Significance
Glyphosate is a nonselective herbicide used around the globe for weed control in glyphosate-resistant (GR) and noncrop situations. The extensive and exclusive use of glyphosate has led to the evolution of herbicide resistance in many crop weeds. The molecular target of glyphosate, the 5-enolpyruvlyshikimate-3-phosphate synthase (EPSPS) gene, confers resistance upon amplification and was first documented in GR Amaranthus palmeri. We now report that amplified EPSPS copies in GR A. palmeri are present in the form of extrachromosomal circular DNA molecules (eccDNAs) with various conformations. We discovered that eccDNAs are transmitted to the next generation by tethering to mitotic and meiotic chromosomes. These results represent a report of extrachromosomal structures that drive rapid adaptive evolution in higher organisms.
Keywords: 5-enolpyruvylshikimate-3-phosphate synthase, eccDNA, glyphosate resistance, adaptive evolution, gene amplification
Abstract
Gene amplification has been observed in many bacteria and eukaryotes as a response to various selective pressures, such as antibiotics, cytotoxic drugs, pesticides, herbicides, and other stressful environmental conditions. An increase in gene copy number is often found as extrachromosomal elements that usually contain autonomously replicating extrachromosomal circular DNA molecules (eccDNAs). Amaranthus palmeri, a crop weed, can develop herbicide resistance to glyphosate [N-(phosphonomethyl) glycine] by amplification of the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene, the molecular target of glyphosate. However, biological questions regarding the source of the amplified EPSPS, the nature of the amplified DNA structures, and mechanisms responsible for maintaining this gene amplification in cells and their inheritance remain unknown. Here, we report that amplified EPSPS copies in glyphosate-resistant (GR) A. palmeri are present in the form of eccDNAs with various conformations. The eccDNAs are transmitted during cell division in mitosis and meiosis to the soma and germ cells and the progeny by an as yet unknown mechanism of tethering to mitotic and meiotic chromosomes. We propose that eccDNAs are one of the components of McClintock’s postulated innate systems [McClintock B (1978) Stadler Genetics Symposium] that can rapidly produce soma variation, amplify EPSPS genes in the sporophyte that are transmitted to germ cells, and modulate rapid glyphosate resistance through genome plasticity and adaptive evolution.
Eukaryotic cells use gene amplification mechanisms to overexpress specific genes for survival under stress. Amplified gene copies are often found as part of autonomously replicating extrachromosomal circular DNA molecules (eccDNAs), including double minutes (DMs) (1–8). The eccDNAs have been widely observed in many drug-resistant and tumor cell lines. The eccDNAs vary in size, ranging from a few hundred kilobases to megabases (9, 10). The eccDNAs may be simple (oligomeric) in structure, derived without any rearrangement from the corresponding chromosome (11, 12), or complex eccDNAs with duplicated copies of the same gene (13–15). EccDNAs containing sequences from different chromosomal loci have also been reported (16), indicating that different mechanisms may drive eccDNA assembly and evolution.
Despite the lack of centromeres, eccDNAs can be transmitted to daughter cells by tethering of their chromatin body to the telomeric region of segregating chromosomes from anaphase to telophase (17, 18). All reported cases of eccDNAs have been studied in cell lines; their genesis, behavior, and inheritance have not been studied in soma and germ cells of living organisms.
Living organisms, including plants and insects, have also evolved resistance to xenobiotics compounds, such as herbicides and insecticides, via gene amplification (19–25). In all reported studies, amplified gene copies were located in specific chromosomes or multiple chromosomal regions, but not in the form of eccDNAs (19, 23, 24, 26). Therefore, gene amplification in these living organisms is thought to have occurred by chromosomal rearrangements, such as inversion and reciprocal exchanges, or in association with transposable elements (23, 27–31).
A 40-fold to >100-fold amplification of the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene is associated with glyphosate resistance in Amaranthus palmeri populations (23). Initial reports suggested that the EPSPS amplicon was at least 30 kb in length and contained miniature inverted-repeat transposable elements (MITEs), which were postulated to disperse the amplicon to all of the glyphosate-resistant (GR) A. palmeri chromosomes at multiple sites (23, 31). More recently, the length of the EPSPS amplicon was extended to 297 kb, and termed the “EPSPS cassette,” by sequencing overlapping large-insert clones derived from a bacterial artificial chromosome (BAC) library (29). These clones flank the EPSPS gene, which was unique to GR A. palmeri across the United States, suggesting a single origin (29, 32). Here, we report that the EPSPS cassette is, in fact, an extrachromosomal circular DNA carrying the EPSPS gene, hereafter referred to as eccDNA. We report on the dynamics of eccDNA structure, variation, and behavior in mitotic and germ cells, as well as possible modes of inheritance, and discuss how they may trigger the plasticity of the GR response.
Results
Copy Number Variation in the EPSPS Gene Is Associated with Unique Chromosome Organization of the EPSPS Cassette.
We identified glyphosate-sensitive/susceptible (GS) and GR isolates of A. palmeri with various EPSPS copy numbers ranging from 1 to 120 based on quantitative PCR (qPCR) assays (Table S1). To study chromosome organization in relation to copy number variation (CNV) of the EPSPS gene, a DNA probe specific to the EPSPS gene and an EPSPS-containing BAC 22F22, or its flanking BACs, 5K07 and 1A02, were co-hybridized to chromosomes from several GS and GR A. palmeri plants (Fig. 1 and Fig. S1). FISH using an EPSPS gene probe on mitotic cells of GS A. palmeri with one copy of EPSPS revealed a tiny hybridization signal in the pericentromeric region of one pair of chromosomes (Fig. S1 C and D). FISH using BAC 22F22, in addition to a tiny signal on one pair of chromosomes, also generated dispersed FISH sites on all chromosomes from the same GS plant (Fig. S1 B and D).
Fig. 1.
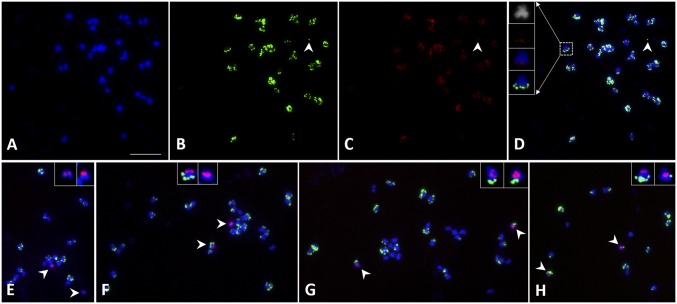
FISH mapping of eccDNAs and the EPSPS gene in mitotic metaphase chromosomes of root meristem cells of GR A. palmeri with 80 EPSPS copies. (A) Mitotic metaphase chromosomes stained by DAPI. (B) EccDNAs showing green signals overlying all chromosomes except one (arrowhead). (C) All eccDNAs seen in B carry EPSPS copies (red signal). (D) Merged image. (Insets) Colocalization of eccDNA (green) and EPSPS (red) signals on the tip of a chromosome, but not as part of the chromosome. Arrowheads in B–D point to eccDNA that is not associated with metaphase chromosomes. (E) Marker chromosome labeled with 5S rDNA (red signals) free of eccDNAs. (F) Another cell showed one marker chromosome free of signals, but its homolog has eccDNA. (G) Both marker chromosomes have eccDNA signals. (H) CNV of eccDNA signals in the two marker chromosomes. (E–H, Insets) 5S rDNA-bearing chromosomes showing random distribution of eccDNAs in different cells. Arrowheads in E–H point to 5S rDNA bearing chromosomes showing random distribution of eccDNAs in different cells. (Scale bar, 10 μm.)
In an A. palmeri plant with 12 EPSPS copies, the EPSPS-FISH signals on one pair of chromosomes were brighter than those from GS plants with a single copy of EPSPS, indicating that the EPSPS gene in this plant was amplified near its original location (Fig. S1 G and H). The BAC 22F22-FISH hybridization signals were faint but dispersed over all of the chromosomes except for the significantly more intense hybridization signals at the amplified EPSPS gene locus (Fig. S1 F and H).
In a GR A. palmeri plant with 80 EPSPS copies, EPSPS-FISH signals were detected on most chromosomes (Fig. 1). However, the FISH signals appeared to be at the edges of or outside the condensed chromosomes (Fig. 1 C and D). Strong and distinct hybridization signals were generated using BAC 22F22 (Fig. 1B) and were colocalized with those of EPSPS signals (Fig. 1D). Hybridization signals not associated with chromosomes were often observed in different metaphase cells of GR A. palmeri plants (arrows in Fig. 1 B–D). These data indicate that these structures were probably part of the EPSPS cassette reported by Molin et al. (29), and we began studies on the structure of the cassette using FISH on extended DNA fibers.
EPSPS Cassette Is an EccDNA Displaying Unique Structural Polymorphisms.
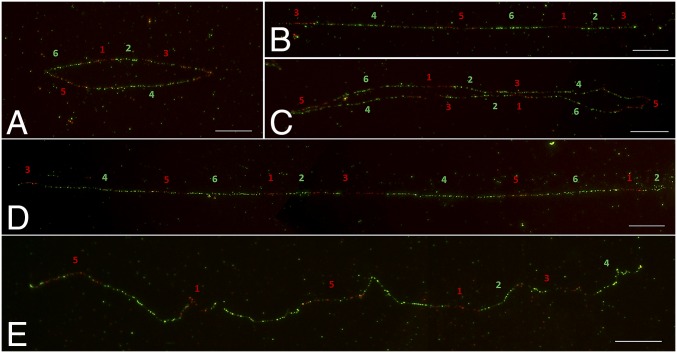
Molin et al. (29) reported the EPSPS cassette, which is 297 kb in length, consisting of seven overlapping BACs. Further selection by sequencing of two additional BACs revealed overlapping sequencing of the free ends indicating a potentially circular orientation of the EPSPS cassette. Six BACs associated with the EPSPS cassette were used in fiber-FISH mapping. These BACs were grouped into two pools, and they were labeled with alternate green/red colors based on their location in the EPSPS cassette (Fig. S2). Surprisingly, we found that ∼50.2% of fibers are in circular form, indicating that the EPSPS cassette is, in fact, an eccDNA (Fig. 2A, Table 1, and Fig. S2C). Based on the proportion of red and green signal tracks in circular molecules, we consider these eccDNAs to be intact and the wild-type form (Fig. 2A). The microscopic size of the circular form of these eccDNAs varied from ∼30 µm to ∼200 µm, which might be due to the variation of DNA fiber extension in the experiments. Therefore, we were unable to use the microscopic measurement data in classifying the eccDNA types. Instead, we scored eccDNAs for structural polymorphisms based on circular or linear structure and the number and proportion of red and green signals. In the circular DNA class, another 11.8% were determined to be a dimerized circular form of wild-type eccDNA with head-to-tail tandem duplication (hereafter, dimeric eccDNA) (Fig. 2C).
Fig. 2.
Fiber-FISH images of eccDNAs in GR A. palmeri with 80 EPSPS copies. (A) Circular form of eccDNA. (B) Linear form of eccDNA. (C) Dimerized circular form of eccDNA with head-to-tail tandem orientation. (D) Linear form of eccDNA with head-to-tail dimer. (E) Atypical fiber representing structural changes. Note: In the relatively long DNA fibers (D, E), two images were captured sequentially with an overlapping region and then they were combined into a single image using Adobe Photoshop. 1, BAC 01G15; 2, BAC 13C09; 3, BAC 22F22; 4, BAC 23A10; 5, BAC 03A06; 6, BAC 08H14. (Scale bars, 10 μm.)
Table 1.
Frequency of different structure polymorphisms of eccDNAs detected by fiber-FISH
| Structure | Circular | Linear | Dimeric circular | Dimeric linear | Atypical* |
| No. of observations | 564 | 245 | 133 | 90 | 92 |
| Frequency, % | 50.2 | 21.8 | 11.8 | 8.0 | 8.2 |
Fiber-FISH patterns that cannot be discriminated from other four types.
The remaining 38% of the eccDNA showed a linear structure (Table 1). Linearized fibers with different breakpoints but similar in composition to wild-type eccDNA were the most frequent (21.8%) class (Fig. 2B and Fig. S2D). Linear forms of dimeric eccDNAs were also detected (8.0%) (Fig. 2D). We also detected atypical fibers where the hybridization patterns deviated from the expected FISH patterns in 8.2% of the total fibers analyzed (Fig. 2E). Overall, our results demonstrated that ∼50% of eccDNAs of GR A. palmeri were structurally diverged due to duplication and deletion events (Table 1).
EccDNAs Display CNV and Random Chromosome Associations in Soma Cells of GR A. palmeri.
The GR A. palmeri plants with 80 EPSPS copies as determined by qPCR displayed surprising CNV in soma cells as revealed by FISH. The hybridization patterns of the eccDNAs on metaphase cells varied from cell to cell in the same plant. To determine whether the hybridization patterns of metaphase chromosomes were random, a 5S rDNA probe was used in FISH, which showed hybridization signals on one chromosome pair of A. palmeri. We observed four different patterns of the eccDNA signals on 5S rDNA-labeled homologous chromosome pairs in different cells (n = 24) from a single root tip meristem: (i) Both chromosomes were without eccDNA signals (16.7%) (Fig. 1E), (ii) one of the two chromosomes was without eccDNA signal (25%) (Fig. 1F), (iii) both chromosomes had similar signal intensity (33.3%) (Fig. 1G), and (iv) the two chromosomes varied in signal intensity (25%) (Fig. 1H). We conclude from these data that most of the eccDNAs are extrachromosomal elements that are randomly anchored to the chromosomes at mitotic metaphases.
EccDNAs Display Unique Behavior and a Chromosome Tethering Mechanism for Inclusion in Daughter Cells During Meiosis.
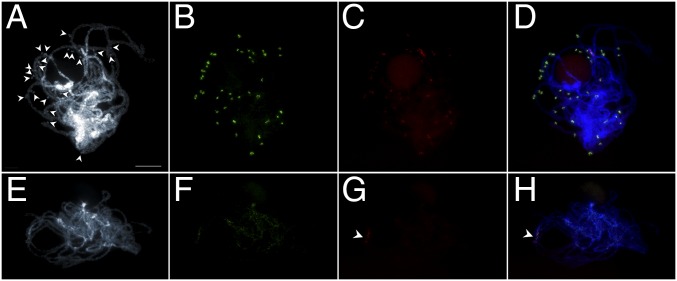
We analyzed the distribution of eccDNAs in meiotic pachytene chromosomes of GS and GR A. palmeri plants (Fig. 3). As expected, the eccDNAs were not observed in GS A. palmeri plants with one to 12 EPSPS copies (Fig. 3 E–H). However, 4′,6-diamidino-2-phenylindole (DAPI)–stained pachytene chromosome of the GR A. palmeri revealed numerous eccDNAs outside the chromosome axis (arrowheads in Fig. 3A), indicating that eccDNAs were not integrated into the chromosomes. The BAC 22F22 and EPSPS gene signals were colocalized (Fig. 3 B–D).
Fig. 3.
FISH mapping of eccDNA (green signals) and the EPSPS gene (red signals) on meiotic pachytene chromosomes of GR A. palmeri with 80 EPSPS copies (A–D) and GS A. palmeri with 12 EPSPS copies (E–H). (A) DAPI-stained pachytene chromosome showing eccDNAs lying outside the pachytene chromosomes (arrowheads). (B) FISH signals with the eccDNA probe. (C) FISH signals with the EPSPS gene probe. (D) Colocalization of eccDNA and EPSPS probes to eccDNAs. (E) DAPI-stained pachytene chromosome of GS A. palmeri did not reveal the eccDNAs. There was faint dispersed signals with eccDNA probe (F), but there was an amplified EPSPS gene signal in the pericentromeric region of one chromosome pair (G and H, arrowheads). (Scale bar, 10 μm.)
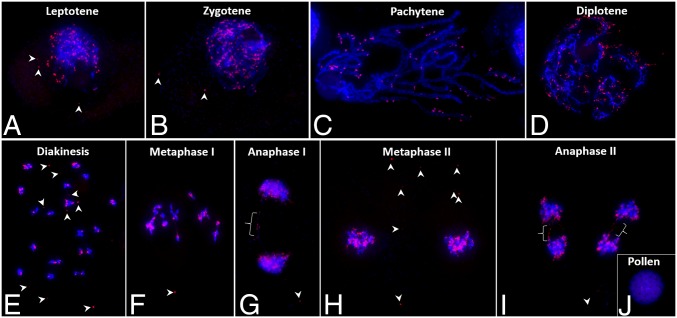
To study this apparent tethering of the eccDNAs to chromosomes during cell division further, we analyzed eccDNA behavior during all stages of meiosis I and II from leptotene to telophase II and also in immature pollen grains (Fig. 4 A–J). Numerous eccDNAs can be seen associated with leptotene and zygotene chromosomes, and a few are lying in the cytoplasm (arrowheads in Fig. 4 A and B). At pachytene stage, homologous chromosomes are fully paired. If eccDNAs were integrated into the chromosomes, then double signals would be observed; however, most signals were not double but, instead, single or in clumps and lying next to the chromosomes (Fig. 4C). Moreover, random and variable association of eccDNAs with different chromosomes was seen in well-spread chromosomes at pachytene (Fig. 4C), diplotene (Fig. 4D), diakinesis (Fig. 4E), and metaphase stages (Fig. 4F). The association of eccDNAs with laggard and stretched chromosomes was clearly observed at anaphase I (Fig. 4G) and anaphase II (Fig. 4I). By metaphase II, a few eccDNAs were seen lying away from the chromosomes in the cytoplasm (Fig. 4H). Pollen from GR plants also showed eccDNA signals, indicating transmission to the gametophyte (Fig. 4J).
Fig. 4.
Distribution of eccDNAs (red signals) on meiotic chromosomes in microsporocytes of GR A. palmeri during progression from the leptotene stage of meiosis I through anaphase of meiosis II (A–I) and pollen (J) detected by FISH (arrowheads point to the eccDNAs that are not associated with chromosomes). Brackets in G and I represent the lagging eccDNAs associated with chromatin bridges at anaphase to telophase stages.
EccDNAs Are Sexually Transmitted to Progeny Plants and Display Dramatic CNV in Soma Cells.
Our meiotic chromosome study indicated the possibility of transmission of the eccDNAs to offspring by a chromosome tethering mechanism. To study sexual transmission, we made crosses between a female GS A. palmeri plant lacking eccDNAs and a male GR A. palmeri plant carrying the eccDNAs, and vice versa. Plants grown from the seed of these crosses were labeled F1 plants (F1). Ten F1 plants from each reciprocal cross were randomly selected for qPCR and FISH analysis (Fig. 5 and Fig. S3). FISH analyses of root tip cells in these 20 F1 plants showed that all of the plants had positive signals associated with their mitotic metaphase chromosomes, indicating transmission of eccDNAs to the offspring (Fig. 5 and Fig. S3).
Fig. 5.
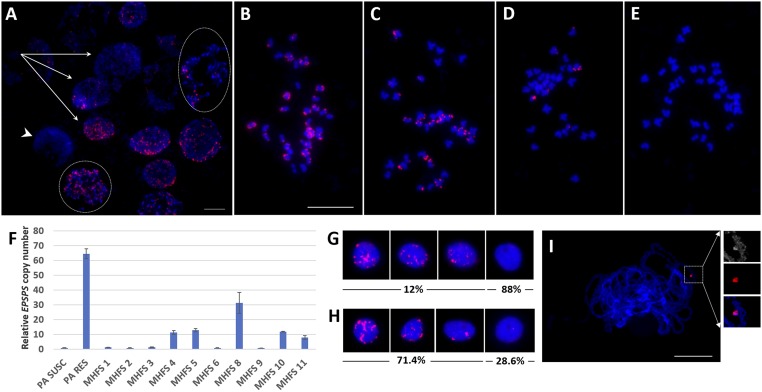
Inheritance and soma cell heterogeneity of eccDNAs and EPSPS copy number in F1 plants of GS A. palmeri x GR A. palmeri, including cells lacking eccDNAs (arrowheads) in a single root tip meristem. (A) Variable number of eccDNA FISH signals (red) in interphase and prometaphase stages (circles). (B–E) Different metaphase cells showing variable numbers of eccDNAs, including no eccDNAs (E). (F) Average EPSPS copy numbers of F1 plants and the controls. The y axis represents the relative β-tubulin/EPSPS gene copy number. (G) FISH analysis on nuclei isolated from leaf tissue of plant MHFS 1. (H) FISH analysis on nuclei isolated from leaf tissue of plant MHFS 8 showing variable eccDNA signals. Note that 12% (n = 100) (G) and 71.4% (n = 70) (H) indicate the percentages of different cells having eccDNA associated with FISH-positive nuclei in plants MHFS 1 and MHFS 8, respectively. (I) Pachytene FISH showing anchoring of an eccDNA on pachytene chromosomes in plant MHFS 1. (Scale bars, 10 μm.) Insets, Top, 2× enlarged and pseudo-colored DAPI image showing association of an eccDNA to pachytene chromosome; Middle, FISH labeled eccDNA; Bottom, merged image of top and middle.
We found that the eccDNA in F1 progeny displayed CNV ranging in number from one to 39 (Fig. 5F and Fig. S3A). All progeny except one harbored eccDNAs, but FISH signals on mitotic metaphase spreads prepared from a single root tip meristem were dramatically variable from cell to cell (Fig. 5). We observed several FISH signal patterns in different cells (n = 50) from a single root preparation of one plant [male highly glyphosate resistant and female glyphosate susceptible (MHFS) 1]: (i) eccDNAs were associated with most of the chromosomes in 50% of the cells similar to the GR parent plant (Fig. 5 A and B), (ii) eccDNAs were associated with half of the chromosomes in 26% of the cells (Fig. 5C), (iii) eccDNAs were associated with only a few of the chromosomes in 16% of the cells (Fig. 5 A and D), and (iv) all chromosomes were free of eccDNAs in 8% of the cells (Fig. 5E). Similar FISH patterns were detected on the metaphase cell spreads in all 20 F1 plants (Fig. 5 A–E and Fig. S3). These results showed that eccDNAs varied in copy number due to missegregation during mitotic divisions and were not integrated into chromosomes.
EccDNAs Display CNV in Different Tissues of a Plant.
Most surprisingly, qPCR analysis using genomic DNA prepared from leaf tissue showed that EPSPS copy number in five (MHFS 1, MHFS 2, MHFS 3, MHFS 6, and MHFS 9) of 10 F1 plants was similar to the copy number found in GS plants (Fig. 5F). However, FISH analysis from root tip meristems indicated that >90% of mitotic metaphase cells in these five plants had positive FISH signals (Fig. 5 A–D).
To resolve this apparent contradiction, we then performed FISH on nuclei isolated from leaf tissue of selected plants used in qPCR analysis (Fig. 5 G and H). The plant MHFS 1 with an estimated one EPSPS copy showed positive FISH signals in 12% (n = 100) of the nuclei isolated from the leaf tissue cells (Fig. 5G). The plant MHFS 8 with an estimated 31 EPSPS copies showed positive FISH signals in 71.4% (n = 70) of the nuclei isolated from the leaf tissue cells (Fig. 5H).
Next, we analyzed eccDNA variation in germ cells of plant MHFS 1 with an estimated one copy of the EPSPS gene. The results revealed that 12 of 20 cells at the pachytene stage of prophase I of meiosis showed eccDNAs ranging in number from one to 15 and eight of 20 of the cells were lacking eccDNAs (Fig. 5I and Fig. S4). In tapetum tissue, seven of 10 of the cells of this plant had eccDNAs. The frequency of eccDNAs in plant MHFS 1 was higher in the mitotic root tip (92%) and meiotic cells (60–70%) than in cells from leaf tissue (12%); the reasons for this variation are not known except that the former are actively dividing cells, while the leaf is a nondividing, differentiated tissue.
Discussion
This is a report on the role of eccDNA-driven gene amplification and rapid adaptive evolution in higher organisms. The lifestyle of higher organisms, including flowering plants and mammals, alternates between the dominant sporophytic phase and short-lived gametophytic phase. The male and female gametophytes produce the gametes and transmit the genetic information, and the zygotes develop into the sporophytes. The Darwinian evolution model acts on random, preexisting genetic variation in individuals and populations. In Darwinian evolutionary theory, there is no role of the life experience of the sporophyte or the inheritance of acquired characters as Lamarck (33) had proposed. McClintock (34, 35), based on her research on chromosome structure and behavior in soma and germ cells of maize, proposed that sporophytic genomes, in fact, can respond to challenges, such as stress, and that this acquired genomic variation is transmitted to the germ cells. McClintock (34) proposed the “…presence of innate systems that are able to restructure a genome… to be triggered into action by form of stress… according to the nature of the challenge…”. We propose that eccDNA elements identified in this research are one component of McClintock’s postulated innate system that rapidly produced soma variation, drove amplification of EPSPS genes in the sporophyte, and were transmitted to germ cells and modulated rapid evolution of glyphosate resistance in A. palmeri.
Wahl (13) proposed a general role of eccDNAs in gene amplification in mammalian and rodent cell lines for many different genes and selective drug agents. He proposed that eccDNAs may originate from chromosomes by deletions or circularization of blocked replicative forks, grow into DMs that are visible under the microscope, undergo unequal segregation during mitotic divisions in the presence of selective agents, and may integrate into the chromosomes to form homogeneously staining regions. Recent work on human cancer lines using combined whole-genome sequencing and cytogenetic analysis has validated the essential role of eccDNA in oncogene amplification, heterogeneity, and the evolution of cancer (1). In yeast, 23% of the genome is represented in eccDNAs ranging in size from 1 to 38 kb, and 80% of eccDNA contained autonomously replicating sequences (36). EccDNAs have been documented in many plant species, ranging in size from 2 to 20 kb and containing tandem repeats, suggesting their origin via intrachromosomal homologous recombination (37). Our results support widespread occurrence of eccDNA and its crucial role in gene amplification and plasticity of the sporophytic genome response to challenge.
Initial reports suggested that the EPSPS amplicon was at least 30 kb in length and contained MITEs, which were postulated to disperse the amplicon to each of the GR A. palmeri chromosomes at multiple sites (23, 31). EPSPS amplicon FISH signals were interpreted as being dispersed and integrated throughout the chromosome complement of A. palmeri in these studies (23, 29, 31). However, somatic metaphase chromosome-based analysis by Gaines et al. (23) did not provide the resolution to detect the tethering of FISH signals to chromosomes. Using a marker chromosome tagged with 5S rDNA-FISH signal, we observed random association of eccDNA signals to the marker chromosome. If the eccDNA was integrated into the chromosome, then they would display uniform signals in all cells, which was not the case (Fig. 1). Moreover, during the pachytene stages of meiosis when chromosomes are highly elongated, the FISH signals were clearly observed as being associated with, rather than part of, the chromosome, and some were not associated with any chromosome at all (Fig. 3). Finally, unequal mitotic segregation of eccDNAs produced variable FISH signals in different cells in the same preparation, and some were lacking the signal (Fig. 5 and Fig. S3). These data indicated that eccDNAs are not integrated into the chromosome, and are autonomously replicating structures that display unequal mitotic segregation, and thereby produce soma cell heterogeneity for resistance evolution.
Molin et al. (29) prepared a BAC library from a GR A. palmeri biotype from Mississippi, and sequencing of overlapping BACs revealed a 297-kb sequence unique to GR A. palmeri, which they termed an EPSPS cassette. The EPSPS cassette consisted of an array of repetitive sequences, 72 putative genes, and an autonomous replication sequence (ARS). EPSPS cassette-specific marker analysis revealed that GR biotypes across the United States had a single origin (29, 32). Using overlapping BACs from the Mississippi biotype, our fiber-FISH analysis of a Kansas GR biotype clearly established that the EPSPS cassette is, in fact, an eccDNA (Fig. 2 and Fig. S2). The shared common structure also supported a single origin of eccDNA of GR biotypes (32). The discovery of ARSs in eccDNA supports our analysis of eccDNA behavior in dividing soma and germ cells leading to CNV. The EPSPS cassette also expressed HSC70 (heat shock protein), which are heat, drought, and salt stress-inducible (29). Thus, a single Georgia GR A. palmeri plant (23) that acquired eccDNA 12 y ago, was indeed a supercharged weed biotype that could not only resist herbicide but also potentially withstand heat, drought, and salt stress, and that it underwent a selection sweep and spread to many states in the United States in a short time frame (29, 32).
Apart from CNV, eccDNAs displayed structural polymorphisms. The monomeric and dimeric circular forms were predominant (62%; 50% monomers and 12% dimers). The second largest population (30%) included linear forms (monomeric and dimeric molecules) with a nearly intact structure, as well as different-sized linear molecules with different breakpoints in the eccDNA or partial deletions. This number is likely overestimated because mechanical force during DNA fiber preparation can break circular molecules into linear forms. Nonetheless, it is possible that some linear molecules were generated from circular molecules associated with replication errors of the eccDNAs, as was shown in the chloroplast genome (38). They could also represent rare chromosome integration events in an evolutionary trajectory toward more stable acquired herbicide resistance. The remaining linear fibers (8%) were atypical eccDNAs with modified hybridization patterns. These may be the result of recombination events or random cleavage and fusion of replication intermediates, which has also been demonstrated in the chloroplast genome (38). These evolutionary dynamics of eccDNAs also suggest that the collection of smaller eccDNAs from different genomics regions can recombine and evolve into large eccDNA organelles under strong selection pressure.
The mechanisms of tethering and replication of eccDNAs are unknown. However, the eccDNAs seem to have evolved a tethering mechanism for transmission to daughter cells during cell division. The eccDNAs were invariably associated with chromosomes, and these associations were clearly observed in meiosis (Fig. 4). The tethering is reminiscent of the behavior of autonomously replicating viruses, such as engineered plasmid vectors derived from Epstein–Barr virus (EBV) and bovine papillomavirus type1 (BPV1) in mammalian cell lines (39, 40). Epstein–Barr nuclear antigen (EBNA1) and E2 proteins, which initiate replication from EBV and BPV cis-acting origin of replications (oriP), mediate anchoring to the host chromosomes (39–43). Mitotic chromosome tethering in mammalian cell lines transfected with engineered plasmid vector containing a mammalian scaffold/matrix-attached region sequence and simian virus 40 oriP have also been described (44–46). These observations raise the strong possibility that eccDNAs may also have cis-acting sequences, such as oriP, that recruit cellular transacting factors to mediate chromosome association.
Hepadnaviruses, including human hepatitis B virus, possess a DNA genome and replicate through reverse transcription of an RNA intermediate, the pregenomic RNA (pgRNA) (47). The pgRNA is transcribed from covalently closed circular DNA (cccDNA). The cccDNA exists as a stable episome, which, in turn, is organized into minichromosomes by histone and nonhistone proteins (48, 49) that are localized in the nuclei of infected hepatocytes. The cccDNAs are reverse-transcribed into the relaxed-circle (RC) form of viral DNAs (50). The RC-DNAs can be reintegrated into the nuclei for amplification of their own cccDNAs (51). The EPSPS cassette sequence harbors a reverse transcriptase gene long enough to encode a functional protein among other genes that may function in DNA replication (29). These products of transcription and reverse transcription may facilitate the RNA intermediates for amplification and assembly into DNA strands. Identifying these molecular mechanisms may facilitate the development of stable plant artificial chromosomes carrying agronomically useful traits. Furthermore, development of compounds that interfere with elements of the tethering mechanism of eccDNAs to chromosomes may provide novel mechanisms of weed control.
Materials and Methods
Sampling of GR A. palmeri and EPSPS Copy Determination.
A. palmeri plants used in this study were generated from seeds collected from a field near Manhattan, Kansas, where there was an incident of lack of control of this population with glyphosate application in the previous season. This field was exposed to frequent applications of glyphosate in Roundup Ready soybeans, grown in rotation. Seed of A. palmeri was randomly sampled from 10 plants and pooled. Sixty seedlings from the above sample, along with a known GS A. palmeri plant, were germinated, and seedlings were transplanted individually into Miracle-Gro potting mix in 10-cm × 10-cm × 10-cm plastic pots and watered from the top in a greenhouse (25/20 °C temperature; 15:9 h of light day/night, supplemented with 120 mmol⋅m−2⋅s−1 illumination using sodium vapor lamps). At least 20 plants (10–12 cm tall) were treated separately with a field use rate of 868 g⋅acid equivalent (ae)⋅ha−1 plus 2% (wt/vol) ammonium sulfate or twice the field use rate of glyphosate. All treatments were applied with a moving single-nozzle, bench-type sprayer (Research Track Sprayer; De Vries Manufacturing) equipped with a flat-fan nozzle tip (80015LP TeeJet tip; Spraying Systems Co.) delivering 168 L⋅ha−1 at 222 kPa in a single pass at 3.2 km⋅h−1. Plant survival was assessed 4 wk after treatment.
In response to glyphosate treatment [868 g⋅ae⋅ha−1 plus 2% (wt/vol) ammonium sulfate], plants showing injury levels high (>80%) and low (<30%) in comparison to untreated checked plants were grouped as GS and GR, respectively. To determine the number of EPSPS gene copies, at least four plants from each category, along with known GS A. palmeri, were selected and genomic DNA (gDNA) was isolated as follows. Fresh leaf tissue was collected from individual plants, flash-frozen, and stored at −80 °C for gDNA isolation. The gDNA was extracted from frozen leaf tissue (100 mg) using a DNeasy Plant Mini Kit (Qiagen, Inc.) following the manufacturer’s instructions. DNA was quantified on a Nanodrop Spectrophotometer.
The qPCR was performed using a CFX96TM Real-Time Detection System from Bio-Rad to determine the EPSPS gene copy number in GR A. palmeri plants. The qPCR reaction mix consisted of 8 μL of SYBR Green Master Mix (Bio-Rad), 2 μL each of forward (F) and reverse (R) primers (5 μM), and 2 μL of gDNA (15 ng⋅μL−1) to make the total reaction volume up to 14 μL. EPSPS gene copy number was measured relative to the β-tubulin gene (reference gene). PCR conditions were 95 °C for 15 min, and 40 cycles of 95 °C for 30 s and 60 °C for 1 min. A melt curve profile was included following the thermal cycling protocol to determine the specificity of the qPCR reaction. The following primer sequences were used: EPSPS F 5′ ATGTTGGACGCTCTCAGAACTCTTGGT 3′ and EPSPS R 5′ TGAATTTCCTCCAGCAACGGCAA3′ (23), and β-tubulin F 5′ ATGTGGGATGCCAAGAACATGATGTG 3′ and β-tubulin R 5′ TCCACTCCACAAAGTAGGAAGAGTTCT 3′ (52). EPSPS gene copy number was measured with three technical replicates. Gene copy number was determined using the 2ΔCT method, where CT is the threshold cycle and ΔCT is CTTarget gene (EPSPS) − CTReference gene (β-tubulin) (52). Several GR and GS A. palmeri plants were selected for molecular cytogenetic mapping.
Reciprocal Hybridizations.
Male and female plants of GR (carrying eccDNA) and GS (lacking eccDNA) A. palmeri were grown individually in Miracle-Gro potting mix in 10-cm × 10-cm × 10-cm plastic pots and watered from the top in a greenhouse (25/20 °C temperature; 15:9 h of light day/night, supplemented with 120 mmol⋅m−2⋅s−1 illumination using sodium vapor lamps). After flower initiation, the inflorescences of female GR and male GS plants, and vice versa, were covered together with plastic bread bags (33 cm × 60 cm) containing microperforations. A total 10 F1 plants for each reciprocal cross were randomly selected for FISH and qPCR analysis.
BAC Clones.
Clones of A. palmeri containing and flanking the EPSPS sequence were prepared as described (29) in a contractual arrangement between W.T.M. (US Department of Agriculture-Agricultural Research Services 6066-21000-060-00-D) and C.A.S. (Clemson University Genomics and Computational Laboratory) and provided to D.-H.K. for the purposes of this study. The clones were prepared from seedlings from a single plant from a Mississippi population demonstrating high glyphosate resistance. The BACs provided were 22F22 (contains EPSPS), 05K07, 01A02, 06D23, 13C09, 01G15, 08H14, and 23A10.
Slide Preparation.
Preparations of mitotic and meiotic chromosomes followed published protocols (53), with minor modifications. Root tips were collected from plants and treated in a nitrous oxide gas chamber for 1.5 h. The root tips were fixed overnight in 3:1 ethanol/glacial acetic acid and then squashed in a drop of 45% acetic acid. Young floral buds, about 1∼2 mm long, were selected for meiotic chromosome preparations. Anthers from a single flower bud were squashed in 45% acetic acid on a slide and checked under a phase microscope. All preparations were stored at −70 °C until use.
Probe Labeling.
Sequences of A. palmeri EPSPS gene (GenBank accession no. JX564536) were used to develop the PCR primers for cloning of the EPSPS gene. The PCR product was cloned in 2.1-TOPO TA vector (Invitrogen), and the clone was labeled with digoxigenin-11-deoxyuridine triphosphate (Roche Diagnostics) using a standard nick translation reaction. The clone, maize 5S rDNA (54), was labeled with biotin-16-dUTP (Roche). The BAC clones were labeled with either biotin-16-dUTP or digoxigenin-11-dUTP using a nick translation reaction. Biotin- and digoxigenin-labeled probes were detected with Alexa Fluor 488 streptavidin antibody (Invitrogen) and rhodamine-conjugated antidigoxigenin antibody (Roche), respectively.
Image Analysis.
Chromosomes were counterstained with DAPI in Vectashield Antifade solution (Vector Laboratories). The images were captured with a Zeiss Axioplan 2 microscope (Carl Zeiss Microscopy LLC) using a cooled CCD camera CoolSNAP HQ2 (Photometrics) and AxioVision 4.8 software. The final contrast of the images was processed using Adobe Photoshop CS5 software.
Fiber-FISH.
Young leaf tissues were collected from fast-growing GR A. palmeri plants. Nuclei isolation, DNA fiber preparation, and fiber-FISH were performed following published protocols (38, 55). Fiber-FISH images were captured and processed as described previously in the FISH procedure.
Supplementary Material
Acknowledgments
We thank John Raupp for critical review of the manuscript and Duane Wilson for technical assistance. This research was supported by grants from the Kansas Wheat Commission and the Kansas Crop Improvement Association, Wheat Genetics Resource Center Industry/University Cooperative Research Center National Science Foundation Contract 1338897, and Department of Agronomy and US Department of Agriculture-Agricultural Research Services Project 6066-21000-060-00-D 9. This is contribution number 18-189-J from the Kansas Agricultural Experiment Station (Kansas State University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719354115/-/DCSupplemental.
References
- 1.Turner KM, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowell JK. Double minutes and homogeneously staining regions: Gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- 3.Hamkalo BA, Farnham PJ, Johnston R, Schimke RT. Ultrastructural features of minute chromosomes in a methotrexate-resistant mouse 3T3 cell line. Proc Natl Acad Sci USA. 1985;82:1126–1130. doi: 10.1073/pnas.82.4.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll SM, et al. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: Functional evidence for a mammalian replication origin. Mol Cell Biol. 1987;7:1740–1750. doi: 10.1128/mcb.7.5.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Bliek AM, Lincke CR, Borst P. Circular DNA of 3T6R50 double minute chromosomes. Nucleic Acids Res. 1988;16:4841–4851. doi: 10.1093/nar/16.11.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz JC, Choi KH, von Hoff DD, Roninson IB, Wahl GM. Autonomously replicating episomes contain mdr1 genes in a multidrug-resistant human cell line. Mol Cell Biol. 1989;9:109–115. doi: 10.1128/mcb.9.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storlazzi CT, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: Origin and structure. Genome Res. 2010;20:1198–1206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennisi E. Circular DNA throws biologists for a loop. Science. 2017;356:996. doi: 10.1126/science.356.6342.996. [DOI] [PubMed] [Google Scholar]
- 9.Stark GR, Wahl GM. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- 10.Hahn PJ, Nevaldine B, Longo JA. Molecular structure and evolution of double-minute chromosomes in methotrexate-resistant cultured mouse cells. Mol Cell Biol. 1992;12:2911–2918. doi: 10.1128/mcb.12.7.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ståhl F, Wettergren Y, Levan G. Amplicon structure in multidrug-resistant murine cells: A nonrearranged region of genomic DNA corresponding to large circular DNA. Mol Cell Biol. 1992;12:1179–1187. doi: 10.1128/mcb.12.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogt N, et al. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci USA. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahl GM. The importance of circular DNA in mammalian gene amplification. Cancer Res. 1989;49:1333–1340. [PubMed] [Google Scholar]
- 14.Schoenlein PV, Shen DW, Barrett JT, Pastan I, Gottesman MM. Double minute chromosomes carrying the human multidrug resistance 1 and 2 genes are generated from the dimerization of submicroscopic circular DNAs in colchicine-selected KB carcinoma cells. Mol Biol Cell. 1992;3:507–520. doi: 10.1091/mbc.3.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A new role for hypoxia in tumor progression: Induction of fragile site triggering genomic rearrangements and formation of complex DMs and HSRs. Mol Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- 16.Gibaud A, et al. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci. Hum Mol Genet. 2010;19:1276–1285. doi: 10.1093/hmg/ddq004. [DOI] [PubMed] [Google Scholar]
- 17.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 18.Kanda T, Otter M, Wahl GM. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- 19.Devonshire AL, Field LM. Gene amplification and insecticide resistance. Annu Rev Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- 20.Field LM, Devonshire AL, Forde BG. Molecular evidence that insecticide resistance in peach-potato aphids (Myzus persicae Sulz.) results from amplification of an esterase gene. Biochem J. 1988;251:309–312. doi: 10.1042/bj2510309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Field LM, Devonshire AL. Structure and organization of amplicons containing the E4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer) Biochem J. 1997;322:867–871. doi: 10.1042/bj3220867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton MG, Karunaratne SH, Giakoumaki E, Roberts N, Hemingway J. Quantitative analysis of gene amplification in insecticide-resistant Culex mosquitoes. Biochem J. 2000;346:17–24. [PMC free article] [PubMed] [Google Scholar]
- 23.Gaines TA, et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA. 2010;107:1029–1034. doi: 10.1073/pnas.0906649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jugulam M, et al. Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia. Plant Physiol. 2014;166:1200–1207. doi: 10.1104/pp.114.242826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heap I. 2017 International survey of herbicide resistant weeds. Available at www.weedscience.org. Accessed November 1, 2017.
- 26.Dillon A, et al. Physical mapping of amplified 5-enolpyruvylshikimate-3-phosphate synthase gene copies in glyphosate-resistant Amaranthus tuberculatus. Plant Physiol. 2017;173:1226–1234. doi: 10.1104/pp.16.01427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackman RL, Spence JM, Field LM, Devonshire AL. Chromosomal location of the amplified esterase genes conferring resistance to insecticides in Myzus persicae (Homoptera: Aphididae) Heredity. 1995;75:297–302. [Google Scholar]
- 28.Blackman RL, Spence JM, Field LM, Devonshire AL. Variation in the chromosomal distribution of amplified esterase (FE4) genes in Greek field populations of Myzus persicae (Sulzer) Heredity. 1999;82:180–186. [Google Scholar]
- 29.Molin WT, Wright AA, Lawton-Rauh A, Saski CA. The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: A repetitive path to resistance. BMC Genomics. 2017;18:91. doi: 10.1186/s12864-016-3336-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jugulam M, Gill BS. Molecular cytogenetics to characterize mechanisms of gene duplication in pesticide resistance. Pest Manag Sci. 2017;74:22–29. doi: 10.1002/ps.4665. [DOI] [PubMed] [Google Scholar]
- 31.Gaines TA, et al. Identification of genetic elements associated with EPSPs gene amplification. PLoS One. 2013;8:e65819. doi: 10.1371/journal.pone.0065819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molin WT, et al. Survey of the genomic landscape surrounding the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene in glyphosate-resistant Amaranthus palmeri from geographically distant populations in the USA. Pest Manag Sci. July 7, 2017 doi: 10.1002/ps.4659. [DOI] [PubMed] [Google Scholar]
- 33.Lamarck J-B. Philosophie Zoologique ou Exposition des Considérations Relatives à l’Histoire Naturelle des Animaux. Dentu; Paris: 1809. pp. 422–450. [Google Scholar]
- 34.McClintock B. Stadler Genetics Symposium. Vol 10. Springer; New York: 1978. Mechanisms that rapidly reorganize the genome; pp. 25–48. [Google Scholar]
- 35.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 36.Møller HD, Parsons L, Jørgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci USA. 2015;112:E3114–E3122. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 2008;53:1027–1034. doi: 10.1111/j.1365-313X.2007.03394.x. [DOI] [PubMed] [Google Scholar]
- 38.Lilly JW, Havey MJ, Jackson SA, Jiang J. Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell. 2001;13:245–254. doi: 10.1105/tpc.13.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marechal V, et al. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanda T, Otter M, Wahl GM. Coupling of mitotic chromosome tethering and replication competence in epstein-barr virus-based plasmids. Mol Cell Biol. 2001;21:3576–3588. doi: 10.1128/MCB.21.10.3576-3588.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanda T, Kamiya M, Maruo S, Iwakiri D, Takada K. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids. J Cell Sci. 2007;120:1529–1539. doi: 10.1242/jcs.03434. [DOI] [PubMed] [Google Scholar]
- 44.Baiker A, et al. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu N, Miura Y, Sakamoto Y, Tsutsui K. Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res. 2001;61:6987–6990. [PubMed] [Google Scholar]
- 46.Jenke AC, et al. Nuclear scaffold/matrix attached region modules linked to a transcription unit are sufficient for replication and maintenance of a mammalian episome. Proc Natl Acad Sci USA. 2004;101:11322–11327. doi: 10.1073/pnas.0401355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summers J, Mason WS. Replication of the genome of a hepatitis B–like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 48.Bock CT, Schranz P, Schröder CH, Zentgraf H. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- 49.Pollicino T, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol. 2007;13:48–64. doi: 10.3748/wjg.v13.i1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godar AS, et al. Physiological and molecular mechanisms of differential sensitivity of Palmer amaranth (Amaranthus palmeri) to mesotrione at varying growth temperatures. PLoS One. 2015;10:e0126731. doi: 10.1371/journal.pone.0126731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koo D-H, Jiang J. Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J. 2009;59:509–516. doi: 10.1111/j.1365-313X.2009.03881.x. [DOI] [PubMed] [Google Scholar]
- 54.Koo D-H, Zhao H, Jiang J. Chromatin-associated transcripts of tandemly repetitive DNA sequences revealed by RNA-FISH. Chromosome Res. 2016;24:467–480. doi: 10.1007/s10577-016-9537-5. [DOI] [PubMed] [Google Scholar]
- 55.Jackson SA, Wang ML, Goodman HM, Jiang J. Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome. 1998;41:566–572. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.