Significance
Indoleamine 2,3-dioxygenase (IDO1) is a heme protein that catalyzes the dioxygenation of tryptophan. Cells expressing this activity are able to profoundly alter their surrounding environment to suppress the immune response. Cancer cells exploit this pathway to avoid immune-mediated destruction. Through a range of kinetic, structural, and cellular assays, we show that two classes of highly selective inhibitors of IDO1 act by competing with heme binding to apo-IDO1. This shows that IDO1 is dynamically bound to its heme cofactor in what is likely a critical step in the regulation of this enzyme. These results have elucidated a previously undiscovered role for the ubiquitous heme cofactor in immune regulation, and it suggests that other heme proteins in biology may be similarly regulated.
Keywords: IDO1, heme, cancer, kynurenine
Abstract
For cancer cells to survive and proliferate, they must escape normal immune destruction. One mechanism by which this is accomplished is through immune suppression effected by up-regulation of indoleamine 2,3-dioxygenase (IDO1), a heme enzyme that catalyzes the oxidation of tryptophan to N-formylkynurenine. On deformylation, kynurenine and downstream metabolites suppress T cell function. The importance of this immunosuppressive mechanism has spurred intense interest in the development of clinical IDO1 inhibitors. Herein, we describe the mechanism by which a class of compounds effectively and specifically inhibits IDO1 by targeting its apo-form. We show that the in vitro kinetics of inhibition coincide with an unusually high rate of intrinsic enzyme–heme dissociation, especially in the ferric form. X-ray crystal structures of the inhibitor–enzyme complexes show that heme is displaced from the enzyme and blocked from rebinding by these compounds. The results reveal that apo-IDO1 serves as a unique target for inhibition and that heme lability plays an important role in posttranslational regulation.
Multicellular life is tasked with the immensely complex process of clearing foreign and aberrant cells while preventing autoimmunity. This finely tuned balance is often mediated by enzymes involved in central metabolism, reflecting the ancient origin and strong selective pressure of immune regulation (1). Indoleamine 2,3-dioxygenase (IDO1) is one such enzyme that oxidizes the essential amino acid tryptophan to produce N-formylkynurenine, which is further hydrolyzed to kynurenine (2, 3). This enzyme is present in many tissues and is up-regulated in response to inflammation, specifically the presence of cytokines, such as IFN-γ, as well as bacterial lipopolysaccharides (4, 5). Cells expressing IDO1 are able to deplete the inflamed environment of the metabolically expensive substrate, tryptophan, inhibiting the proliferation of immune targets (6, 7).
IDO1 is also able to serve as an immunosuppressive enzyme. In a seminal paper, IDO1, which was known to be highly expressed in placental tissue, was shown to be essential for the protection of embryos from maternal immune responses. An IDO1 inhibitor, 1-methyl-tryptophan, caused rejection of fetuses capable of provoking the maternal immune response, whereas fetuses closely related to the mother, and thus less likely to provoke an immune response, survived (8). This remarkable ability of a single enzyme to mediate such complex immune behavior has since been studied in depth, revealing a rich variety of mechanisms by which this enzyme affects immune regulation.
Expression of IDO1 inactivates surrounding immune cells through the combined effects of low tryptophan and high concentrations of kynurenine (1, 9). T cells are especially sensitive to low tryptophan concentrations, where they undergo cell cycle arrest (10). Additionally, the downstream metabolites of the product of IDO1 are potent activators of the aryl hydrocarbon receptor through which apoptosis of immune cells can be initiated (11). IDO1 has further been shown to provide protection from oxidative stress caused by inflammatory processes (12).
The importance of IDO1 in precise immune regulation is highlighted by its effects in a variety of processes and disease states, including autoimmune disorders, response to infection, tolerance in transplantation, HIV infection, and blood pressure regulation (13–15). This regulation/dysregulation is perhaps most sinisterly manifested in cancer cells that co-opt the immunosuppressive ability of IDO1 to evade immune destruction (16–18). The transcriptional control of IDO1 in cancers is often altered through mutation of the Bin1 repressor that allows for vastly up-regulated levels of IDO1 (19). Tumors displaying high IDO1 activity are associated with poor prognoses (20). Accordingly, IDO1 is a prime target in the treatment of almost all cancers for which IDO1 inhibition could restore the ability of the immune system to remove these cancer cells on its own or in combination with other treatments (17). The immunosuppressive effects of IDO1 are also implicated in persistent bacterial infection, wherein IDO1 inhibition has been shown to aid in clearing the infection (21).
The mechanism of IDO1 proceeds through a sequential oxygen insertion reaction, where molecular oxygen binds to the ferrous heme and then adds across the C2-C3 double bond of tryptophan through an alkylperoxo intermediate to afford the epoxide with the second atom of molecular oxygen left on the heme in a ferryl compound II structure. This IDO1 ferryl is then proposed to attack the epoxide, breaking the oxygen–iron bond and reducing the iron back to the ferrous resting state (22–24). This surprising ferryl attack on an epoxide has not been fully explored, as it is not the rate-determining step, but it is consonant with the recent revelations of the reactivity and versatility of compound II in other enzymes (25–27).
Typical of other heme proteins, IDO1 is vulnerable to competitive inhibitors, such as substrate mimics or compounds with heme binding capabilities (28–31). This strategy, however, has suffered from a paucity of inhibitors capable of the specificity and nanomolar binding affinity desirable in a pharmaceutical (32). Exceptions to this long-standing paradigm have been reported recently that bind to the enzyme without binding directly to the heme iron (33). We report here an examination of the unique inhibitory modes of two classes of inhibitors that display favorable specificity and robust inhibition of IDO1 (HeLa cell IC50 = 4.2 and 0.50 nM for compounds 1 and 2, respectively) (SI Materials and Methods) (34).
Heme Cofactor of IDO1 Is Labile
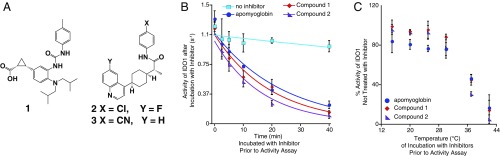
The mechanism of IDO1 inhibition by compounds 1 and 2 (Fig. 1A) was interrogated using activity assays with recombinant human IDO1. Compounds 1 and 2, the latter currently being investigated in phase II clinical trials, are structurally disparate and lack any significant similarity to the substrate tryptophan or any obvious heme binding moieties that could explain their mode of action (Fig. S1). In both cases, these compounds showed an intriguingly slow onset of inhibition, which is inconsistent with a simple competitive mechanism (Fig. 1B). Both compounds also showed a clear temperature dependence, only prompting inhibition above 30 °C (Fig. 1C and Fig. S2).
Fig. 1.
Time and temperature dependence of IDO1 inhibition. (A) Structure of three IDO1 inhibitors. (B) IDO1 (2.4 µM) was incubated with compounds 1 (20 µM), 2 (20 µM), or equine apomyoglobin (20 µM) at 37 °C for various times (x axis), after which this inhibition was tested using a standard activity assay (y axis). (C) IDO1 (2.4 µM) was incubated with compounds 1, 2, or equine apomyoglobin (20 µM) for 15 min at various temperatures (x axis), after which inhibition was tested using a standard activity assay following the rate of N-formylkynurenine production (y axis). Error bars represent SD. N ≥ 3.
As a test for IDO1 heme loss as the key initiatory step of inhibition, IDO1 was incubated with apo-myoglobin, which is capable of binding to free heme rapidly and essentially irreversibly, with a binding constant of 1014 M−1 (35). Remarkably, IDO1 with added apo-myoglobin was seen to lose activity, interpreted as heme loss to apo-myoglobin, with nearly the same time and temperature dependence as it does with 1 and 2 (t1/2 = 17, 14, and 11 min, respectively). This behavior is in contrast to the steadily maintained activity of IDO1 incubated in the absence of inhibitor or apo-myoglobin, indicating that heme binding to IDO1 is a dynamic, reversible process (Fig. 1B).
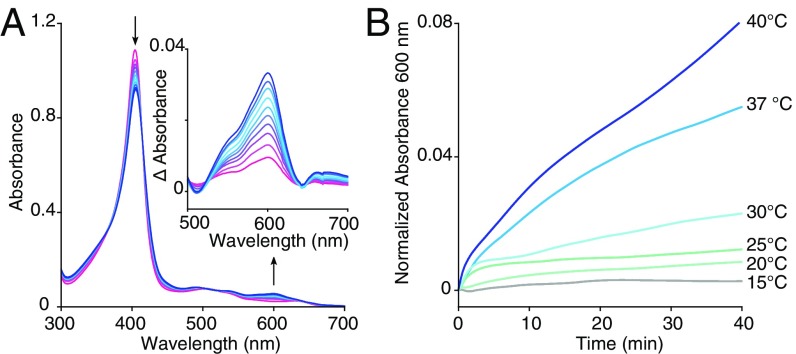
For a spectroscopic test of heme loss, IDO1 was incubated with a modified version of sperm whale apo-myoglobin, H64YV68F, that, when bound to heme, possesses a unique green color (36). IDO1 heme loss to apo-myoglobin can thus be conveniently monitored by an increase in absorbance at 600 nm. This confirmed that IDO1 readily loses its heme cofactor (Fig. 2A and Fig. S3) and that this unusual heme lability mirrors the temperature dependence of inhibition by 1 and 2 (Fig. 2B). In this way, IDO1 heme loss was shown to be inhibitor-independent and likely the shared rate-determining step of inhibition by 1 and 2, accounting for the striking similarity of inhibitory profiles for these two distinct inhibitors.
Fig. 2.
Transfer of heme from IDO1 to apomyoglobin. (A) The UV-visible spectra of heme transfer from IDO1 (5 µM) to apo-myoglobin H64YV68F (95 µM) over 40 min at 37 °C. (B) Heme dissociation from IDO1 (5 µM) to apo-myoglobin H64YV68F (95 µM) at various temperatures as indicated by the increase in absorbance at 600 nm.
1 and 2 Bind to IDO1 with Varying Off Rates
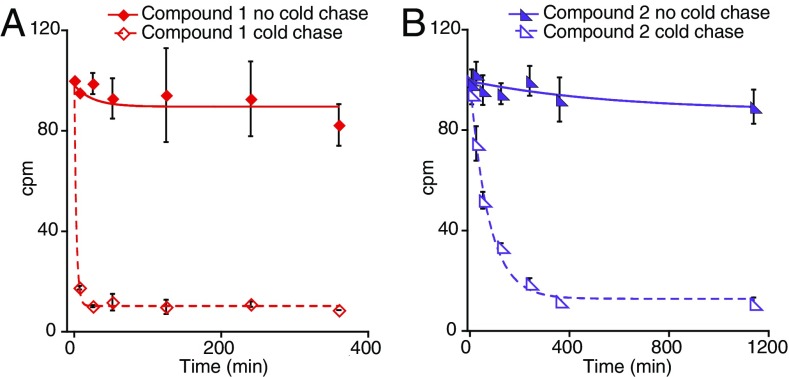
The mechanism by which these inhibitors exploit IDO1 heme lability was further explored using 14C-radiolabeled inhibitors in cold chase experiments. To test whether and how strongly these inhibitors bind to IDO1, the protein was incubated with 14C-labeled 1 and 2. These incubations then had natural abundance versions of the inhibitors added, and at various times, the buffer and any unbound inhibitor were exchanged away from the protein using centrifugal concentrators. In this way, any residual inhibitor must be bound to IDO1 and can be quantified using its 14C radiation. This showed that IDO1 does bind to these compounds, but it does so with very large differences in off rate: t1/2 of 2 and 50 min for 1 and 2, respectively (Fig. 3). This, despite nearly identical inhibition kinetics, strongly supports heme loss to be the shared and rate-determining step of inhibition.
Fig. 3.
Cold chase experiments with radiolabeled inhibitors. Radiolabeled versions of 1 (A) and 2 (B) were incubated with IDO1 followed by introduction of natural abundance 1 and 2, buffer exchange, and quantification of the residual, IDO1-bound, 14C-labeled 1 and 2. Error bars represent SD. N = 2.
Inhibition Does Not Show a Linear Relationship with Inhibitor Concentration
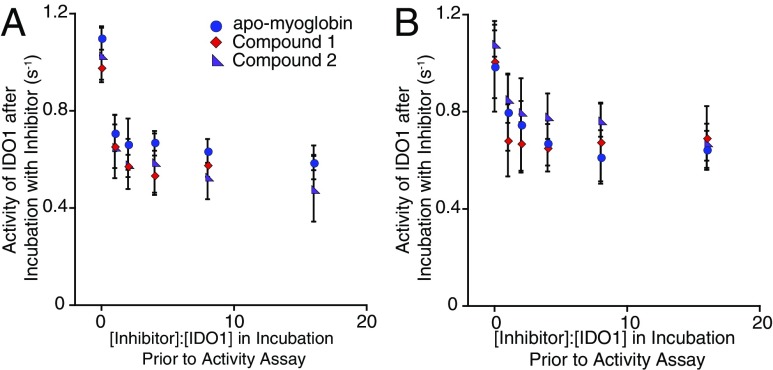
The concentration dependence of inhibition by 1 and 2 was then examined to illuminate any differences that may indicate a binding event before heme loss. In incubations preceding activity assays, the concentrations of the inhibitors, including apo-myoglobin, were varied from 1 to 16 times that of IDO1, and despite these large differences, the rates of inhibition were nearly identical and certainly far from showing a linear relationship (Fig. 4A). These results show that apo-IDO1 formed from intrinsic heme loss is the target for these inhibitors, which must act in some way to block heme from rebinding.
Fig. 4.
Concentration dependence of IDO1 inhibition. (A) IDO1 (2.4 µM) was incubated with various concentrations from 0 to 40 µM (x axis) of compounds 1, 2, or equine apomyoglobin at 37 °C, and for 15 min after, this inhibition was tested using a standard activity assay (y axis). (B) The experiment was repeated but with conditions that allow for turnover in the incubation with IDO1 before the activity assay. In this case, IDO1 (2.4 µM) was incubated with 2.4–40 µM 1, 2, or equine apomyoglobin at 37 °C for 15 min in the presence of 500 µM l-tryptophan, 10 mM ascorbate, 10 µM methylene blue, and 10 µg/mL catalase. The rate of N-formylkynurenine production was then tested using a standard activity assay (y axis). Error bars represent SD. N ≥ 3.
Inhibition Is Independent of IDO1 Turnover
This mechanism predicts that inhibition is independent of activity. To further confirm this hypothesis, we incubated IDO1 with various concentrations of inhibitor and the natural substrate, l-tryptophan. Activity was then assessed subsequently. As with all activity experiments described herein, d-tryptophan was used as the substrate, which unlike l-tryptophan, does not cause substrate inhibition and thus provides for a more convenient, repeatable means of determining activity (37–39). In these experiments, any l-tryptophan not converted to N-formylkynurenine will have been carried over from the incubations and acted as a substrate in addition to the 10 mM d-tryptophan (maximum final concentration of 15 μM l-tryptophan with a 33× dilution from incubation with inhibitor to activity assay solution). The inhibition profiles were nearly identical, regardless of turnover conditions during incubation with 1 and 2 (Fig. 4B). This observation confirms that the mechanism of inhibition is independent of turnover, supporting apo-IDO1 as the target for inhibition and its formation via heme dissociation as the rate-limiting step for this process.
Crystal Structures of IDO1–Inhibitor Complexes Reveal Their Mode of Action
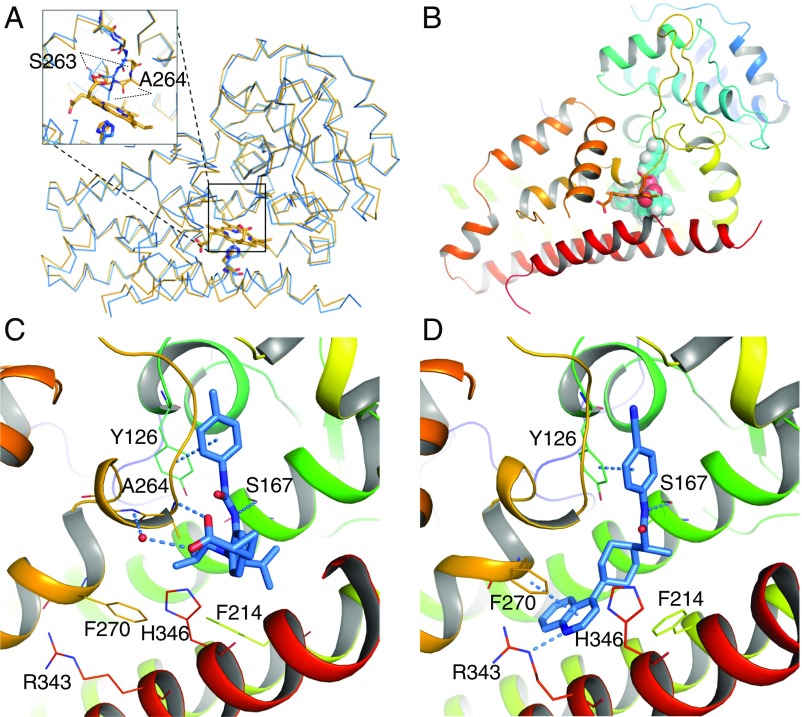
The interaction of these inhibitors with IDO1 was clearly elucidated by X-ray crystallographic structures obtained from cocrystallization with 1 and 3, the latter being a close analog of 2 (Fig. 5). (Compound 1 is BMS-978587, 2 is BMS-986205, and 3 is BMS-116.) Both inhibitors bound in a manner that displaced the heme cofactor, with each occupying different, although overlapping, space in the vacated heme pocket (Figs. S4 and S5). Remarkably, the overall structure of IDO1 bound to each inhibitor was largely unperturbed compared with a heme-containing IDO1 structure (0.61 and 0.44 Å rmsd from 2D0T for cocrystal structures with 1 and 3, respectively). The carboxylate of 1 forms a hydrogen bond with the backbone amide of Ala-264 and with His-346, which ordinarily coordinates with the heme iron on the proximal side. Binding of 1 also led to a shift in the flexible loop made of residues 260–265, an event previously observed with the binding of phenylimidazole (40). The loop shift revealed the putative substrate binding site to the distal face of heme, where the phenylurea group in 1 binds via edge-to-face π-interactions with Tyr-126 and hydrogen bonds with Ser-167. The quinoline of 3 occupies an additional pocket made available by side-chain rearrangements of Phe-270, Phe-214, His-346, and Arg-343 that are stabilized by edge-to-face π-interactions with Phe-270 and a hydrogen bond with Arg-343.
Fig. 5.
Structure of IDO1–inhibitor complexes. (A) Superposition of the backbone atoms of the crystal structures of IDO1 holoenzyme (orange) over that of IDO1/3 (blue). The overall structure of IDO1 undergoes little change on ligand binding. The loop corresponding to residues 260–265 is shifted, resulting in significant displacement (measured on Cα) of Ser-263 and Ala-264. Moreover, heme-coordinating residue His-346 is slightly rotated to interact with the ligand. (B) Sphere representation of 1 overlaid with heme shows that they bind IDO1 in a mutually exclusive manner. Binding geometries are shown for 1 in C and 3 in D. Structures were prepared from crystallographic coordinates using Maestro software to add hydrogens, determine protonation state of amino acid side chains, and minimize heavy atom deviation to a maximum of 0.3 Å from their crystallographic coordinates. Helix S (residues 383–399) is hidden in C and D for clarity. Figs. S4 and S5 depict the pocket rendering of holo-IDO1 in comparison with inhibitor-bound structures (Fig. S4), the electron density maps of inhibitors 1 and 3 bound to apo-IDO1 (Fig. S5), and 2D ligand interaction maps for 1 and 3 (Fig. S5).
Apo-IDO1 Is Present in Cells
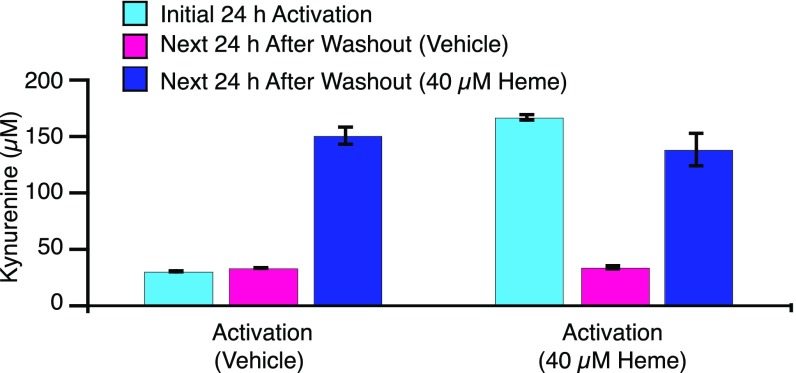
The physiological relevance of apo-IDO1 and its potential as a target for inhibition are supported by previous studies that have shown that IDO1 exists in the apo-form within cells and is capable of activation on addition of exogenous heme (41). To further support apo-IDO1 as the authentic target of 1 and 2, additional cellular assays were performed under conditions in which IDO1 was overexpressed but heme concentrations were varied. Briefly, human ovarian cancer cells were treated with IFN-γ to induce IDO1 expression. The cells were then tested for IDO1 activity after addition of the ribosomal inhibitor cycloheximide to prevent interference from newly translated IDO1. Cellular IDO1 activity was then assessed after further incubation in the presence or absence of added heme. Despite evidence that IDO1 protein levels were similar (Fig. S6), the activity of those cells with added heme was approximately fivefold higher than that of cells not treated with additional heme, indicating that at least 85% of IDO1 exists in these cells in an apo-form that is capable of activation by exogenously added heme (Fig. 6). Thus, the target of these inhibitors as revealed in the crystal structures is confirmed to be not only physiologically relevant but also, the predominant form of this enzyme in these cells.
Fig. 6.
Kynurenine production by human SKOV3 ovarian tumor cells. Left depicts results from cells stimulated for 24 h with IFN-γ in the absence of added hemin. Right depicts cells stimulated with IFN-γ in the presence of 40 µM hemin. Blue bars depict kynurenine production after 24 h of IFN-γ stimulation. Cells were washed for 1 h with fresh media containing cycloheximide to halt protein synthesis. Kynurenine production was measured 24 h after addition of fresh media devoid of heme (pink) or containing 40 µM hemin (dark blue). Error bars represent SD.
Having established that 1 and 2 compete with heme for apo-IDO1, it was of interest to determine the effect of added heme in the inhibition assays. When 40 µM heme was added to HeLa cells concurrently with stimulation of the cells with IFN-γ and inhibition was measured after 20 h of incubation, IC50 values were shifted less than twofold compared with the same experiment with no added heme (1, IC50 ± heme = 7.1/4.2 nM; 2, IC50 ± heme = 1.1/0.5 nM). The lack of a dramatic heme-dependent potency shift for compound inhibition during the prolonged induction period could result from direct competition for newly synthesized apo-IDO1 between inhibitor and heme, whereby the higher affinity and/or slower off rate of the inhibitors compensate for the higher heme concentration.
Heme Lability Is Dependent on Redox State
The cellular results show that IDO1 heme loss is a major factor in its normal regulation. We found that IDO1 heme lability is strongly dependent on the redox status of iron in the heme cofactor. Using compound 2 to capture apo-IDO1 as it forms from heme loss, we found that ferrous IDO1, which is the resting state of the enzyme in its active form, binds at least 10-fold more tightly to heme than enzyme does in the ferric state, which is catalytically inactive (Fig. 7 and Fig. S7). This was measured by fitting the loss in absorbance at the corresponding Soret maxima to a single exponential and comparing the half-lives. The kinetics of this loss show that this process is complex and may involve multiple states of heme binding, which will require additional study. The overall difference in heme retention, however, is significant.
Fig. 7.
Redox state dependence of heme dissociation from IDO1. IDO1 (1.1 µM) was incubated for 40 min at 37 °C in the presence of 10 µM 2. Heme dissociation from IDO1 was tracked by the loss of absorbance at the IDO1 λmax of the Soret peak: 404 nm for ferric IDO1 and 425 nm for ferrous IDO1. (A) Spectra of ferric IDO1 at various times. Inset shows a comparison of the loss in absorbance for both ferric and ferrous IDO1 as a function of time. (B) Spectra of ferrous IDO1 at various times. IDO1 was reduced using 5 mM sodium dithionite in a nitrogen-flushed sealed cuvette, and the decay of dithionite can be seen by its decreasing absorption below 375 nm. Inset is a scheme showing inhibitor binding to IDO1 in place of heme, causing the observed reduction in absorbance.
A stretch of 20 amino acids missing from electron density of all IDO1 structures has been predicted to form a “closed” state on substrate binding, effectively blocking the solvent-exposed face of heme and introducing an intermolecular hydrogen bond with a heme propionate (42). It is, therefore, plausible that intracellular tryptophan concentrations also play a role in heme recruitment and retention in IDO1. We tested heme loss from IDO1 to apo-myoglobin H64YV68F in the presence of 0–10 mM l-tryptophan (the Kd of l-tryptophan and ferric IDO1 is reported to be 5.8 mM) and found that the rate of heme loss was seen to decrease with increasing substrate concentration (Fig. S8) (37). This effect on ferric IDO1 may not be relevant, as intracellular concentrations of tryptophan are unlikely to approach this Kd (43); however, the Kd of ferrous IDO1 and l-tryptophan is significantly lower, 13 µM (37), and therefore, it is possible that tryptophan exerts this heme retention effect more strongly on the active, ferrous form of IDO1.
Discussion
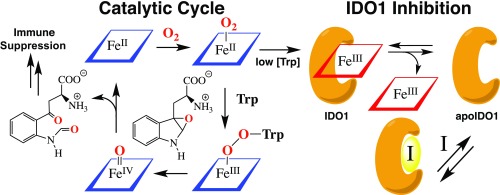
The goal of effective and specific inhibition of IDO1 has been intensively pursued since the discovery that this enzyme is critical to cancer’s evasion of immune responses (17). Whereas most inhibitors to date act as competitive inhibitors either by mimicking the substrate tryptophan or by binding to the heme cofactor (28), 1 and 2 are capable of inhibiting IDO1 by taking advantage of an entirely separate strategy of competing with the heme cofactor itself in binding to the apo-form of the enzyme (Fig. 8). In the effort to understand the mechanism of this inhibition, the importance of heme lability in IDO1 and its potential as a regulatory mechanism have been more fully revealed.
Fig. 8.
Catalytic mechanism of tryptophan catabolism by IDO1 and its inhibition via heme dissociation.
The formation of such a large pool of apo-IDO1 is still not fully understood, although it has previously been shown that IDO2, a homolog of IDO1, is capable of down-regulating the activity of IDO1 through heme transfer and sequestration (44), and it is known that there is significant cross-talk between IDO1 and heme catabolism through heme oxygenase-1, leading to loss of IDO1 activity via heme starvation (45). It has also been shown that ferrous IDO1 can bind nitric oxide in the absence of tryptophan, inducing rupture of the heme iron-proximal histidine bond to form apo-IDO1 (46). This transformation bears similarity to the heme receptor of soluble guanylyl cyclase that, although it is part of a signaling process and is not catalytic, uses heme lability in its regulation and is a target for compounds capable of binding to its apo-form (47, 48).
We have here shown that heme lability is likely to play a significant role in posttranslational control of IDO1 activity in cells, and the fact that this lability is strongly dependent on the redox state of the heme provides a potential regulatory mechanism to control IDO1 activity. Raven and coworkers (49) have shown that IDO1 accumulates in the catalytically inactive ferric state during turnover when tryptophan concentrations are low in vitro, and IDO1 is capable of autoxidizing to the ferric state in vivo (50). Together, these data suggest that IDO1 is self-regulating; under low tryptophan conditions, activity can decrease immediately by conversion to the catalytically inactive ferric state from which it can proceed to the apo-form through heme loss (Fig. 8). Such a process would contribute to the large pool of apo-IDO1 that serves as the target for these inhibitors.
Additionally, the complexity seen in the heme loss kinetics, a fast phase followed by a slow phase evident in both assays of heme loss to apomyoglobin H64YV68F and heme loss in the presence of 2 (Figs. 2 and 7), suggests that a simple model of only holo- and apo-IDO1 may not fully encompass this heme loss phenomenon and that there may be intermediate states of heme binding that will require additional study.
In conclusion, we have shown that IDO1 readily loses heme in a dynamic, reversible, and oxidation state-dependent manner. Targeting the pool of apo-IDO1 provides a means by which a class of inhibitors is capable of effectively inhibiting IDO1 with the potential to restore normal immune clearing of cancerous cells. These results also reveal more fully the intricate metabolic control that can be achieved by modulating the affinity of IDO1 for its heme cofactor and suggest that many other such enzymes could be similarly regulated and mechanistically rich.
Materials and Methods
Compounds 1, 2, and 3 were prepared at Bristol-Myers Squibb Co. by procedures described in published patent applications (51, 52). The 14C-radiolabeled versions of 1 and 2 were provided by Bristol-Myers Squibb Co.
Protein was expressed and purified using standard methods. Cell-free activity assays were performed using standard methods with an Agilent 8453 diode array spectrometer and temperature-controlled Fisher Scientific isotemp 1016s recirculating chiller. Cold chase experiments using 14C-radiolabeled 1 and 2 were performed as described in SI Materials and Methods. Crystallization conditions used to obtain inhibitor-bound and holo-IDO1 are described in SI Materials and Methods. Cell-based activity assays using HeLa and SKOV3 cells were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dianlin Xie and Mian Gao for construction of expression vectors; Frank Marsilio, Susan E. Kiefer, and Nicolas Szapiel for early IDO expression and purification work; Yuval Blat, Hao Lu, and Litai Zhang for helpful discussions; Kathy Johnston and Joseph Naglich for assay design in Hela cells; and Alfred Lammens and Stefan Steinbacher (Proteros Biostructures GmbH) for cocrystal structural work with 3. P.A.K. thanks BMS, Inc. for fellowship support. Support of this work was provided by NIH Grant 2R37 GM036298 (to J.T.G.).
Footnotes
Conflict of interest statement: M.T.N., P.A.K., and J.T.G. declare no conflict of interest. X.Z. and L.A. are former employees of Bristol-Myers Squibb Co. J.T.H., J.A.N., A.B., D.M., A.A., R.B., H.L., Z.L., S.P.S., and C.Y. are employees of Bristol-Myers Squibb Co.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 6AZU (holoenzyme), 6AZV (1), and 6AZW (3)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719190115/-/DCSupplemental.
References
- 1.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto S, Hayaishi O. Tryptophan pyrrolase of rabbit intestine. D- and L-tryptophan-cleaving enzyme or enzymes. J Biol Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- 3.Suzuki T, et al. Comparison of the sequences of Turbo and Sulculus indoleamine dioxygenase-like myoglobin genes. Gene. 2003;308:89–94. doi: 10.1016/s0378-1119(03)00467-0. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki F, Kuroiwa T, Takikawa O, Kido R. Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem J. 1985;230:635–638. doi: 10.1042/bj2300635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 6.Däubener W, et al. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect Immun. 2001;69:6527–6531. doi: 10.1128/IAI.69.10.6527-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curti A, Trabanelli S, Salvestrini V, Baccarani M, Lemoli RM. The role of indoleamine 2,3-dioxygenase in the induction of immune tolerance: Focus on hematology. Blood. 2009;113:2394–2401. doi: 10.1182/blood-2008-07-144485. [DOI] [PubMed] [Google Scholar]
- 8.Munn DH, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 9.van Baren N, Van den Eynde BJ. Tryptophan-degrading enzymes in tumoral immune resistance. Front Immunol. 2015;6:34. doi: 10.3389/fimmu.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munn DH, et al. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metz R, et al. IDO inhibits a tryptophan sufficiency signal that stimulates mTOR: A novel IDO effector pathway targeted by D-1-methyl-tryptophan. OncoImmunology. 2012;1:1460–1468. doi: 10.4161/onci.21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keskin DB, Marshall B, Munn D, Mellor AL, Gearhart DA. Decreased protein nitration in macrophages that overexpress indoleamine 2, 3-dioxygenase. Cell Mol Biol Lett. 2007;12:82–102. doi: 10.2478/s11658-006-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platten M, et al. Treatment of autoimmune neuroinflammation with a synthetic tryptophan metabolite. Science. 2005;310:850–855. doi: 10.1126/science.1117634. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16:279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favre D, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munn DH, et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science. 2002;297:1867–1870. doi: 10.1126/science.1073514. [DOI] [PubMed] [Google Scholar]
- 17.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 18.Muller AJ, et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc Natl Acad Sci USA. 2008;105:17073–17078. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 20.Munn DH, Mellor AL. IDO in the tumor microenvironment: Inflammation, counter-regulation, and tolerance. Trends Immunol. 2016;37:193–207. doi: 10.1016/j.it.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautam US, et al. In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2018;115:E62–E71. doi: 10.1073/pnas.1711373114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis-Ballester A, et al. Evidence for a ferryl intermediate in a heme-based dioxygenase. Proc Natl Acad Sci USA. 2009;106:17371–17376. doi: 10.1073/pnas.0906655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basran J, et al. The mechanism of formation of N-formylkynurenine by heme dioxygenases. J Am Chem Soc. 2011;133:16251–16257. doi: 10.1021/ja207066z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Groves JT. Oxygen activation and radical transformations in heme proteins and metalloporphyrins. Chem Rev. December 29, 2017 doi: 10.1021/acs.chemrev.7b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basran J, Booth ES, Lee M, Handa S, Raven EL. Analysis of reaction intermediates in tryptophan 2,3-dioxygenase: A comparison with indoleamine 2,3-dioxygenase. Biochemistry. 2016;55:6743–6750. doi: 10.1021/acs.biochem.6b01005. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Ullrich R, Hofrichter M, Groves JT. Heme-thiolate ferryl of aromatic peroxygenase is basic and reactive. Proc Natl Acad Sci USA. 2015;112:3686–3691. doi: 10.1073/pnas.1503340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moody PCE, Raven EL. The nature and reactivity of ferryl heme in compounds I and II. Acc Chem Res. January 12, 2018 doi: 10.1021/acs.accounts.7b00463. [DOI] [PubMed] [Google Scholar]
- 28.Röhrig UF, Majjigapu SR, Vogel P, Zoete V, Michielin O. Challenges in the discovery of indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors. J Med Chem. 2015;58:9421–9437. doi: 10.1021/acs.jmedchem.5b00326. [DOI] [PubMed] [Google Scholar]
- 29.Lewis-Ballester A, et al. Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1. Nat Commun. 2017;8:1693. doi: 10.1038/s41467-017-01725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malachowski WP, et al. O-alkylhydroxylamines as rationally-designed mechanism-based inhibitors of indoleamine 2,3-dioxygenase-1. Eur J Med Chem. 2016;108:564–576. doi: 10.1016/j.ejmech.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prendergast GC, Malachowski WP, DuHadaway JB, Muller AJ. Discovery of IDO1 inhibitors: From bench to bedside. Cancer Res. 2017;77:6795–6811. doi: 10.1158/0008-5472.CAN-17-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markwalder JA, et al. Identification and optimization of a novel series of indoleamine 2,3-dioxygenase inhibitors. Bioorg Med Chem Lett. 2017;27:582–585. doi: 10.1016/j.bmcl.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Crosignani S, et al. Discovery of a novel and selective indoleamine 2,3-dioxygenase (IDO-1) inhibitor 3-(5-Fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and its characterization as a potential clinical candidate. J Med Chem. 2017;60:9617–9629. doi: 10.1021/acs.jmedchem.7b00974. [DOI] [PubMed] [Google Scholar]
- 34.Williams DK, et al. Development of a series of novel o-phenylenediamine-based indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors. Bioorg Med Chem Lett. 2018;28:732–736. doi: 10.1016/j.bmcl.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Hargrove MS, Barrick D, Olson JS. The association rate constant for heme binding to globin is independent of protein structure. Biochemistry. 1996;35:11293–11299. doi: 10.1021/bi960371l. [DOI] [PubMed] [Google Scholar]
- 36.Hargrove MS, et al. His64(E7) → Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J Biol Chem. 1994;269:4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 37.Sono M, Taniguchi T, Watanabe Y, Hayaishi O. Indoleamine 2,3-dioxygenase. Equilibrium studies of the tryptophan binding to the ferric, ferrous, and CO-bound enzymes. J Biol Chem. 1980;255:1339–1345. [PubMed] [Google Scholar]
- 38.Lu C, Lin Y, Yeh SR. Inhibitory substrate binding site of human indoleamine 2,3-dioxygenase. J Am Chem Soc. 2009;131:12866–12867. doi: 10.1021/ja9029768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efimov I, et al. The mechanism of substrate inhibition in human indoleamine 2,3-dioxygenase. J Am Chem Soc. 2012;134:3034–3041. doi: 10.1021/ja208694g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugimoto H, et al. Crystal structure of human indoleamine 2,3-dioxygenase: Catalytic mechanism of O2 incorporation by a heme-containing dioxygenase. Proc Natl Acad Sci USA. 2006;103:2611–2616. doi: 10.1073/pnas.0508996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas SR, et al. Antioxidants inhibit indoleamine 2,3-dioxygenase in IFN-γ-activated human macrophages: Posttranslational regulation by pyrrolidine dithiocarbamate. J Immunol. 2001;166:6332–6340. doi: 10.4049/jimmunol.166.10.6332. [DOI] [PubMed] [Google Scholar]
- 42.Álvarez L, et al. Structural study of a flexible active site loop in human indoleamine 2,3-dioxygenase and its functional implications. Biochemistry. 2016;55:2785–2793. doi: 10.1021/acs.biochem.6b00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heidemann R, Lütkemeyer D, Büntemeyer H, Lehmann J. Effects of dissolved oxygen levels and the role of extra- and intracellular amino acid concentrations upon the metabolism of mammalian cell lines during batch and continuous cultures. Cytotechnology. 1998;26:185–197. doi: 10.1023/A:1007917409455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metz R, et al. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int Immunol. 2014;26:357–367. doi: 10.1093/intimm/dxt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill M, et al. Heme oxygenase-1 inhibits rat and human breast cancer cell proliferation: Mutual cross inhibition with indoleamine 2,3-dioxygenase. FASEB J. 2005;19:1957–1968, and erratum (2006) 20:1573. doi: 10.1096/fj.05-3875com. [DOI] [PubMed] [Google Scholar]
- 46.Samelson-Jones BJ, Yeh SR. Interactions between nitric oxide and indoleamine 2,3-dioxygenase. Biochemistry. 2006;45:8527–8538. doi: 10.1021/bi060143j. [DOI] [PubMed] [Google Scholar]
- 47.Martin F, et al. Structure of cinaciguat (BAY 58-2667) bound to Nostoc H-NOX domain reveals insights into heme-mimetic activation of the soluble guanylyl cyclase. J Biol Chem. 2010;285:22651–22657. doi: 10.1074/jbc.M110.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fritz BG, et al. Oxidation and loss of heme in soluble guanylyl cyclase from Manduca sexta. Biochemistry. 2011;50:5813–5815. doi: 10.1021/bi200794c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Booth ES, Basran J, Lee M, Handa S, Raven EL. Substrate oxidation by indoleamine 2,3-dioxygenase: Evidence for a common reaction mechanism. J Biol Chem. 2015;290:30924–30930. doi: 10.1074/jbc.M115.695684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vottero E, et al. Cytochrome b(5) is a major reductant in vivo of human indoleamine 2,3-dioxygenase expressed in yeast. FEBS Lett. 2006;580:2265–2268. doi: 10.1016/j.febslet.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 51.Balog JA, et al. 2014. Patent Cooperation Treaty Int Appl WO 2014150677.
- 52.Beck HP, et al. 2016. Patent Cooperation Treaty Int Appl WO 2016073774.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.