the accuracy and simplify the registration of the frozen PET/CT scan to the computer model of the holder, we will produce dedicated PET/CT beds that fit the holders in a given position. When using a defined bed position during the PET/CT scan, we can make use of an optimized registration matrix that will reduce individual registration errors. For easier and more accurate registration of the alive scans to the frozen PET/CT, we will design a dedicated fixation for the mice that allows us to perform the alive PET, CT, and MRI scans as well as the freezing process without moving the animal. Therefore, differences in the alive and frozen PET/CT will mainly be the result of thermal deformations, with minimal movement of the animal. This is especially important for alive MR data, since these first have to be registered to alive PET/CT images and then need to be corrected for thermal deformations.
Conclusion
We have presented an approach for the accurate, image-guided acquisition of tissue samples from mice. The tissue remains frozen during the entire procedure, minimizing any degradation of the samples. High-quality samples could be obtained for metabolomics, transcriptomics, proteomics, and histology with high spatial accuracy.
Supplementary Material
Acknowledgments
We thank Sandro Aidone and Maren Harant for support during the imaging experiments; Daniel Bukala for the liver dissections; Armin Kolb for the temperature sensors and accompanying software; Dr. Kerstin Fuchs for support during the optical imaging experiment; Dr. Ursula Kohlhofer for H&E staining and microscopy; Hans Lindner, Richard Kessell, Dominik Kaltenbacher, Axel Wechsler, Walter Kühborth, and Timo Cuntz for construction of the IGMM; and Nicolas Casadei and Sven Poths for RNA isolation and analysis. The research leading to these results received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Advanced Grant Agreement 323196.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714178115/-/DCSupplemental.
References
- 1.Dazert E, et al. Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc Natl Acad Sci USA. 2016;113:1381–1386. doi: 10.1073/pnas.1523434113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanoun S, et al. 18F-choline positron emission tomography/computed tomography and multiparametric magnetic resonance imaging for the detection of early local recurrence of prostate cancer initially treated by radiation therapy: Comparison with systematic 3-dimensional transperineal mapping biopsy. Int J Radiat Oncol Biol Phys. 2017;97:986–994. doi: 10.1016/j.ijrobp.2016.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehrl HF, et al. Multimodal elucidation of choline metabolism in a murine glioma model using magnetic resonance spectroscopy and 11C-choline positron emission tomography. Cancer Res. 2013;73:1470–1480. doi: 10.1158/0008-5472.CAN-12-2532. [DOI] [PubMed] [Google Scholar]
- 7.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 8.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor JPB, et al. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21:249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longo DL, et al. In vivo imaging of tumor metabolism and acidosis by combining PET and MRI-CEST pH imaging. Cancer Res. 2016;76:6463–6470. doi: 10.1158/0008-5472.CAN-16-0825. [DOI] [PubMed] [Google Scholar]
- 11.Zinnhardt B, et al. Combined PET imaging of the inflammatory tumor microenvironment identifies margins of unique radiotracer uptake. Cancer Res. 2017;77:1831–1841. doi: 10.1158/0008-5472.CAN-16-2628. [DOI] [PubMed] [Google Scholar]
- 12.Moore CM, et al. Image-guided prostate biopsy using magnetic resonance imaging-derived targets: A systematic review. Eur Urol. 2013;63:125–140. doi: 10.1016/j.eururo.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Wehrl HF, et al. Assessment of murine brain tissue shrinkage caused by different histological fixatives using magnetic resonance and computed tomography imaging. Histol Histopathol. 2015;30:601–613. doi: 10.14670/HH-30.601. [DOI] [PubMed] [Google Scholar]
- 14.Shah V, et al. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum. 2009;80:104301. doi: 10.1063/1.3242697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priester A, et al. Magnetic resonance imaging underestimation of prostate cancer geometry: Use of patient specific molds to correlate images with whole mount pathology. J Urol. 2017;197:320–326. doi: 10.1016/j.juro.2016.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rommel D, et al. Alginate moulding: An empirical method for magnetic resonance imaging/positron emission tomography co-registration in a tumor rat model. Nucl Med Biol. 2008;35:571–577. doi: 10.1016/j.nucmedbio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Strobel K, Bergmann R, Meister S, van den Hoff J, Pietzsch J. Improved multimodality imaging using alginate molding in xenograft tumor models. J Magn Reson Imaging. 2010;31:747–752. doi: 10.1002/jmri.22090. [DOI] [PubMed] [Google Scholar]
- 18.Dauguet J, et al. Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J Neurosci Methods. 2007;164:191–204. doi: 10.1016/j.jneumeth.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin RJA, Dungworth JC, Cobb SR, Pitt AR. Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics. 2008;8:3801–3808. doi: 10.1002/pmic.200800201. [DOI] [PubMed] [Google Scholar]
- 20.Roy D, Steyer GJ, Gargesha M, Stone ME, Wilson DL. 3D cryo-imaging: A very high-resolution view of the whole mouse. Anat Rec (Hoboken) 2009;292:342–351. doi: 10.1002/ar.20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julien S, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 22.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 23.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 24.Worley B, Powers R. MVAPACK: A complete data handling package for NMR metabolomics. ACS Chem Biol. 2014;9:1138–1144. doi: 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaton JW, Bateman D, Hauberg S, Wehbring R. 2007 GNU Octave version 4.2.1 manual: A high-level interactive language for numerical computations. Available at https://www.gnu.org/software/octave/doc/v4.2.1/. Accessed February 22, 2018.
- 26.Torgrip RJO, Åberg KM, Alm E, Schuppe-Koistinen I, Lindberg J. A note on normalization of biofluid 1D 1H-NMR data. Metabolomics. 2008;4:114–121. [Google Scholar]
- 27.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78:4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 28.De Meyer T, et al. NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem. 2008;80:3783–3790. doi: 10.1021/ac7025964. [DOI] [PubMed] [Google Scholar]
- 29.Steinhart JS, Hart SR. Calibration curves for thermistors. Deep-Sea Res Oceanogr Abstr. 1968;15:497–503. [Google Scholar]
- 30.Gallego Romero I, Pai AA, Tung J, Gilad Y. RNA-seq: Impact of RNA degradation on transcript quantification. BMC Biol. 2014;12:42. doi: 10.1186/1741-7007-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed KA, Chinnaiyan P. Applying metabolomics to understand the aggressive phenotype and identify novel therapeutic targets in glioblastoma. Metabolites. 2014;4:740–750. doi: 10.3390/metabo4030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sepiashvili L, et al. Novel insights into head and neck cancer using next-generation “omic” technologies. Cancer Res. 2015;75:480–486. doi: 10.1158/0008-5472.CAN-14-3124. [DOI] [PubMed] [Google Scholar]
- 33.Sharma K, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson P, et al. Loss of neprilysin alters protein expression in the brain of Alzheimer’s disease model mice. Proteomics. 2015;15:3349–3355. doi: 10.1002/pmic.201400211. [DOI] [PubMed] [Google Scholar]
- 35.Yokoi N, et al. Identification of putative biomarkers for prediabetes by metabolome analysis of rat models of type 2 diabetes. Metabolomics. 2015;11:1277–1286. doi: 10.1007/s11306-015-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haukaas TH, et al. Impact of freezing delay time on tissue samples for metabolomic studies. Front Oncol. 2016;6:17. doi: 10.3389/fonc.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ungerstedt J, Nowak G, Ungerstedt U, Ericzon B-G. Microdialysis monitoring of porcine liver metabolism during warm ischemia with arterial and portal clamping. Liver Transpl. 2009;15:280–286. doi: 10.1002/lt.21690. [DOI] [PubMed] [Google Scholar]
- 38.Gladden LB, Crawford RE, Webster MJ. Effect of lactate concentration and metabolic rate on net lactate uptake by canine skeletal muscle. Am J Physiol. 1994;266:R1095–R1101. doi: 10.1152/ajpregu.1994.266.4.R1095. [DOI] [PubMed] [Google Scholar]
- 39.Visser EP, et al. Spatial resolution and sensitivity of the Inveon small-animal PET scanner. J Nucl Med. 2009;50:139–147. doi: 10.2967/jnumed.108.055152. [DOI] [PubMed] [Google Scholar]
- 40.Galbán S, et al. MRI-guided stereotactic biopsy of murine GBM for spatiotemporal molecular genomic assessment. Tomography. 2017;3:9–15. doi: 10.18383/j.tom.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sköld K, et al. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: Stathmin 2-20 and peptides as sample quality indicators. Proteomics. 2007;7:4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 42.Leung JY, Andrechek ER, Cardiff RD, Nevins JR. Heterogeneity in MYC-induced mammary tumors contributes to escape from oncogene dependence. Oncogene. 2012;31:2545–2554. doi: 10.1038/onc.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill R, Song Y, Cardiff RD, Van Dyke T. Heterogeneous tumor evolution initiated by loss of pRb function in a preclinical prostate cancer model. Cancer Res. 2005;65:10243–10254. doi: 10.1158/0008-5472.CAN-05-1579. [DOI] [PubMed] [Google Scholar]
- 44.Borri M, et al. Characterizing heterogeneity within head and neck lesions using cluster analysis of multi-parametric MRI data. PLoS One. 2015;10:e0138545. doi: 10.1371/journal.pone.0138545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Divine MR, et al. A population-based Gaussian mixture model incorporating 18F-FDG PET and diffusion-weighted MRI quantifies tumor tissue classes. J Nucl Med. 2016;57:473–479. doi: 10.2967/jnumed.115.163972. [DOI] [PubMed] [Google Scholar]
- 46.Katiyar P, et al. A novel unsupervised segmentation approach quantifies tumor tissue populations using multiparametric MRI: First results with histological validation. Mol Imaging Biol. 2017;19:391–397. doi: 10.1007/s11307-016-1009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dogdas B, Stout D, Chatziioannou AF, Leahy RM. Digimouse: A 3D whole body mouse atlas from CT and cryosection data. Phys Med Biol. 2007;52:577–587. doi: 10.1088/0031-9155/52/3/003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaye EA, et al. Consistency of signal intensity and T2* in frozen ex vivo heart muscle, kidney, and liver tissue. J Magn Reson Imaging. 2010;31:719–724. doi: 10.1002/jmri.22029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Disselhorst JA, Bezrukov I, Kolb A, Parl C, Pichler BJ. Principles of PET/MR imaging. J Nucl Med. 2014;55:2S–10S. doi: 10.2967/jnumed.113.129098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.