Significance
Replicative DNA polymerases (DNAPs) combine two processes to ensure the extremely faithful synthesis of DNA, nucleotide selectivity, and editing of misinserted nucleotides by a 3′-5′ exonuclease activity. The 3′-5′ exonuclease activity is governed by four universally conserved aspartate residues that coordinate the two metal ions responsible for the hydrolysis of the last phosphodiester bond of the primer strand. In this work, we have identified an additional conserved aspartate residue that establishes multiple interactions with residues responsible for the exonucleolysis and for the forward movement of the DNAP along the DNA after inserting a nucleotide, acting as a master organizer for the processive intramolecular proofreading of polymerization errors.
Keywords: DNA polymerase, 3′-5′ exonuclease, site-directed mutagenesis
Abstract
Most replicative DNA polymerases (DNAPs) are endowed with a 3′-5′ exonuclease activity to proofread the polymerization errors, governed by four universally conserved aspartate residues belonging to the Exo I, Exo II, and Exo III motifs. These residues coordinate the two metal ions responsible for the hydrolysis of the last phosphodiester bond of the primer strand. Structural alignment of the conserved exonuclease domain of DNAPs from families A, B, and C has allowed us to identify an additional and invariant aspartate, located between motifs Exo II and Exo III. The importance of this aspartate has been assessed by site-directed mutagenesis at the corresponding Asp121 of the family B ϕ29 DNAP. Substitution of this residue by either glutamate or alanine severely impaired the catalytic efficiency of the 3′-5′ exonuclease activity, both on ssDNA and dsDNA. The polymerization activity of these mutants was also affected due to a defective translocation following nucleotide incorporation. Alanine substitution for the homologous Asp90 in family A T7 DNAP showed essentially the same phenotype as ϕ29 DNAP mutant D121A. This functional conservation, together with a close inspection of ϕ29 DNAP/DNA complexes, led us to conclude a pivotal role for this aspartate in orchestrating the network of interactions required during internal proofreading of misinserted nucleotides.
To guarantee faithful DNA synthesis, most replicative DNA polymerases (DNAPs) are associated with a 3′-5′ exonuclease activity contained in an independent domain of the same polypeptidic chain (with the exception of the multisubunit DNAP III of Escherichia coli, where the editing activity resides on the separate subunit epsilon) that removes the misinserted nucleotides before their extension (1), contributing around two orders of magnitude to the global fidelity of the replication process (2). The exonuclease domain is evolutionarily conserved among those DNAPs from families A, B, and C endowed with a proofreading activity (3) and consists of a central core of β-sheets surrounded by six α-helices (4–13). The exonuclease active site is constituted of four invariant aspartates belonging to motifs Exo I, Exo II, and Exo III (3), which coordinate the two metal ions responsible for the hydrolysis of the last phosphodiester bond, and a conserved tyrosine residue of the Exo III motif, which binds and orients a water molecule, as the attacking nucleophile. To allow efficient removal of any misinserted nucleotide, the mismatched primer 3′-end has to be firmly stabilized at the 3′-5′ exonuclease active site. Such a stabilization relies on a widely conserved threonine residue of the Exo I motif that interacts with the 3′-OH at the primer terminus, and on the invariant asparagine and phenylalanine residues at the Exo II motif that makes contacts with the two 3′-terminal nucleotides (14, 15).
The 30- to 40-Å separation between the polymerization and exonuclease active sites imposes a fine-tuned functional coordination between both activities during polymerization to ensure faithful and efficient DNA synthesis. Under favorable conditions (balanced dNTP pool, moderate temperature, and ionic strength), the 3′-end of the growing strand must be preferentially stabilized at the polymerization active site to allow multiple elongation cycles. Eventually, misinsertion of a 2′-deoxynucleoside-5′-monophosphate (dNMP) provokes a drastic drop of the elongation catalytic rate, mainly due to the defective nucleolytic attack of the mismatched 3′-end on the next incoming nucleotide. In addition, the terminal mispair provokes a disruption of contacts with minor groove hydrogen acceptors at the polymerization site, favoring the melting of the 3′-end and its further transference to and stabilization at the 3′-5′ exonuclease active site, where the hydrolysis reaction occurs and only the mismatched nucleotide must be removed (16). Such a transference of the primer terminus from the pol site to the exo site (and vice versa) can take place intramolecularly (without dissociation of the DNAP/DNA complex), as described in the replicative DNAPs from bacteriophages T4 (17) and T7 (18) and in adenovirus DNAP (19). In other DNAPs, the proofreading activity requires a previous dissociation of the DNAP/DNA complex and further binding of the 3′-terminus at the exonuclease active site (intermolecular proofreading), as described in E. coli Pol I (20) and mitochondrial Pol γ (21).
Bacteriophage ϕ29 DNAP belongs to the family B of DNA-dependent DNAPs (22, 23), is the sole enzyme responsible for viral DNA replication (24–26), and has served for decades as a model to determine the role of conserved residues responsible for both the polymerization and exonuclease activities (27–29). This monomeric 67-kDa enzyme consists of an N-terminal exonuclease domain (residues 1–189) that contains the catalytic residues involved in proofreading and a C-terminal polymerization domain (residues 190–572) (27, 28). The ϕ29 DNAP has an intrinsic high processivity and strand displacement capacity, two features that allow the enzyme to replicate the entire genome from a single binding event without requiring the assistance of processivity and unwinding factors (30). The replication fidelity of this extraordinarily processive enzyme rests on a robust nucleotide selectivity (31) and on an efficient intramolecular proofreading of misinserted nucleotides (32).
In this work, we analyze the biochemical role of an invariant aspartate other than the catalytic ones present in the exonuclease domain of the DNAPs from families A, B, and C. The analysis of the biochemical activities of ϕ29 DNAP variants at the corresponding Asp121 leads us to infer an essential role for this aspartate in coordinating both exonucleolysis and translocation during intramolecular proofreading.
Results and Discussion
The Invariant Aspartate of the 3′-5′ Exonuclease Domain.
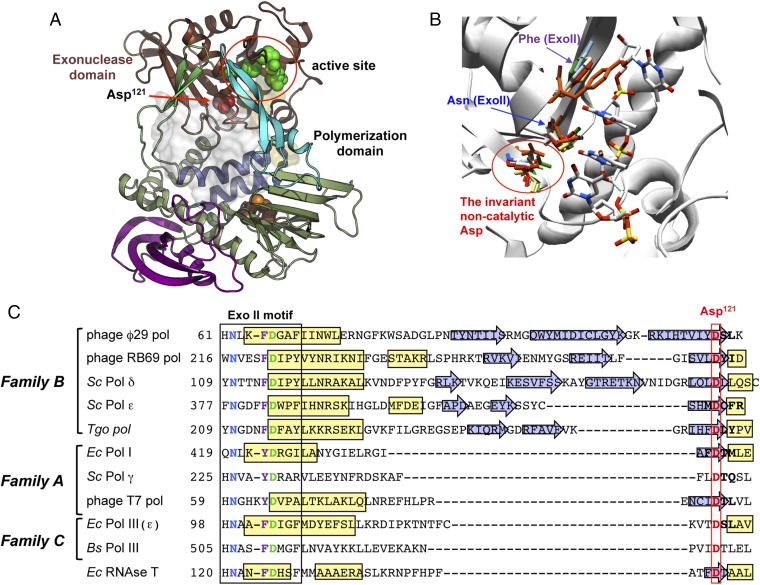
Previous alignments of the conserved exonuclease domain of family B DNAPs, such as viral DNAPs from Herpes simplex virus; Epstein–Barr virus; adenovirus; bacteriophage T4, ϕ29, or RB69; bacterial DNAP II from E. coli or Thermus litoralis; and cellular DNAPs α, δ, and ε from yeast and humans, showed the presence of an additional and invariant aspartate, between the Exo II and Exo III motifs that would correspond to Asp121 in ϕ29 DNAP (33, 34) (Fig. 1 A and C). Crystallographic structure of ϕ29 DNAP with ssDNA bound at the 3′-5′ exonuclease active site shows Asp121 placed at the end of a β-sheet, and despite its proximity to catalytic active site residues, it does not contact either the DNA substrate or the catalytic metal ions.
Fig. 1.
(A) Structural view of ϕ29 DNAP and Asp121 location. Cartoon representation of ϕ29 DNAP [Protein Data Bank (PDB) code 2PYJ]. The color code is brown and green for the exonuclease and polymerase domains, respectively. Subdomains in polymerase domain are highlighted in different colors (TPR1 and TPR2 regions in purple and lime-green colors, respectively; fingers in blue; and thumb in cyan). The Asp121 loop is highlighted in red, with Asp121 being shown as spheres. Catalytic residues for exonuclease and polymerase activities are shown as green and orange spheres, respectively. DNA locations are shown by transparent surface representation, in light orange for ssDNA (PDB ID code 2PY5) and in white for primer template DNA (PDB ID code 2PYJ). (B) Structural superposition of the exonuclease active site of ϕ29 DNAP (PDB ID code 1XHX), RB69 DNAP (PDB ID code 2P5G), S. cerevisiae Pol δ (PDB ID code code 3IAY) and ε (PDB ID code 4M8O), T. gorgonarius DNAP (PDB ID code 2XHB), E. coli DNAP I (PDB ID code 1DPI), T7 DNAP (PDB ID code 1TK8), and E. coli DNAP III (PDB ID code 2HNH). (C) Multiple sequence alignment of the Exo II motif and the noncatalytic aspartate in family A, B, and C members. DNAP sequence references were compiled by Braithwaite and Ito (34), with the exception of bacteriophage DNAP RB69 (National Center for Biotechnology Information reference sequence YP_009100595.1); S. cerevisiae (Sc) Pol ε (GenBank accession no. AJT09723.1); T. gorgonarius (Tgo) DNAP (NCBI reference sequence WP_088885078.1); Sc Pol γ (GenBank accession no. 854508); and RNase T of E. coli (Ec) (NCBI reference sequence WP_074465944.1), which belongs to the DNAQ-like superfamily. The α-helices are represented in yellow, and the β-sheets are indicated with blue arrows. The conserved aspartate of the Exo II motif and the noncatalytic aspartate are shown in green and red, respectively, and the conserved asparagine and aromatic residue (phenylalanine or tyrosine) from the Exo II motif are colored in blue and purple, respectively. Bs, B. subtilis.
Structural superposition of the 3′-5′ exonuclease active site of representative members not only from family B (DNAPs from bacteriophages ϕ29 and RB69, DNAP from Thermococcus gorgonarius, and DNAPs δ and ε from Saccharomyces cerevisiae) but also from families A (E. coli DNAP I, S. cerevisiae DNAP γ, and bacteriophage T7 DNAP) and C (E. coli DNAP III) allowed the identification in all cases of the aspartate structurally homologous to ϕ29 DNAP Asp121 (Fig. 1B). The amino acid sequence alignment shown in Fig. 1C, adjusted by superposition of the 3D structures, outlines the great variability of both the sequence and length of the intervening protein region between the Exo II motif and the mentioned aspartate, explaining why the correct alignment and identification of such a residue among the three DNAP families has remained elusive so far (15, 35). The aspartate mentioned above would correspond to the aspartic residue described to be present between the Exo II and Exo III motifs of the DNAQ-like exoribonucleases of the DEDD family as E. coli RNase T (36), which plays a critical role in the ribonucleolytic activity of these enzymes (37).
To analyze the role of the above-mentioned residue in the catalytic activities of the DNAPs, we have carried out the biochemical analysis of ϕ29 DNAP variants at the corresponding Asp121. To do that, Asp121 was changed into alanine (D121A) as a neutral change, into glutamate (D121E) to conserve the negative charge, and into asparagine (D121N) to maintain the structure of the side chain. The mutant derivatives were overexpressed and purified as described in Materials and Methods, and their involvement in the activity of the DNAP was analyzed by in vitro biochemical assays.
Mutations at ϕ29 DNAP Residue Asp121 Affect the Exonuclease Activity.
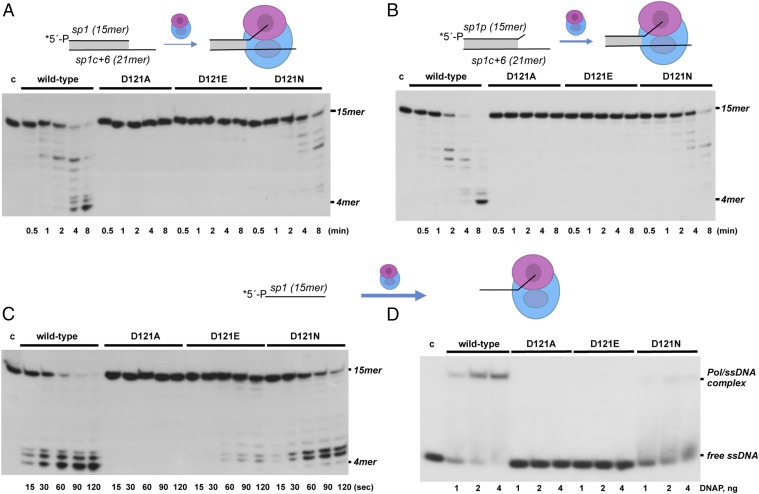
The location of Asp121 in the exonuclease domain of the DNAP led us first to analyze its involvement in the 3′-5′ exonucleolytic degradation of a primer strand. To do that, the enzyme variants were evaluated in their ability to degrade the primer/template structure sp1/sp1c+6 (Materials and Methods and Table 1). This hybrid (15mer/21mer) contains a 6-nt 5′-protruding end, and can therefore be used as substrate for the exonuclease activity (dsDNA) and also for DNA-dependent DNA polymerization (discussed below). The sp1/sp1c+6 molecule is stabilized at the polymerization active site by direct contacts between polymerase ligands and the primer 3′-terminus, and by base pairing with the template strand. However, without adding nucleotides, the binding strength required to position the primer terminus at the exonuclease active site must be sufficient to overcome such a stabilization at the polymerization site. As shown in Fig. 2A, whereas the wild-type enzyme efficiently degraded the primer strand in a processive way until its length was shortened to a 4/6mer, the substitutions at Asp121 either reduced (mutant D121N) or abolished (mutants D121A and D121E) the exonuclease activity of the enzyme.
Table 1.
Structures and sequences of DNA substrates
| Name | Sequences of DNA substrates* | Assay |
| sp1 (15mer) | 5′-32P-GATCACAGTGAGTAC | 3′-5′ exonuclease and EMSA on ssDNA |
| Hybrid | 5′-32P-GATCACAGTGAGTAC | 3′-5′ exonuclease, polymerization and EMSA on dsDNA |
| sp1/sp1c+6 (15mer/21mer) | 3′-CTAGTGTCACTCATGTTATCT | |
| Hybrid | 5′-32P-GATCACAGTGAGTAG | Nucleotide misincorporation |
| sp1p/sp1c+6 (15mer/21mer) | 3′-CTAGTGTCACTCATGTTATCT | |
| Hybrid | 5′-32P-GATCACAGTGAGTAC | Polymerization |
| sp1/sp1c+18 (15mer/33mer) | 3′-CTAGTGTCACTCATGTTATCTTGCTGCCGGTCA | |
| Hybrid | 5′-32P-GATCACAGTGAGTAC | Translocation |
| sp1/sp1c+13A (15mer/28mer) | 3′-CTAGTGTCACTCATGATCTATGTGAAGA |
In all hybrid molecules, the primer oligonucleotide is 5′-phosphorylated.
Fig. 2.
Assays of 3′-5′ exonucleolytic activity of mutants in residue Asp121 of ϕ29 DNAP, using as templates a perfectly matched dsDNA (A), a 3′-mismatched dsDNA molecule (B), and an ssDNA molecule (C). The assays were performed as described in Materials and Methods. After incubation for the indicated times at 25 °C, degradation of the labeled DNA was analyzed by electrophoresis in 7 M urea/20% polyacrylamide gels and autoradiography. The position of the 4mer degradation intermediate is indicated. (D) ssDNA binding of wild-type or mutant ϕ29 DNAPs. The EMSA assay was carried out as described in Materials and Methods, using the 5′-labeled sp1 (15mer) as substrate, in the presence of the indicated amounts of either wild-type or mutant ϕ29 DNAPs. After nondenaturing gel electrophoresis, the bands corresponding to free ssDNA and to the DNAP/DNA complex were detected by autoradiography. c, control lane without enzyme. Asterisks indicate the 5′-32P-labeled end of the primer strand.
To evaluate if the low exonuclease activity exhibited by the protein derivatives was due to an impaired ability to melt the primer terminus, we used as substrate a primer/template hybrid harboring a mismatch at the 3′-end of the primer strand (sp1p/sp1c+6; Materials and Methods and Table 1). This is the physiological substrate of the 3′-5′ exonuclease activity, where the presence of a terminal mismatch favors melting of the primer/template and repositioning of the primer strand, whose 3′-end has to reach the exonuclease active site. As can be observed in Fig. 2B, whereas both the wild-type and mutant D121N DNAPs displayed an exonuclease activity on this substrate stronger than on the perfectly matched 3′-end molecule, the presence of a frayed 3′-end did not favor its removal by mutants D121A and D121E.
From a catalytic point of view, the optimal substrate for the exonuclease activity is an ssDNA molecule, whose 3′-end can bind the catalytic site directly, with no need for previous melting and further transference from the polymerization site. Thus, we assessed the ability of the mutant proteins to degrade the 15mer ssDNA molecule of heterogeneous sequence sp1 (Materials and Methods and Table 1). As shown in Fig. 2C, the wild-type enzyme degraded the oligonucleotide until the length was shortened to 4mer. Whereas mutant D121N also degraded the ssDNA molecule, although to a lower extent than the wild-type enzyme, the exonuclease activity of mutants D121A and D121E was either strongly diminished (D121E) or fully inactive (D121A) with this substrate.
The Changes Introduced at ϕ29 DNAP Residue Asp121 Impair the Stabilization of ssDNA Substrates at the 3′-5′ Exonuclease Active Site.
To ascertain if the impaired exonuclease activity displayed by ϕ29 DNAP mutants D121A and D121E was the consequence of a defective stabilization of ssDNA at the exonuclease active site, an electrophoretic mobility shift assay (EMSA) using oligonucleotide sp1 was carried out (Materials and Methods). In this assay, the presence of a retarded band depends on the absence of divalent metal ions to prevent exonucleolytic degradation of the ssDNA substrate. Thus, binding of the polymerase molecule to ssDNA will rely exclusively on the interaction with specific protein ligands at the exonuclease active site, as ϕ29 DNAP residues Thr15, Asn62, and Phe65 (14, 15). As shown in Fig. 2D, the wild-type enzyme produced a single retardation band interpreted to be a stable enzyme/DNA complex where the 3′-terminus would be located at the 3′-5′ exonuclease active site (14). While mutant D121N also gave rise to a faint retardation band, DNAP variants D121A and D121E were unable to stably bind the oligonucleotide substrate. Therefore, the hindered ssDNA binding could account for the low activity in mutant D121E and the complete absence of exonucleolysis in mutant D121A.
The marked effect that substitutions introduced in ϕ29 DNAP Asp121 have in the exonuclease activity is supported by previous mutagenesis studies performed in the corresponding Asp127 of the family B DNAP from bacteriophage PRD1 (35) and in the Asp533 of family C DNAP III from Bacillus subtilis (38). In addition, substitution of the homologous Asp150 residue in E. coli RNase T also reduced the ribonuclease activity of the enzyme by more than 95% (37). In the latter case, it was surmised that Asp150 was playing a role in maintaining the local fold of the protein.
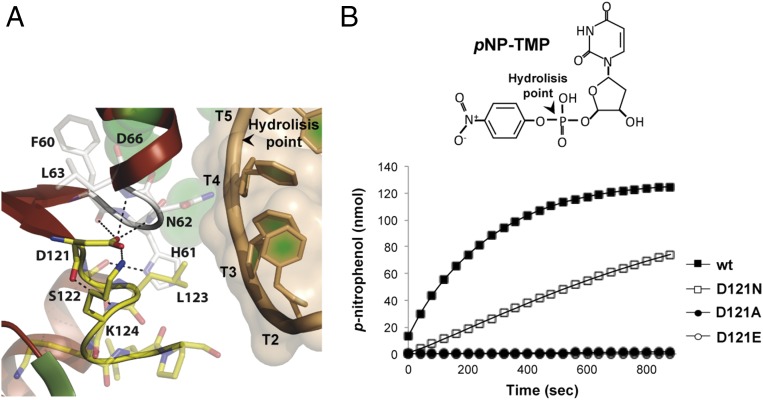
Notwithstanding that the results would indicate a role for Asp121 in the stabilization of the ssDNA substrate, the crystallographic structures of ϕ29 DNAP complexed with oligonucleotide (dT)5 at the exo site showed no direct contacts between this residue and ssDNA (39). Asp121 establishes a net of hydrogen bonds with multiple residues, standing out the interactions with His61 and Asn62 (Fig. 3A). Both residues were first described as ssDNA ligands required for a stable binding of the melted primer terminus at the 3′-5′ exonuclease active site of proofreading DNAPs (14, 15, 40). Whereas the effect of His61 in ssDNA stabilization could be indirect as it interacts with Ser122 and Phe128 (39), both biochemical and crystallographic studies confirmed the direct role of Asn62 in contacting the penultimate 3′-end nucleotide, an essential interaction to allow exonucleolytic activity (14, 39). Mutants D121A and D121E were also unable to hydrolyze the 5′-p-nitrophenyl ester of thymidine 5′-monophosphate (pNP-TMP), a minimal substrate for the exonuclease activity whose binding for further hydrolysis relies on ligands responsible for the stabilization of the 3′-terminal nucleotide (41) (Fig. 3B), and also on the interaction with Asn62 (this study).
Fig. 3.
(A) Detailed view of the interactions established between Asp121 and ssDNA ligands of the exonuclease active site. Residues involved in the interaction are shown as sticks. An ssDNA fragment is shown as a light-orange cartoon inside a transparent surface representation. (B) Hydrolysis of pNP-TMP. The assay was performed as described in Materials and Methods. Catalytic efficiency for the hydrolysis of pNP-TMP catalyzed by the wild-type (wt) ϕ29 DNAP (■) and mutants D121E (○), D121A (●), and D121N (□) was determined spectrophotometrically by monitoring p-nitrophenol production at 420 nm at 25 °C at the indicated times.
These results strongly suggest that the interaction between Asp121 and Asn62 is decisive to ensure a proper orientation/stabilization of the last phosphodiester bond at the exo site for its cleavage during intramolecular proofreading.
Mutations at ϕ29 DNAP Residue Asp121 Hamper Nucleotide Polymerization.
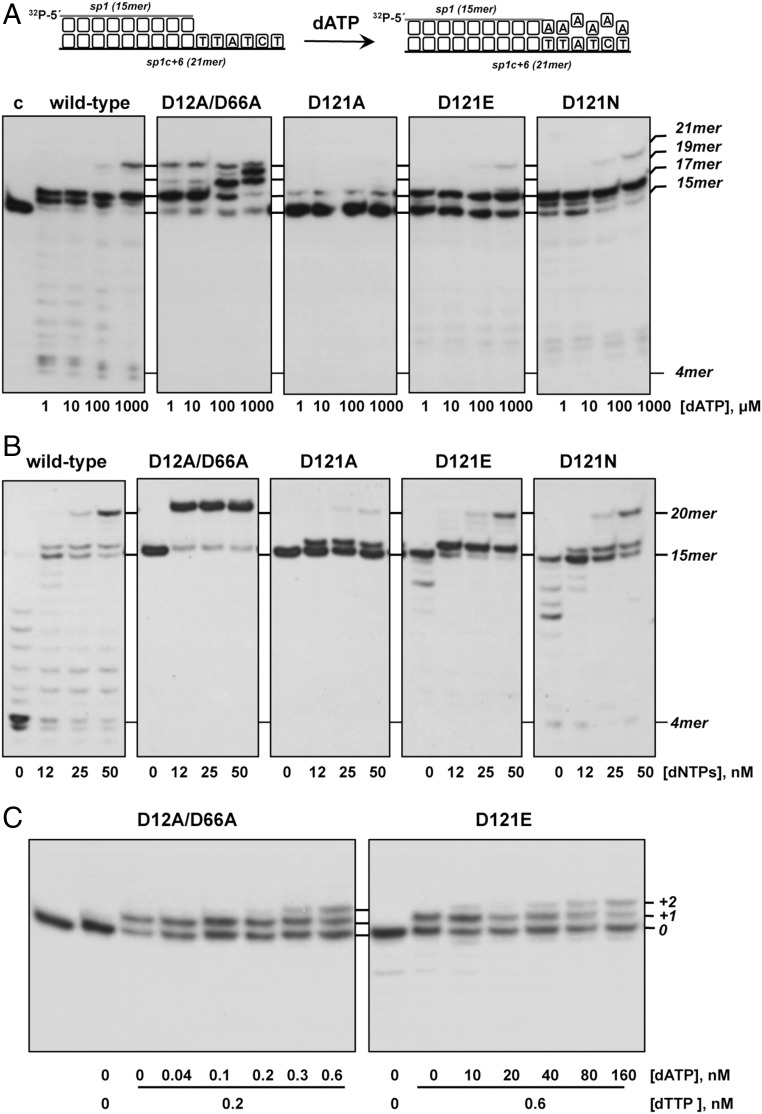
As mentioned above, the 3′-5′ exonuclease activity of replicative DNAPs prevents the fixation of polymerization errors by the efficient proofreading of misincorporated nucleotides. The reduced exonuclease activity displayed by the mutant variants could be compromising the editing function of the polymerase, allowing the stable incorporation of incorrect nucleotides and reducing the fidelity of the polymerization process. To address this issue, the ability to promote stable nucleotide misincorporation by the mutants was studied by analyzing the insertion of 2′-deoxyadenosine-5′-monophosphate (dAMP) opposite noncomplementary positions. To do that, we performed a misincorporation assay (Materials and Methods) using the sp1/sp1c+6 molecule as substrate and increasing amounts of dATP as the sole deoxynucleotide. Under these conditions, the dAMP can be correctly incorporated opposite the two consecutive thymine bases at the 16mer and 17mer positions of the template (scheme in Fig. 4A). As shown, the wild-type enzyme only extended the primer molecule to the 16mer and 17mer positions, producing stable incorporation of dAMP opposite dAMP and 2′-deoxycytidine-5′-monophosphate (dCMP) only at the highest concentration of nucleotides assayed. As a control of the maximum misincorporation that could be reached in this assay, the experiment was performed in parallel with the exonuclease-deficient ϕ29 DNAP mutant D12A/D66A, which lacks two catalytic residues of the 3′-5′ exonuclease active site (3). As can be observed, both the absence of proofreading capacity and the presence of a wild-type polymerization activity led mutant D12A/D66A to stably misinsert dAMP opposite noncomplementary positions, even at the lowest dNTP concentration assayed. Unexpectedly, and despite their low exonucleolytic activity, mutants D121E and D121N showed a nucleotide insertion pattern similar to that displayed by the wild-type enzyme, indicating that the fidelity was not compromised. Mutant D121A exhibited a poor elongation of the primer strand even at the highest dATP concentration assayed, pointing to an additional polymerization defect.
Fig. 4.
(A) Misincorporation during DNA replication by mutants at residue Asp121 of ϕ29 DNAP. The assay was performed as described in Materials and Methods, using as substrate the 5′-labeled hybrid sp1/sp1c+6 (depicted on top of the figure) and the indicated concentration of dATP. After incubation for 5 min at 25 °C, samples were analyzed by 7 M urea/20% polyacrylamide gel electrophoresis and autoradiography. The position corresponding to the unextended primer (15mer) and to either extended (17mer, 19mer, and 21 mer) or degraded (4mer) products is indicated. (B) Polymerization/exonuclease-coupled assay. The assay was performed as described in Materials and Methods using the 5′-labeled hybrid sp1/sp1c+6 as substrate and the indicated concentration of the four dNTPs. After incubation for 5 min at 25 °C, samples were analyzed by 7 M urea/20% polyacrylamide gel electrophoresis and autoradiography. Polymerization or 3′-5′ exonucleolysis is detected as an increase or decrease, respectively, in the size (15mer) of the 5′-labeled primer. (C) Translocation assay. The assay was performed as described in Materials and Methods using the 5′-labeled hybrid sp1/sp1c+13A (15mer/28 mer). After incubation for 5 min at 25 °C, samples were analyzed by 7 M urea/20% polyacrylamide gel electrophoresis and autoradiography. The position corresponding to the primer (0) and +1 and +2 extension products is indicated.
These unforeseen results led us to evaluate the polymerization capability of the Asp121-mutant DNAP variants better, as well as to ascertain whether the mutations introduced affect the dynamic equilibrium between their 3′-5′ exonuclease and 5′-3′ polymerization activities. To do that, we analyzed the functional coupling between synthesis and degradation on the molecule sp1/sp1c+6 (15mer/21mer) as a function of the concentration of the four dNTPs (Materials and Methods and Table 1). Without nucleotides, the only products that can be detected are those generated by the exonucleolytic degradation of the primer terminus. Both the extent and degradation pattern obtained reflect the level of exonuclease activity of the variants with respect to the wild-type and the D12A/D66A polymerases. As the concentration of dNTPs increases, the exonuclease activity is progressively overcome by the 5′-3′ polymerization and net dNMP incorporation can be observed as an increase in the size of the labeled primer (Fig. 4B), allowing one to define the concentration of dNTPs needed to obtain an efficient elongation of the primer for each mutant derivative [polymerase/exonuclease (Pol/Exo) ratio]. As observed in Fig. 4B, mutant D121N showed a Pol/Exo equilibrium similar to that displayed by the wild-type enzyme, reaching the 20mer position at 25 nM dNTPs. Although the reduced exonuclease activity allowed mutants D121A and D121E to efficiently elongate the primer strand at the lowest concentration of dNTPs used (12 nM), polymerization was blocked after incorporation of the first nucleotide in both cases.
Previous kinetic studies (42–44), as well as structural data of family A polymerases (9, 45, 46), allowed the definition of the sequential steps that take place during DNA polymerization. Thus, to allow a next dNMP addition, the DNAP has to move forward to position the new 3′-OH end from the postinsertion site to the preinsertion site, a process called translocation, which is triggered by the release of the pyrophosphate moiety and the concomitant opening of the fingers subdomain (47). The blockage observed after the insertion of the first nucleotide with mutants D121A and D121E led us to ascertain whether substitution of Asp121 affects the translocation step. Accordingly, and as shown in Fig. 4C, whereas mutant D12A/D66A required a similar nucleotide concentration to give the +1 and +2 elongation products (0.2 nM and 0.3 nM, respectively), catalysis of the second nucleotide addition by mutant D121E required 80 nM nucleotide concentration, 127-fold higher than that needed to add the first nucleotide (0.6 nM), pointing to a key role for Asp121 in the translocation step after nucleotide incorporation. Additionally, mutants D121A and D121E exhibit an impaired ability to stabilize a primer/template molecule at their polymerization active site (Fig. S1).
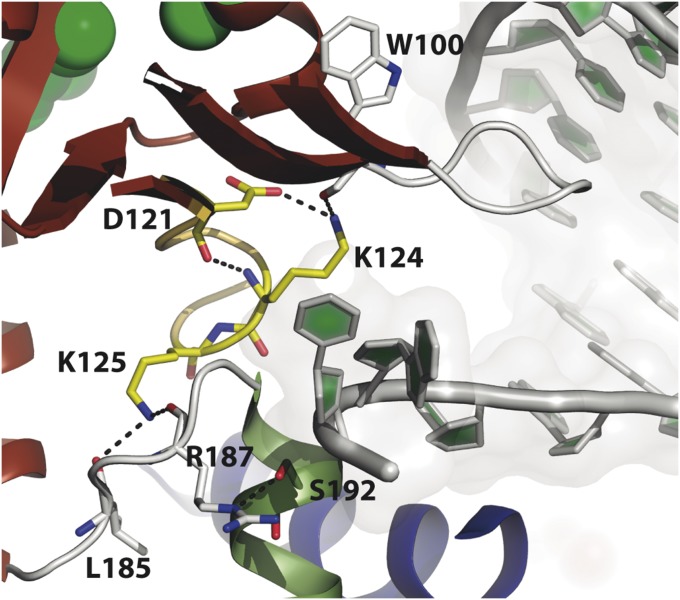
Considering that the global structure of the DNAP is not affected by the mutations introduced (Fig. S2), the question that arises is how Asp121 can contribute to the translocation step. Close inspection of the ϕ29 DNAP ternary complexes (48) shows a direct interaction between Asp121 and Lys124 (Fig. 5). Such a contact would stabilize the conformation of the loop formed by residues Asp121-Lys125. Lys125 makes contacts with a protein region constituted by residues (Gln183-Thr189) that connects both protein domains (exonuclease and polymerase) and makes direct interactions with the ssDNA downstream template. The loss of such interactions could be responsible for the block of the forward movement of the DNAP after the insertion of the first nucleotide (i.e., translocation). This hypothesis could also account for the reduced polymerization activity observed with B. subtilis DNAP III mutants D533A/G (38) and PRD1 DNAP variant D127A (35).
Fig. 5.
Detailed view of the interaction network between Asp121 loop residues (in yellow sticks) and protruding template strand ligands (in white sticks). Template strands are shown as light-gray cartoons.
The Aspartic Residue Has a Critical Role in both Exonucleolysis and Polymerization also in Family A DNAPs.
DNAPs from families A and B exhibit a bipartite architecture with an N-terminal 3′-5′ exonuclease domain and a C-terminal polymerization one. Whereas the crystallographic structures of members from both families show that the distance between the active sites is about 30–40 Å, their editing domains lie on opposite sides with respect to the polymerization active site depending on the specific DNAP family (6, 8, 9, 16) [compare Figs. 1A (family B ϕ29 DNAP) and 6A (family A T7 DNAP)].
Fig. 6.
(A) Crystal structure of T7 DNAP with the N-terminal 3′-5′ exonuclease domain (residues 1–187) colored in pink, the C-terminal polymerization domain (residues 188–698) colored in green, and thioredoxin colored in orange. Catalytic active site residues from the polymerization and exonuclease domains are shown as orange spheres, whereas Asp90 is represented as yellow spheres (PDB ID code 1TK8) (9). (B) Polymerization and exonuclease activities of wild-type and mutant T7 DNAPs. The assay was performed as described in Materials and Methods using the 5′-labeled primer/template molecule sp1/sp1c+18 (15mer/33 mer) depicted on top of the figure, in the presence (+) or absence (−) of dNTPs and thioredoxin. Polymerization or 3′-5′ exonucleolysis is detected as an increase or decrease, respectively, in the size (15mer) of the 5′-labeled primer. After incubation for 10 min at 30 °C, samples were analyzed by 7 M urea/20% polyacrylamide gel electrophoresis and autoradiography. Asterisk indicates the 5′-32P-labeled end of the primer strand.
Such dissimilarities in the relative orientation between both domains also make DNA accession to the polymerization active site different. Thus, in family B DNAPs, the template/primer substrate is bound to the groove formed by the thumb and palm subdomains, whereas in family A, members’ stabilization of the DNA at the polymerization domain is also contributed to by the exonuclease domain. The different orientation of the domains imposes a divergent proofreading mechanism. Thus, in family B DNAPs, the primer terminus diffuses in one dimension from the polymerization to the exonuclease site by a 40° rotation in the helix of the DNA (16). In family A DNAPs, the DNA must helically translocate backward by three bases to allow sufficient unwinding of the primer strand to leave the polymerization domain and to reach the exonuclease active site (6). Both the movement of the primer terminus and the helical translocation of the DNA make internal proofreading more exigent in this type of DNAP. To evaluate the effect that substitutions of the aspartate residue have in the biochemical activities of family A DNAPs, we performed site-directed mutagenesis at the corresponding Asp90 of bacteriophage T7 DNAP (alignment in Figs. 1 and 6A), obtaining the D90A variant.
Both the exonuclease and polymerization activities of the wild-type and D90A mutant T7 DNAP were evaluated using the hybrid molecule sp1/sp1c+18 (Fig. 6B). As expected, in the absence of nucleotides, the wild-type enzyme exerted its potent 3′-5′ exonuclease activity, whereas addition of nucleotides led to a net DNA synthesis showing a distributive polymerization pattern. The presence of thioredoxin, the host-encoded processivity factor of T7 DNAP, stimulated the polymerization activity due to the higher stability of the enzyme/DNA complex. Unlike the wild-type enzyme, the mutant protein lacking the aspartate (Asp90) was exonuclease-deficient, with degradation of the primer strand not being detectable even in the presence of thioredoxin. Regarding the polymerization reaction, the T7 DNAP mutant D90A was blocked after one nucleotide insertion (Fig. 6B), displaying the same behavior as the ϕ29 DNAP mutant D121A. The effect of the mutation was not overcome by the addition of thioredoxin, as only a minor fraction of the +1 products was fully elongated in its presence.
Collectively, the results presented here, together with previous mutagenesis studies performed in E. coli RNAseT (36, 37), family B PRD1 DNAP (35), and family C B. subtilis DNAP III (38), allow us to propose the involvement of the universally conserved aspartate residing at the 3′-5′ exonuclease domain in the two main activities of proofreading DNAPs. Its contact with multiple residues from both domains of the enzyme leads us to envision it as the keystone of a Romanesque vault, maintaining simultaneously the correct orientation of ssDNA ligands at the exonuclease active site to allow internal proofreading and of template-binding residues to promote the translocation step after nucleotide insertion. Those contacts most probably prevent the dissociation of the polymerase/DNA complex, guaranteeing the processivity of the replication process.
Materials and Methods
Materials.
Unlabeled nucleotides were supplied by GE Healthcare. The [α-32P]dATP (3,000 Ci/mmol) and [γ-32P]ATP (3,000 Ci/mmol) were supplied by PerkinElmer. T4 polynucleotide kinase was purchased from New England Biolabs. The pNP-TMP was from Sigma. The thioredoxin was supplied by BioNova. The sequences of the oligonucleotides used are shown in Table 1. Primer oligonucleotides were 5′-labeled with 32P using [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase. The hybrid molecules shown in Table 1 were obtained by annealing the primer to template (1:1 ratio) in the presence of 50 mM Tris⋅HCl (pH 7.5) and 0.2 M NaCl, with heating to 90 °C for 10 min before slowly cooling to room temperature overnight. All oligonucleotides were from Sigma–Aldrich.
Site-Directed Mutagenesis of ϕ29 and T7 DNAPs.
DNAP mutants were obtained using the QuikChange Site-Directed Mutagenesis Kit provided by Stratagene, using as a template the plasmid pJLPM (a derivative of pT7-4w2) (49) containing the viral gene 2 that encodes the wild-type ϕ29 DNAP and the plasmid pGP5-3 (50) that contains the viral gene 5 encoding the wild-type T7 DNAP. The presence of the desired mutations, as well as the absence of additional ones, was determined by sequencing the entire gene. All mutant derivatives, as well as the wild-type ϕ29 and T7 DNAPs, were expressed in E. coli BL21(DE3) cells and further purified essentially as described for the wild-type ϕ29 DNAP (51) or the wild-type T7 DNAP (50).
3′-5′ Exonuclease Assays.
The incubation mixture contained (in a final volume of 12.5 μL) 50 mM Tris⋅HCl (pH 7.5), 1 mM DTT, 4% (vol/vol) glycerol, 0.1 mg/mL BSA, 10 mM MgCl2, and 5 nM of either the wild-type or the indicated ϕ29 DNAP variant. As DNA substrates, 1.2 nM 5′-labeled molecule sp1 (ssDNA), sp1/sp1c+6 (matched dsDNA), or sp1p/sp1c+6 (mismatched dsDNA; Table 1) was used. Samples were incubated at 25 °C for the indicated times and quenched by adding EDTA up to a final concentration of 10 mM. Reactions were analyzed by electrophoresis in 7 M urea/20% polyacrylamide gels and autoradiography.
EMSA.
The interaction of either the wild-type or the ϕ29 DNAP mutants with ssDNA was assayed using as substrate the 5′-labeled sp1 oligonucleotide (Table 1). The incubation mixture contained (in a final volume of 20 μL) 12 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 20 mM ammonium sulfate, 0.1 mg/mL BSA, 10 mM MgCl2, 0.7 nM ssDNA, and the indicated amount of wild-type or mutant DNAP. After incubation for 5 min at 4 °C, the samples were subjected to electrophoresis in precooled 4% (wt/vol) polyacrylamide gels [80:1 acrylamide/bis-acrylamide (wt/wt)] containing 12 mM Tris⋅acetate (pH 7.5) and 1 mM EDTA, and run at 4 °C in the same buffer at 8 V/cm (52). After autoradiography, ϕ29 DNAP/DNA stable interaction was detected as a shift (retardation) in the migrating position of the labeled DNA.
Hydrolysis of pNP-TMP.
The incubation mixture contained (in a final volume of 300 μL) 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1 mM DTT, 1 mM MnCl2, 3 mM pNP-TMP [dissolved in 50 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl], and 500 nM wild-type or indicated mutant ϕ29 DNAP. Hydrolysis was studied by monitoring p-nitrophenol production at 420 nm with an Hitachi U-200 spectrophotometer at 25 °C, essentially as described elsewhere (41). Production of p-nitrophenol was plotted against time.
Polymerase/3′-5′ Exonuclease (Pol/Exo)–Coupled Assay.
The primer/template sp1/sp1c+6 contains a 6-nt 5′-protruding end, and therefore can be used as substrate for DNA-dependent DNA polymerization and also for the exonuclease activity (Table 1). The incubation mixture contained (in a final volume of 12.5 μL) 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 4% (vol/vol) glycerol, 0.1 mg/mL BSA, 1.2 nM 5′-labeled sp1/sp1c+6 substrate, 30 nM wild-type or mutant ϕ29 DNAP, and the indicated increasing concentrations of the four dNTPs. After incubation for 5 min at 25 °C, the reaction was stopped by adding EDTA up to a final concentration of 10 mM. Samples were analyzed by electrophoresis in 7 M urea/20% polyacrylamide gels and autoradiography. Polymerization or 3′-5′ exonucleolysis is detected as an increase or decrease, respectively, in the size (15mer) of the 5′-labeled primer.
In the case of the T7 DNAP, the assay was performed using as DNA substrate 1.2 nM hybrid sp1/sp1c+18 (Table 1); 23 nM wild-type or the D90A mutant T7 DNAP; and, when indicated, 2 μM thioredoxin and 1 mM dNTPs. After incubation for 10 min at 30 °C, samples were processed as described above.
DNA Replication Misincorporation Assay.
Conditions were essentially as described for the polymerization/exonuclease-coupled assay on spl/splc+6, in the presence of 30 nM wild-type or mutant ϕ29 DNAP and the indicated concentrations of only dATP, complementary to template positions 1, 2, 4, and 6. Mutant D12A/D66A was used as a control of the maximum misincorporation that can be reached in the assay. To prevent exonucleolytic degradation of the primer terminus, 10 μM dCTP was added. After incubation for 5 min at 30 °C, samples were analyzed by 7 M urea/20% polyacrylamide gels. After autoradiography, misinsertion of dAMP at noncomplementary positions is observed as the appearance of extension products of the 5′-labeled spl primer (15mer) longer than the correct 17mer extension product.
Translocation Assay.
The incubation mixture contained (in a final volume of 12.5 μL) 50 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 4% (vol/vol) glycerol, 0.1 mg/mL BSA, 1.3 nM 5′-labeled sp1/sp1c+13A (15mer/28mer) substrate, and 30 nM ϕ29 DNAP mutants D12A/D66A and D121E, in the presence of the indicated concentration of dTTP to achieve the +1 elongation of 50% of the primer molecules. After incubation for 5 min at 25 °C, the indicated concentration of dATP was added. Samples were incubated for additional 5 min at 25 °C, after which the reactions were stopped by adding EDTA up to a final concentration of 10 mM. Samples were analyzed by electrophoresis in 7 M urea/20% polyacrylamide gels and autoradiography.
Supplementary Material
Acknowledgments
We thank Dr. Stanley Tabor (Harvard Medical School) for providing us with plasmid pGP5-3 that contains the viral gene 5 encoding the wild-type T7 DNAP and José M. Lázaro for his technical assistance in the purification of the proteins. This work was supported by Grants BFU2014-52656-P from the Spanish Ministry of Economy and Competitiveness (to M.S.), BFU2014-53791-P (to M.d.V.), BFU2015/65880-P (to L.B.), and BFU2014-53762-P (to B.G.). Institutional grants from the Fundación Ramón Areces and Banco de Santander to the Centro de Biología Molecular “Severo Ochoa” are also acknowledged.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718787115/-/DCSupplemental.
References
- 1.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 2.Kunkel TA. DNA replication fidelity. J Biol Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- 3.Bernad A, Blanco L, Lázaro JM, Martín G, Salas M. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 4.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 5.Freemont PS, Friedman JM, Beese LS, Sanderson MR, Steitz TA. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci USA. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Yu P, Lin TC, Konigsberg WH, Steitz TA. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry. 1996;35:8110–8119. doi: 10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 9.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 10.Hopfner KP, et al. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, et al. Crystal structure of an archaebacterial DNA polymerase. Structure. 1999;7:1189–1199. doi: 10.1016/s0969-2126(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez AC, Park HW, Mao C, Beese LS. Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J Mol Biol. 2000;299:447–462. doi: 10.1006/jmbi.2000.3728. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto H, et al. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J Mol Biol. 2001;306:469–477. doi: 10.1006/jmbi.2000.4403. [DOI] [PubMed] [Google Scholar]
- 14.de Vega M, Lázaro JM, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of φ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 15.de Vega M, Lázaro JM, Salas M, Blanco L. Mutational analysis of φ29 DNA polymerase residues acting as ssDNA ligands for 3′-5′ exonucleolysis. J Mol Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 16.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 17.Reddy MK, Weitzel SE, von Hippel PH. Processive proofreading is intrinsic to T4 DNA polymerase. J Biol Chem. 1992;267:14157–14166. [PubMed] [Google Scholar]
- 18.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 19.King AJ, Teertstra WR, Blanco L, Salas M, van der Vliet PC. Processive proofreading by the adenovirus DNA polymerase. Association with the priming protein reduces exonucleolytic degradation. Nucleic Acids Res. 1997;25:1745–1752. doi: 10.1093/nar/25.9.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyce CM. How DNA travels between the separate polymerase and 3′-5′-exonuclease sites of DNA polymerase I (Klenow fragment) J Biol Chem. 1989;264:10858–10866. [PubMed] [Google Scholar]
- 21.Olson MW, Kaguni LS. 3′→5′ exonuclease in Drosophila mitochondrial DNA polymerase. Substrate specificity and functional coordination of nucleotide polymerization and mispair hydrolysis. J Biol Chem. 1992;267:23136–23142. [PubMed] [Google Scholar]
- 22.Blanco L, Salas M. Effect of aphidicolin and nucleotide analogs on the phage φ 29 DNA polymerase. Virology. 1986;153:179–187. doi: 10.1016/0042-6822(86)90021-8. [DOI] [PubMed] [Google Scholar]
- 23.Bernad A, Zaballos A, Salas M, Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987;6:4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco L, Salas M. Characterization and purification of a phage φ 29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA. 1984;81:5325–5329. doi: 10.1073/pnas.81.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco L, Salas M. Replication of phage φ 29 DNA with purified terminal protein and DNA polymerase: Synthesis of full-length φ 29 DNA. Proc Natl Acad Sci USA. 1985;82:6404–6408. doi: 10.1073/pnas.82.19.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salas M, Blanco L, Lázaro JM, de Vega M. The bacteriophage phi29 DNA polymerase. IUBMB Life. 2008;60:82–85. doi: 10.1002/iub.19. [DOI] [PubMed] [Google Scholar]
- 27.Blanco L, Salas M. Mutational analysis of bacteriophage φ 29 DNA polymerase. Methods Enzymol. 1995;262:283–294. doi: 10.1016/0076-6879(95)62024-9. [DOI] [PubMed] [Google Scholar]
- 28.Blanco L, Salas M. Relating structure to function in φ29 DNA polymerase. J Biol Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 29.Salas M, de Vega M. Bacteriophage protein-primed DNA replication. In: Hefferon KL, editor. Recent Advances in DNA Virus Replication. Research Signpost; Ithaca, NY: 2006. pp. 259–288. [Google Scholar]
- 30.Blanco L, et al. Highly efficient DNA synthesis by the phage φ 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 31.Esteban JA, Salas M, Blanco L. Fidelity of φ 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 32.de Vega M, Blanco L, Salas M. Processive proofreading and the spatial relationship between polymerase and exonuclease active sites of bacteriophage φ29 DNA polymerase. J Mol Biol. 1999;292:39–51. doi: 10.1006/jmbi.1999.3052. [DOI] [PubMed] [Google Scholar]
- 33.Henninger EE, Pursell ZF. DNA polymerase ε and its roles in genome stability. IUBMB Life. 2014;66:339–351. doi: 10.1002/iub.1276. [DOI] [PubMed] [Google Scholar]
- 34.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W, Ito J. Family A and family B DNA polymerases are structurally related: Evolutionary implications. Nucleic Acids Res. 1994;22:5177–5183. doi: 10.1093/nar/22.24.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo Y, Deutscher MP. Mechanism of action of RNase T. I. Identification of residues required for catalysis, substrate binding, and dimerization. J Biol Chem. 2002;277:50155–50159. doi: 10.1074/jbc.M207706200. [DOI] [PubMed] [Google Scholar]
- 38.Barnes MH, Spacciapoli P, Li DH, Brown NC. The 3′-5′ exonuclease site of DNA polymerase III from gram-positive bacteria: Definition of a novel motif structure. Gene. 1995;165:45–50. doi: 10.1016/0378-1119(95)00530-j. [DOI] [PubMed] [Google Scholar]
- 39.Kamtekar S, et al. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 40.de Vega M, Lázaro JM, Salas M. Phage φ 29 DNA polymerase residues involved in the proper stabilisation of the primer-terminus at the 3′-5′ exonuclease active site. J Mol Biol. 2000;304:1–9. doi: 10.1006/jmbi.2000.4178. [DOI] [PubMed] [Google Scholar]
- 41.Kumar JK, Chiu ET, Tabor S, Richardson CC. A unique region in bacteriophage t7 DNA polymerase important for exonucleolytic hydrolysis of DNA. J Biol Chem. 2004;279:42018–42025. doi: 10.1074/jbc.M406103200. [DOI] [PubMed] [Google Scholar]
- 42.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 43.Joyce CM, Benkovic SJ. DNA polymerase fidelity: Kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 44.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 45.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: Structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 48.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Prado A, et al. Insights into the determination of the templating nucleotide at the initiation of φ29 DNA replication. J Biol Chem. 2015;290:27138–27145. doi: 10.1074/jbc.M115.682278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 51.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage φ 29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 52.Carthew RW, Chodosh LA, Sharp PA. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985;43:439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.